Abstract
The LDL receptor (LDL-R) mediates cholesterol metabolism in humans by binding and internalizing cholesterol transported by LDL. Several different molecular mechanisms have been proposed for the binding of LDL to LDL-R at neutral plasma pH and for its release at acidic endosomal pH. The crystal structure of LDL-R at acidic pH shows that the receptor folds back on itself in a closed form, obscuring parts of the ligand binding domain with the epidermal growth factor (EGF)-precursor homology domain. We have used a structure-based site-directed mutagenesis approach to examine 12 residues in the extracellular domain of LDL-R for their effect on LDL binding and release. Our studies show that the interface between the ligand binding domain and the EGF-precursor homology domain seen at acidic pH buries residues mediating both LDL binding and release. Our results are consistent with an alternative model of LDL-R whereby multiple modules of the extracellular domain interact with LDL at neutral pH, concurrently positioning key residues so that at acidic pH the LDL-R:LDL interactions become unfavorable, triggering release. After LDL release, the closed form of LDL-R may target its return to the cell surface.
Keywords: cholesterol metabolism, cell surface receptor, receptor:ligand interaction
LDL-R FUNCTION
The LDL receptor (LDL-R) plays a crucial role in lipoprotein and cholesterol metabolism in the liver (1). LDL-R binds LDL at neutral pH on the cell surface of hepatocytes. The ligand-receptor complex internalizes through receptor-mediated endocytosis at clathrin-coated pits, releasing its cargo in the endosomes upon exposure to acidic pH (2). After ligand release, LDL-R recycles to the cell surface, while LDL continues to the lysosomes where it is degraded. If the cargo fails to release, the entire complex undergoes lysosomal degradation, profoundly depleting receptors from the cell surface over time (3–6). LDL binding and release by LDL-R is essential to maintain healthy plasma lipoprotein levels; patients with familial hypercholesterolemia (FH) as a result of mutations in LDL-R present with elevated plasma LDL levels, premature atherosclerosis, and attendant heart disease (1, 7).
LDL-R EXTRACELLULAR DOMAIN
The extracellular domain of LDL-R contains seven cysteine-rich repeats (R1–R7, also called LA1–LA7), two epidermal growth factor (EGF)-like repeats (EGF-A and EGF-B), a β-propeller domain, and a third EGF-like repeat (EGF-C) tethered to the cell surface by a single transmembrane segment (8–10). Each cysteine-rich repeat (∼40 amino acids) contains two loops (or lobes) stabilized by three disulfide bridges and one Ca2+ ion (11). The side chains of Asp196, Asp200, Asp206, and Glu207 and the carbonyl oxygens of Trp193 and Gly198 coordinate the Ca2+ ion within R5, a cysteine-rich repeat essential for all lipoprotein binding (11). The analogous Ca2+-binding residues in R4, Asp147, Asp151, Asp157, and Glu158 provide side chains, and Trp144 and Asp149 provide backbone carbonyl oxygens.
LDL-R is thought to adopt a compact closed conformation at acidic pH and an elongated open conformation at neutral pH. Size exclusion chromatography and analytical ultracentrifugation studies show that LDL-R has a much larger hydrodynamic radius at neutral pH than at acidic pH (12–16). Furthermore, the fragment R1-R4 associates with the rest of the molecule in solution at pH 6, but not at pH 8 (12, 13). In the crystal structure at pH 5.3, LDL-R kinks between R7 and EGF-A, closing the molecule back on itself enabling R4 and R5 to dock with their Ca2+-binding sites on the surface of the β-propeller (Fig. 1) (12). Taken together, biophysical and structural data suggest that at neutral pH, LDL-R is in a binding-competent conformation as an elongated molecule, but at acidic pH adopts a compact conformation compatible with LDL release.
Fig. 1.
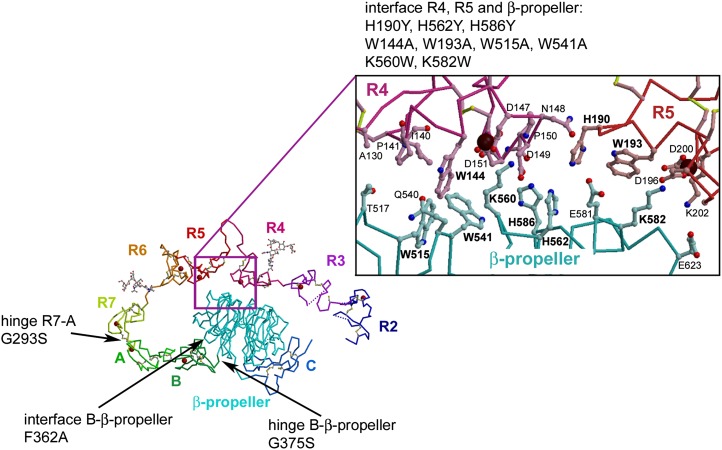
Extracellular domain of LDL-R at pH 5.3. The extracellular domain of LDL-R contains two domains, an N-terminal ligand-binding domain (R1-R7) and a C-terminal EGF-precursor homology domain (EGF-A-EGF-B-β-propeller-EGF-C). Subdomains visible in the crystal structure are labeled, and Ca2+ ions are shown as red spheres. R2–R7 do not interact with each other, except via their linkers, unlike the modules EGF-A, EGF-B, the β-propeller, and EGF-C that form a rigid assembly stabilized by interactions between the modules (12). A close-up of the residues mediating the interface between R4, R5, and the β-propeller is shown in ball-and-stick representation (oxygen, red; nitrogen, blue; Ca2+, maroon; carbon atoms in R4, pink, and in R5, red; β-propeller, cyan). Five salt bridges and numerous hydrophobic interactions stabilize the interface. The 12 mutations designed to probe LDL binding and release in this study are indicated in the structure.
LDL-R LIGAND BINDING AND RELEASE
LDL-R binds LDL with 1:1 stoichiometry, interacting with apolipoprotein B (apoB), the sole 550 kDa protein of LDL (17). The extent of the interactions between LDL-R and LDL and the residues mediating binding are not known, although the modules R3-R7 are critical (18–20). According to one model, negatively charged residues surrounding the Ca2+ ions in the cysteine-rich repeats bind two stretches of positively charged residues on apoB in LDL and a related protein apoE on VLDL (11, 21–23). However, according to another model, negatively charged residues far away from the Ca2+-binding sites mediate LDL binding (14). Release of lipoprotein particles requires the EGF-precursor homology domain (Gly293-Thr692). Removal of these residues produces receptor variants that still bind ligand and undergo endocytosis but are resistant to acid-mediated ligand release and accompany their ligand to the lysosomes (3–6). Since the structure determination of LDL-R, mutagenesis studies have shown that the β-propeller domain is required for ligand release (6, 24) and that replacement of individual or multiple histidines of a three histidine cluster found at the R4, R5, β-propeller interface (H190, H562, and H586) disrupts ligand release (6, 14, 24, 15). Three different models of LDL release have been proposed: at acidic pH, the regions surrounding the Ca2+-binding sites of R4 and R5 prefer to interact intramolecularly with the β-propeller instead of LDL promoting release (model 1) (12, 13); at acidic pH, the β-propeller docks onto the regions surrounding the Ca2+-binding sites of R4 and R5 causing allosteric release of LDL, which is bound elsewhere (model 2) (14); or, lastly, acidic pH results in loss of Ca2+ ions from the cysteine-rich repeats of LDL-R causing LDL to release (model 3) (25). Two issues lie at the heart of the controversy: the exact binding site of LDL on LDL-R and the mechanism by which acidic pH mediates LDL release.
SCOPE OF THIS STUDY
We embarked on structure-based mutagenesis of LDL-R to probe the effects of single amino acids on LDL binding and release. We focused on residues that appear important to maintain the closed conformation of LDL-R seen in the crystal structure at acidic pH and examined their effect on LDL binding and release. Our panel of mutants reveals that 1) residues involved in LDL binding are buried in the R4,R5,β-propeller interface; 2) residues required for LDL binding only partially overlap those mediating release; 3) mutations that reduce LDL release segregate in two groups, those defective only at highly acidic pH (pH 5–5.5) and those defective even at mildly acidic pH (pH 6–7); and, finally, 4) mutations targeting a hinge region between EGF-B and the β-propeller may affect LDL release but not its binding. Our results support a model whereby the open form of LDL-R interacts with LDL using multiple modules, including R4, R5, and the β-propeller; at acidic pH, conformational changes to LDL-R and/or LDL promote LDL release, upon which the closed conformation of LDL-R is adopted as it recycles through highly acidic environments back to the cell surface.
MATERIALS AND METHODS
DNA mutagenesis
Mutants of the human LDL receptor extracellular domain (residues 1–699) in pFastBac1 were generated with the Quickchange kit (Stratagene). The C-terminal hexahistidine-tagged mutants were expressed in insect cells and secreted proteins purified as described (12). Purified proteins were concentrated to approximately 5 mg/ml, buffer exchanged into 2.5 mM sodium phosphate, pH 8, 50 mM NaCl, and 0.5 mM CaCl2, and their purity assessed via SDS-PAGE electrophoresis (see supplementary Fig. I). Protein concentrations were determined using the protein assay (Bio-Rad).
Radiolabeling LDL
LDL was isolated from normal human plasma by sequential flotation at d = 1.006 and 1.063 g/ml. Analysis by SDS-PAGE revealed a single protein corresponding to apoB-100; size exclusion chromatography over Superose HR 6 (GE Healthcare) revealed a single peak. LDL was iodinated with I125 using a protocol similar to (26). After radiolabeling, free iodine was removed by size exclusion chromatography over Sephadex G-25, and the I125-LDL buffer exchanged into 20 mM Tris, pH 8.0, 150 mM NaCl, and 0.3 mM EDTA. The protein concentration of the labeled material was determined using the Bio-Rad protein assay. Preparations of I125-LDL routinely had specific activities in the range 625–850 cpm/ng.
Solid phase binding assay
Solid phase binding assays were carried out using a modified version of (17). The monoclonal antibody HL1 directed against a 10 amino acid linker between R4 and R5 was used to immobilize LDL-Rs. This antibody does not appear to restrict LDL binding or release and tethers LDL-R stably in the range pH 5–8. Immulon 96-well Removawell plates (Thermo Fisher) were incubated with HL1 (2 μg/well) overnight at 4°C in buffer A (20 mM Tris, pH 8, and 100 mM NaCl), blocked with 1% BSA in buffer A for 1 h at 37°C, washed three times at 4°C, and then incubated with 50 ng receptor in buffer A + 1% BSA overnight at 4°C. Wells were then incubated for 3 × 10 min in Buffer A, 1% BSA, 2 mM CaCl2, or Buffer A, 1% BSA, and 20 mM EDTA to render calcium-containing or metal-free receptors. I125-LDL was subsequently added in a series of concentrations (0, 0.9, 1.8, 3.6, 7.3, 10.9, 14.6, 21.8, and 29.1 nM) in 2 mM Ca2+ or 20 mM EDTA containing buffer A + 1% BSA for 1 h at 4°C. Unbound I125-LDL was removed by suction, and the plates washed three times with 50 mM Tris, pH 8, 2 mM CaCl2, 0.5% BSA, or 50 mM Tris, pH 8, 20 mM EDTA, and 0.5% BSA. Individual wells were counted for 5 min in a Cobra II auto γ counter (Packard). Specific binding was calculated as I125-LDL bound in presence of Ca2+ minus I125-LDL bound (nonspecially) in the presence of EDTA. Each data point was performed in triplicate. Binding data were processed and plotted using Prism 3 (Graphpad Software).
Solid phase release assay (pH curve)
HL1 and receptors were coated and washed as described for the binding assay. Wells were then incubated for 3 × 10 min in Buffer A, 1% BSA, and 2 mM CaCl2 to render calcium-containing receptors or with buffer containing 20 mM EDTA instead of CaCl2 to render calcium-free receptors. Wells were incubated with 18.2 nM I125-LDL (10 μg/ml) for 1 h at 4°C in Buffer A + 1% BSA containing 2 mM CaCl2 or 20 mM EDTA. Unbound I125-LDL was removed by suction and the plates washed three times with 2 mM CaCl2-containing or CaCl2-free buffer (50 mM Tris, pH 8, and 0.5% BSA). I125-LDL was released from each well by a single 4°C incubation of 200 μl 20 mM Tris maleate, 20 mM NaCl, 5 mM CaCl2, and 0.5% BSA (pH 5, 5.5, 6.0, 6.3, 7, or 8) for 30 min. Buffer containing released I125-LDL was harvested as was the well containing the unreleased fraction of I125-LDL and counted for 5 min. The total counts (well + buffer) consistently reconstituted the signal of wells not subject to ligand release (within 10%). All data were collected in triplicate and were processed and plotted with Prism3.
Time course release of bound I125-LDL at 4°C
I125-LDL was bound to LDL-R as described in the binding assay and released by a single 4°C incubation of 200 μl of 20 mM Tris maleate, 20 mM NaCl, 5 mM CaCl2, and 0.5% BSA at pH 5 or 8 in a time course. All data were collected in triplicate and were processed and plotted with Prism3.
Gel filtration analysis
The hydrodynamic behavior of wild-type LDL-R and 12 mutants was characterized using an analytical Superdex 200 PC 3.2/30 column (GE Biomedical; bed volume 2.4 ml). Each receptor was eluted at 30 μl/min in 20 mM Tris-HCl and 250 mM NaCl, pH 8, and 20 mM NaAc and 250 mM NaCl, pH 5.5, and the wild type was also run in 20 mM NaAc and 250 mM NaCl, pH 6.3. The high salt concentration minimizes potential interactions between the proteins and the stationary phase. Fractions (25 μl) were collected manually immediately after passing the in-line UV-monitor and analyzed on 10% SDS-PAGE gels, followed by subsequent immunoblotting using a penta-His HRP conjugate antibody (Qiagen). The column was calibrated with standards (Sigma) comprising cytochrome c (12,400 Da), BSA (66,000 Da), and β-amylase (200,000 Da) in 20 mM Tris-HCl and 250 mM NaCl, pH 8, or 20 mM NaAc and 250 mM NaCl, pH 5.5, respectively.
RESULTS
LDL-R mutants
We designed a panel of mutants around salient features of the compact form of LDL-R seen in the crystal structure at pH 5.3 (PDB id: 1N7D). We first focused on the interface between R4, R5, and the β-propeller, which obscures potential ligand-binding epitopes from the solvent, i.e., residues surrounding the Ca2+-binding sites (Trp144, Asp147, Asp149, and Asp151 from R4 and Trp193, Asp196, Gly198, and Asp200 from R5) (Fig. 1). A striking cluster of tryptophan residues, Trp144, Trp515, and Trp541, stabilizes the interaction between R4 and the β-propeller, while one tryptophan, Trp193, mediates the interaction between R5 and the β-propeller. Mutations W144A, W515A, W541A, and W193A were designed to disrupt the R4, R5, β-propeller interface because tryptophan side chains typically can provide extensive hydrophobic surfaces and binding energy to protein interfaces unlike the much smaller alanine side chain. A three-histidine cluster (His190, His562, and His586) sits at the junction between R4, R5, and the β-propeller. The individual histidines were replaced with tyrosine (H190Y, H562Y, and H586Y), a hydrophobic side chain of similar size that maintains its neutral charge between pH 8 and pH 5 (unlike histidines) and reproduces the mutations H190Y and H562Y found in FH patients (12, 27, 28). The β-propeller residues Lys560 and Lys582 interact electrostatically with negatively charged residues at the Ca2+-binding site of R4 and R5, respectively, very similar to the way lysine residues from the escort protein receptor-associated protein (RAP) D3 domain interact with the Ca2+-binding sites in LDL-R R3-R4 and other LDL-R family members (29, 30). Mutations K560W and K582W were chosen to replace the positively charged lysine side chains by a similarly long hydrophobic tryptophan side chain. We reasoned that if the hydrophobic component of Lys560 and Lys582 was important for LDL binding or release, addition of tryptophan residues might stimulate these functions, whereas if the electrostatic component was crucial (as suggested by the interaction mode with RAP), binding or release would be diminished. Overall, the mutants would enable us to assess which of the proposed models for LDL-R function is consistent with the effects of the mutants on LDL binding and release.
We also examined the crystal structure for residues that could mediate conversion between the elongated and the compact form of LDL-R to examine their effect on LDL binding and release. Two regions are of interest: the linker between EGF-B and the β-propeller, and the linker between R7 and the EGF-A. EGF-B packs against the β-propeller in full-length LDL-R at acidic pH but not at neutral pH in the fragment EGF-B-β-propeller-EGF-C (9). To probe this interaction, G375S and F362A were designed. A serine residue at position 375 does not allow the tight kink of polypeptide backbone conformation between EGF-B and the β-propeller in the closed conformation due to its side chain, and G375S is also reported in FH patients (27, 28). F362A should weaken the interaction between EGF-B and the β-propeller by replacing a large hydrophobic residue with a much smaller side chain, creating an unfavorable cavity at the interface. The mutation G293S was designed to sterically hinder conformational flexibility between R7 and EGF-A.
Care was taken to design mutations that were subtle enough to maintain proper folding of the modules (permitting efficient secretion into the medium during overexpression), yet structurally drastic enough to produce measurable changes in LDL binding and release. The locations of the mutations in the extracellular domain are shown in Fig. 1 and summarized in Table 1.
TABLE 1.
Structure-based mutagenesis of LDL-R
| Class | Location in Structure | Comment |
|---|---|---|
| Linker/Hinge mutations: | ||
| G293S | Hinge R7, EGF-A | |
| F362A | Interface EGF-B, β-propeller | |
| G375S | Hinge EGF-B, β-propeller | FH mutationa |
| Histidine cluster: | ||
| H190Y | R5 | FH mutation |
| H562Y | β-propeller | FH mutation |
| H586Y | β-propeller | |
| R4, R5, β-propeller interface: | ||
| W144A | R4 | |
| W193A | R5 | |
| W515A | β-propeller | |
| W541A | β-propeller | |
| K560W | β-propeller | |
| K582W | β-propeller | |
Mutation found in FH patients.
LDL binding studies
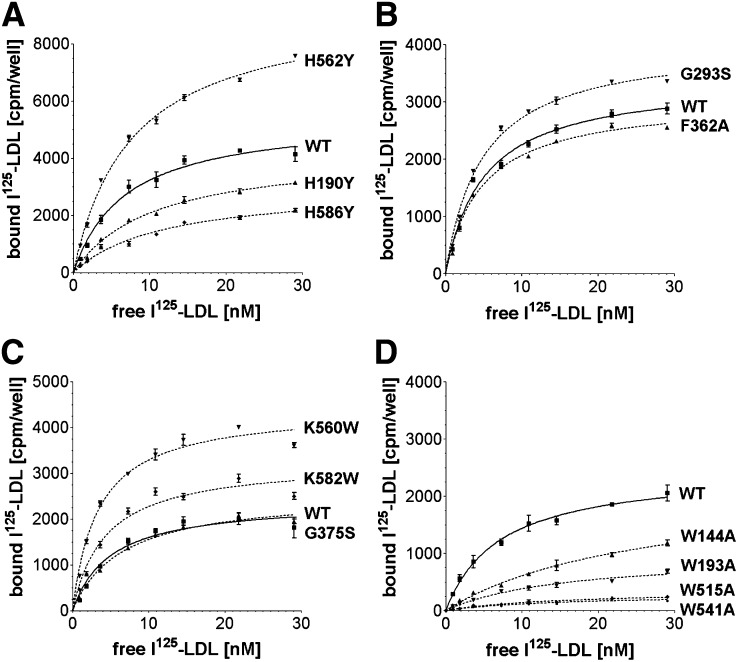
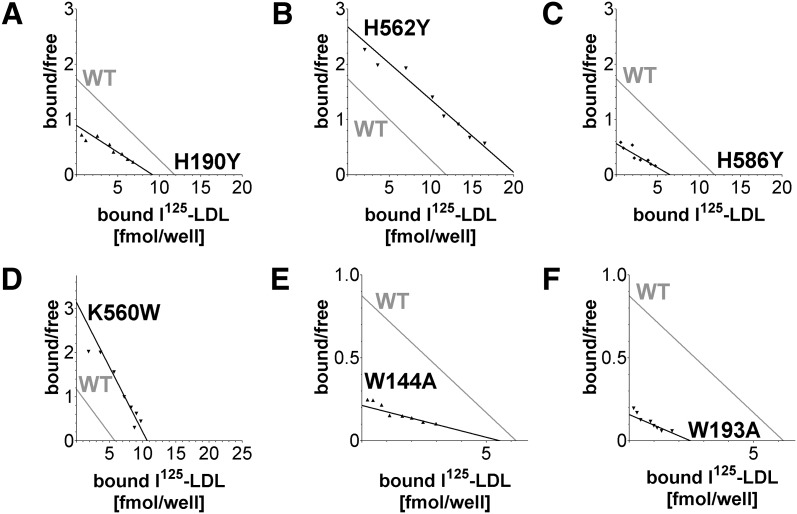
LDL receptors were evaluated for I125-LDL binding using a solid phase binding assay (Fig. 2). Nonlinear regression calculations indicate a Kd for I125-LDL of ∼4 nM for the high affinity component of wild-type (WT) LDL-R, in good agreement with the previously reported value of 4.5 nM at 4°C (31). Saturation curves revealed that all 12 mutants bound I125-LDL, though in different amounts (Fig. 3). Because differences in the amount of bound I125-LDL could be attributed to differences in the number of ligand-binding sites available, Scatchard plot analysis was used to compare the I125-LDL affinity between mutants and WT (Fig. 4; see supplementary Fig. II). The histidine cluster mutants H190Y and H586Y bound less I125-LDL than WT LDL-R (Fig. 3A) and displayed slightly reduced affinity (Fig. 4A, C). In contrast, while H562Y bound more I125-LDL than WT (Fig. 3A), its affinity was normal (Fig. 4B). The mutations designed to probe interdomain interactions, G293S, G375S, and F362A, all bound I125-LDL similarly to WT (Fig. 3B, C) with similar affinity (see supplementary Fig. IIB, C). Mutations designed to increase hydrophobic interactions at the R4, R5, β-propeller interface, K560W and K582W, bound more I125-LDL than WT (Fig. 3C), and in the case of K560W, possibly enhanced affinity (Fig. 4D). The tryptophan mutants (W144A, W193A, W515A, and W541A) bound much less I125-LDL compared with WT (Fig. 3D), and the affinity for I125-LDL was clearly reduced for W144A and W193A (Fig. 4E, F). The ligand affinity for W515A and W541A was not assessed due to the low level of I125-LDL bound. The 12 mutants in our panel each bound significant amounts of I125-LDL (permitting subsequent analysis of their ligand release), though four mutants (H190Y, H586Y, W144A, and W193A) showed somewhat reduced binding and affinity for I125-LDL, and two mutants (W515A and W541A) significantly reduced binding to I125-LDL. It is unknown if these differences in LDL binding would cause changes to LDL metabolism in the context of the whole organism, especially in the case of H190Y, H586Y, and K560W, where the differences compared with WT are quite small; nevertheless, for the purpose of mapping potentially functional regions on the surface of LDL-R, these mutants are useful.
Fig. 2.
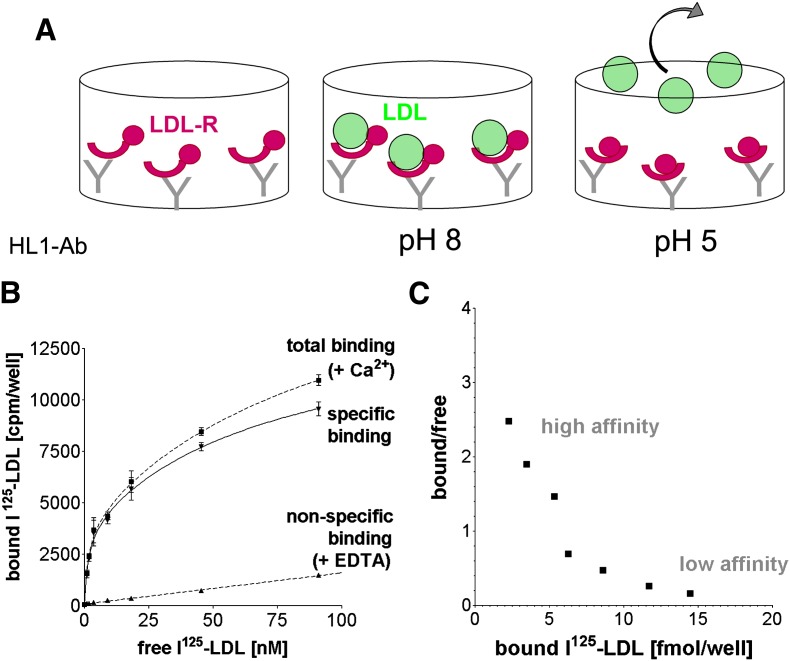
Solid phase LDL binding and release assay. A: Recombinant purified receptors (magenta crescents and spheres) are tethered to 96-well plates with the monoclonal anti-body HL1 (gray Y shapes), which binds the long linker between R4 and R5. After incubation with increasing amounts of I125-LDL (green spheres), saturation binding at pH 8 is observed; a subsequent wash at an acidic pH releases I125-LDL from the wells enabling ligand release to be studied as well. Upon transition from basic to acidic pH, the receptor undergoes a conformational change (see Introduction), causing the LDL-R ligand binding domain (magenta crescents) and the EGF-precursor homology domain (magenta spheres) to change conformations with respect to each other. B: Specific binding between LDL-R and I125-LDL is expressed as total binding (in presence of Ca2+) minus nonspecific I125-LDL binding (in presence of EDTA to disrupt receptors) expressed in counts per minute. Each data point represents triplicate measurements, and the error bars show the SD. C: Scatchard plot analysis of specific I125-LDL binding in B reveals a high affinity (steep slope) and a low affinity (shallow slope) interaction in the range 0–100 nM I125-LDL. Bound/free is defined as bound I125-LDL [(fmol/well)/free I125-LDL [nM]. To focus on the high affinity binding mode, the binding assays were carried out using I125-LDL concentrations below 30 nM. LDL release was measured after incubating receptors in presence of 18 nM I125-LDL, a concentration above half-maximal binding for the different variants but less than saturation, yet still high enough to yield sufficient radioactive counts above background for samples of both released (buffer) as well as bound I125-LDL (well).
Fig. 3.
Solid phase binding assays. Saturation binding curves were determined for WT and 12 mutants using I125-LDL as a ligand. The specific binding is shown. Each data point represents triplicate measurements and the error bars show the SD. The panel of mutants was divided in four sets and assayed together with WT. A: WT, H190Y, H562Y, and H586Y; B: WT, G293S, and F362A; C: WT, G375S, K560W, and K582W; D: WT, W144A, W193A, W515A, and W541A.
Fig. 4.
Scatchard plot analysis of selected binding data from Fig. 3. Each Scatchard plot visualizes a mutant (in black) as well as superimposed WT (in gray) permitting visual assessment of LDL affinity loss (decreased slope) or LDL affinity gain (increased slope) compared with WT. The slopes were calculated using nonlinear regression of the specific binding curves using a one-binding site model. Bound/free is defined as bound I125-LDL [(fmol/well)/free I125-LDL [nM]. A: H190Y; B: H562Y; C: H586Y; D: K560W; E: W144A; and F: W193A. For data and Scatchard plot analysis of all 12 mutants and their accompanying WT controls, see supplementary Fig. II.
LDL release
I125-LDL release by mutants and WT was monitored as a function of pH (Fig. 5). In a separate experiment, the acid-specific portion of I125-LDL release was examined as a function of time (Fig. 6; numerical values are given in supplementary Table I). WT receptor released a large fraction of I125-LDL already at pH ∼7, which corresponds to the barely acidic, very early endosomal compartments. Release was rapid, with 50% of the I125-LDL release occurring within 1–2 min, and was essentially complete by 5 min.
Fig. 5.
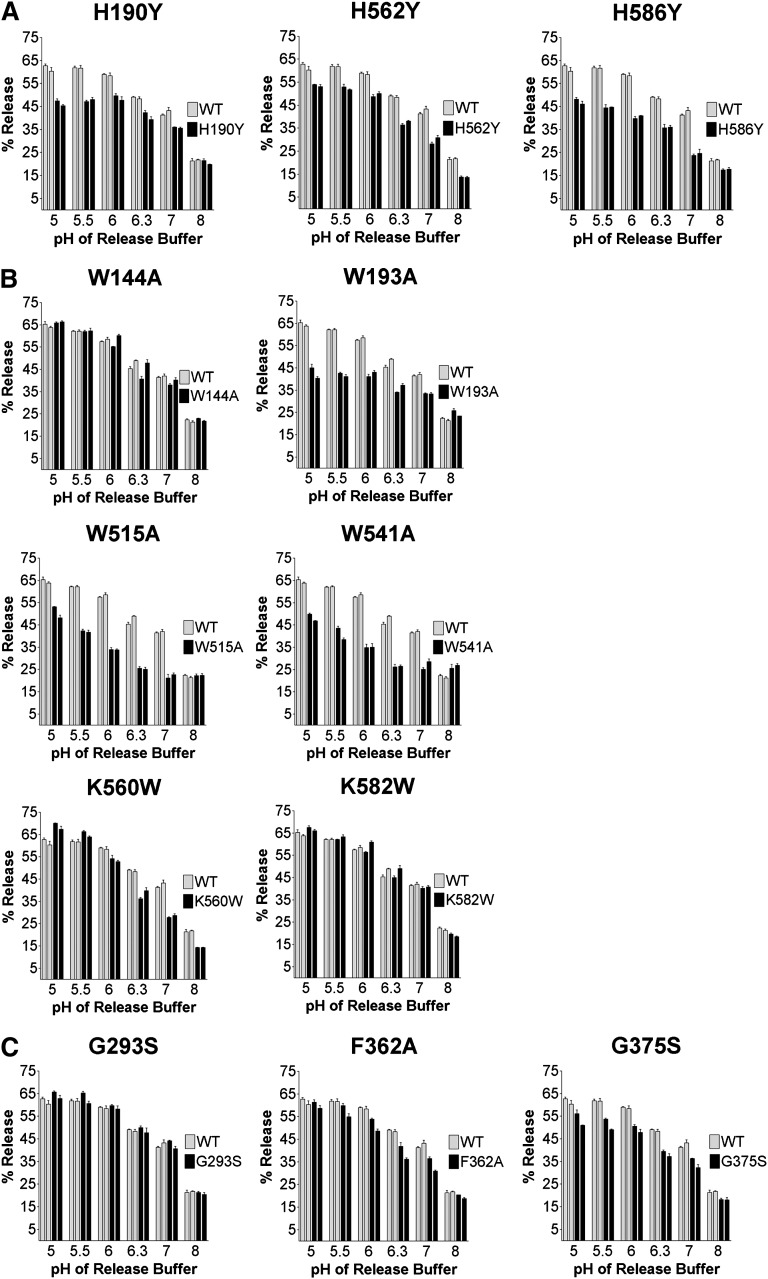
Release of I125-LDL as a function of pH. Twelve mutant and WT receptors were immobilized and prebound with I125-LDL using the solid phase binding assay. Ligand release was measured after 30 min at 4°C in buffers spanning a pH range pH 5–8 (see Materials and Methods). The % Release is expressed as (counts released into buffer)/(counts left in well + counts released into buffer) × 100%. Release by each mutant and WT was assayed in triplicate for each pH; error bars show the SD. The entire release experiment was repeated after 2 weeks and is displayed as the second set of bars. A: H190Y, H562Y, and H586Y; B: W144A, W193A, W515A, W541A, K560W, and K582W; C: G293S, F362A, and G375S. Statistical analysis using paired t-tests shows that I125-LDL release meets the criteria “extremely significantly” different from WT for the mutants H190Y, H562Y, H586Y, W515A, and G375S (P < 0.0001); W541A (P = 0.0002); W193A (P = 0.0004); and F362A (P = 0.0006). The mutant K560W has a P value of 0.08, indicating that its I125-LDL release properties may or may not be different from WT, while the mutants K582W, W144A, and G293S, all with P > 0.3, show release essentially indistinguishable from WT.
Fig. 6.
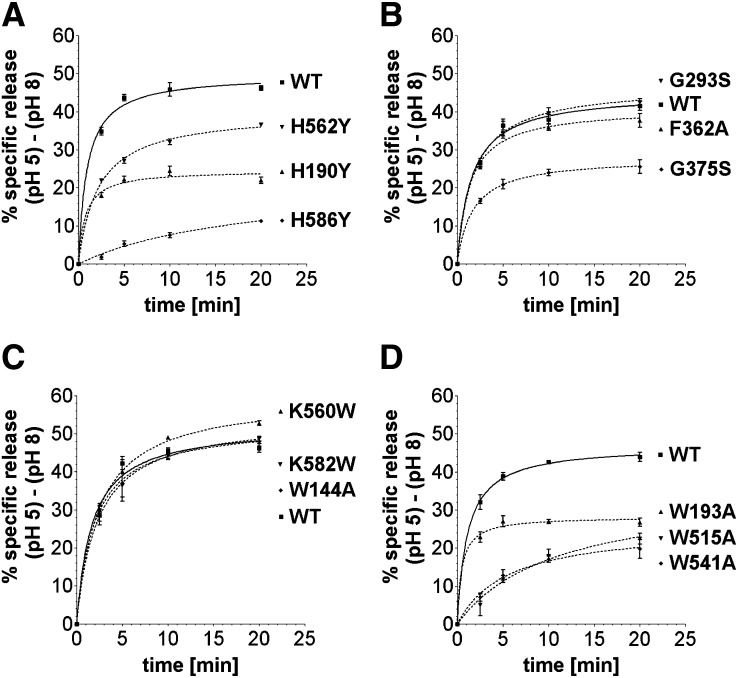
Release of I125-LDL as a function of time. Twelve mutant and WT receptors were immobilized and prebound with I125-LDL using the solid phase binding assay. To take into account that mutants might bind I125-LDL differently or display varying amounts of nonspecific release, the acid-specific portion of ligand release was calculated, i.e., specific release is (% release at pH 5) – (% release at pH 8). Ligand release was measured after 2.5, 5, 10, and 20 min at 4°C using release buffers at pH 5 or pH 8 (see Materials and Methods). Release by each mutant and WT was assayed in triplicate for each pH, and error bars show the SE of the mean.
All three histidine mutants, H190Y, H562Y, and H586Y, showed decreased release at acidic pH compared with WT, but each manifested their defect in distinctly different pH-dependent ways (Fig. 5A). H586Y showed greatly diminished release already under very mildly acidic conditions (pH 7), while H190Y showed a similar level of impairment only under very acidic conditions (pH 5 and pH 5.5). However, H562Y showed slightly reduced I125-LDL release over the complete pH range (pH 5–8), suggesting a more general impact on ligand release. The defects also manifested themselves in different ways kinetically. H190Y released roughly as fast as WT (t1/2max= 1 min), but a portion of receptors did not release ligand at all (Fig. 6A). In contrast, H562Y and H586Y released not only less I125-LDL but also at a slower rate (t1/2 max = 2 min and t1/2 max > 20 min, respectively).
The four tryptophan mutants showed striking differences in their release profiles as well. Though W515A and W541A bound significantly less I125-LDL than WT, both mutants still bound I125-LDL at levels 9 and 6 times greater, respectively, than nonspecifically bound LDL, justifying the examination of their release profiles. Three out of four tryptophan mutants (W193A, W515A, and W541A) showed large defects in pH dependent release that were at least as severe as those observed for the histidine mutants (Fig. 5B). W515A and W541A displayed inefficient I125-LDL release already under mildly acidic conditions (pH 7) but recovered somewhat at acidic pH 5. W515A and W541A also released I125-LDL 5 to 10 times slower compared with WT (t1/2 max = 10.8 min and t1/2 max = 5.5 min, respectively, compared with t1/2 max = 1 min for WT) (Fig. 6D). In contrast, W193A (just like H190Y) manifested a defect in I125-LDL release at highly acidic pH (pH 5 and pH 5.5) and released as fast as WT receptor, though only a subset of the receptors were able to release their ligand (Fig. 6D). Given its strategic location in the R4,R5,β-propeller interface, it is striking that W144A showed no defect in pH dependent LDL release at all (Fig. 5B) and released ligand as fast as WT (Fig. 6C).
The other mutants of the panel showed more subtle effects. K560W (but not K582W) generated a slightly more stable complex at neutral pH, as might be expected for a variant with increased affinity for LDL, that surprisingly was slightly less stable at pH 5–5.5 compared with WT (Fig. 5B). Both, K582W and K560W showed very quick ligand release similar to WT (Fig. 6C). The linker mutants (G293S, G375S, and F362A) showed subtle changes in I125-LDL release as a function of pH (Fig. 5C). A release defect in G375S was much more apparent as a function of time, with two pools of receptors, one that released similarly as WT and another pool that did not appear to release ligand (Fig. 6B). In total, 8 out of 12 mutants showed significant differences in the release of I125-LDL compared with WT, pinpointing the functional importance of these residues.
Hydrodynamic radius of LDL-R mutants
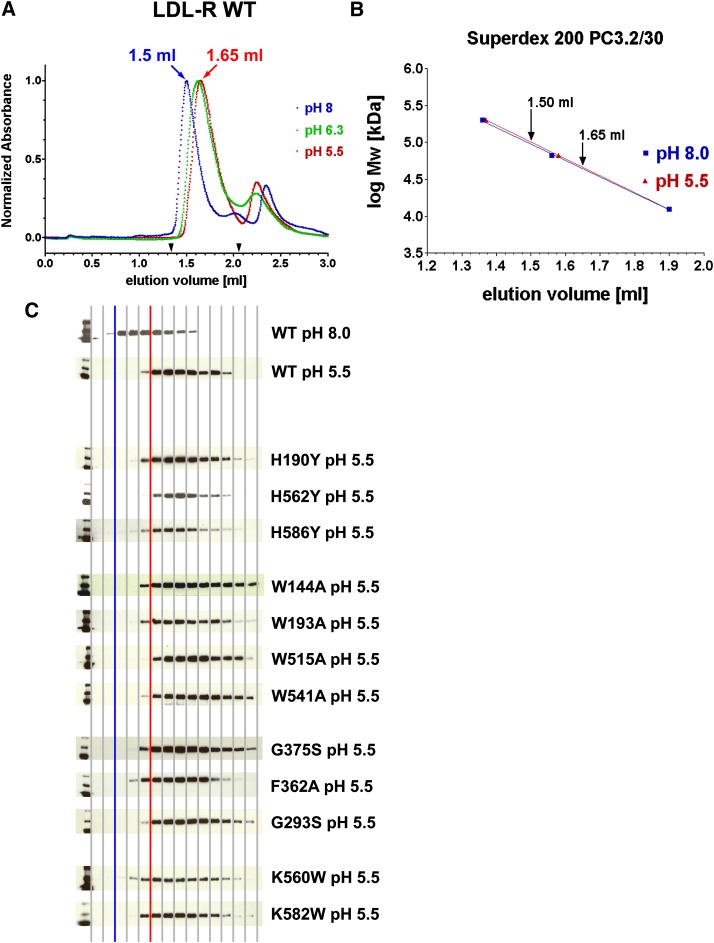
To assess if the mutant receptors displayed similar hydrodynamic behavior in solution as WT, their conformations were characterized on a specialized small-scale analytical size exclusion column at pH 8 and pH 5.5. In agreement with previous studies (12, 14–16), the elution volume for WT was significantly larger at pH 5.5 (suggesting a more compact molecule) than at pH 8 (suggesting a more elongated molecule), a shift that took place already at pH 6.3 (Fig. 7A, B). All 12 mutants displayed similar elution profiles to WT at pH 8 (data not shown) and at pH 5.5 (Fig. 7C). Therefore, all of the point mutations were able to adopt an elongated form at pH 8, even those with LDL binding defects; and none of the mutations were sufficient to prevent formation of the compact form at acidic pH, even those with ligand release defects. In fact, removal of Ca2+ ions from the cysteine-rich repeats of WT was insufficient to convert the receptor to an open form at pH 5.5 (see supplementary Fig. III), suggesting drastic mutations are needed to prevent the compact conformation. Indeed, the more radical variants H562K/H586K and H190K/H562K/H586K do prevent formation of a compact form (15). We conclude that I125-LDL binding defects observed for our mutants are not due to obstruction of the ligand-binding domain at neutral pH; rather, they are due to the disruption of specific binding epitopes for LDL. Furthermore, the ability to form a compact conformation does not appear sufficient for normal I125-LDL release.
Fig. 7.
Hydrodynamic characterization of 12 LDL-R mutants and WT using size exclusion chromatography. A: WT receptor migrates with a larger hydrodynamic radius (smaller elution volume) at pH 8 than at pH 5.5 or pH 6.3. B: The shift in hydrodynamic radius at pH 8 and pH 5.5 is specific to LDL-R as elution of gel filtration calibration markers are not affected. Shown are β-amylase (200 kDa), BSA (66 kDa), and cytochrome c (12.4 kDa). C: Western blot analysis visualizing the elution profiles of the 12 mutants and WT at pH 5.5; for comparison, WT at pH 8 is shown as well. Every second fraction in the elution range 1.33–2.059 ml (see arrowheads in A) was subject to SDS-PAGE electrophoresis, electroblotting, and visualization with an antihistidine tag antibody. For ease of interpretation, a blue vertical line indicates the start of WT elution at pH 8 and a red vertical line the start of WT elution at pH 5.5.
DISCUSSION
We designed a panel of mutations that our studies show affect LDL binding and release. In solution, the mutants show similar conformations of their extracellular domains as WT without aggregation, suggesting no gross misfolding. We were able to design mutants that disrupt binding and release simultaneously (H190Y, H586Y, W193A, W515A, and W541A), as well as mutants that affect only binding or release (W144A and H562Y). Therefore, residues in LDL-R that bind LDL only partially overlap residues mediating ligand release. We can't exclude, however, that certain mutations induce changes in ligand binding/release indirectly (for example, as a result of local misfolding, differential glycosylation, or altered calcium binding). Nor can we exclude that some mutations alter the tethering of the receptor by the anti-body HL1, potentially affecting LDL binding and/or release.
Modules in the extracellular domain of LDL-R express distinct characteristics in terms of LDL binding and release. For R4 and R5, we show that residues surrounding the Ca2+-binding sites are involved in LDL binding (W144 in R4 and W193 in R5). W144 and W193, located in analogous positions in R4 and R5, respectively, each provide a backbone carbonyl to a Ca2+ ion, while their side chains are solvent exposed. The W193A mutation likely does not disrupt protein folding of R5 because a variant containing W193A folds as single disulfide-bonded isomer and binds Ca2+ with similar affinity as wild-type R5 (32), and the isostructural W144A mutation is expected to be tolerated as well. The reduced LDL binding of H190Y suggests, furthermore, that in R5 the surface to one side of the Ca2+ ion formed by His 190, Ser 191, and Trp 193 is involved in LDL binding, agreeing with previous speculations (15, 32). R5 also contributes to ligand release given the impaired LDL release of H190Y and W193A. Strikingly, the release defect in H190Y and W193A is seen only at very acidic pH (pH 5.5 and below). In contrast and amazingly, W144A in R4, which just like W193A is defective in LDL binding, shows completely normal LDL release unlike W193A, demonstrating that R4 and R5 have different roles in the mechanism of LDL release.
β-Propeller
The β-propeller appears involved in both LDL binding and release. H586Y, W515A, and W541A display both decreased binding and release of LDL, while H562Y manifests only a defect in release. All four mutants show their largest defect at pH 6–7 (recovering somewhat at very acidic pH 5–5.5) and release LDL much slower than WT. The β-propeller mutant K560W appears to positively affect LDL binding and stimulate acidic release. The positive charges of Lys560 and Lys582 are therefore not necessary for either LDL binding or release, unlike the interaction between lysine residues of RAP and LDL-R R3-R4. The hinge residues G293S, G375S, and F362A have no effect on LDL binding affinity. G293S in the hinge between R7 and EGF-A shows no effect on LDL release. NMR studies of the fragment R7-EGF-A at pH 6.5 reveal the same kinked conformation as seen in the full-length LDL-R extracellular domain in the crystal structure at pH 5.3, and it has been suggested that R7 and EGF-A remain fixed with respect to each other over a broad pH range spanning neutral as well as acidic pH (24). The mild release defect of G375S suggests that EGF-B and the β-propeller may instead change their orientation with respect to each other at neutral versus acidic pH promoting efficient LDL release, but more drastic acid substitutions are needed to confirm this.
The notion that the β-propeller plays a role in LDL binding and release is indeed consistent with previous data. For example, the double mutation H562K/H586K has significantly reduced ligand binding (15). In the current study, we show that H586Y has reduced affinity for LDL, but H562Y does not. The single mutants H190Y, H562Y, H562N, and H586Y as well as the triple mutants (H190Y/H562Y/H586Y or H190A/H562A/H586A) have been shown to be defective in LDL release at acidic pH (6, 14, 15, 24). In the current study, we show that these three histidine residues touch different aspects of the LDL release mechanism because the defect in release for the β-propeller residues H562Y and H586Y is apparent already at pH 7 and slows release, whereas the release defect for H190Y in R5 (just like W193A in R5) is most apparent at highly acidic pH and does not slow release. Intriguingly, both the tryptophan mutations and the histidine mutations in the β-propeller reduce LDL release in a nearly identical manner as a function of pH and time.
Toward a model for LDL binding and release
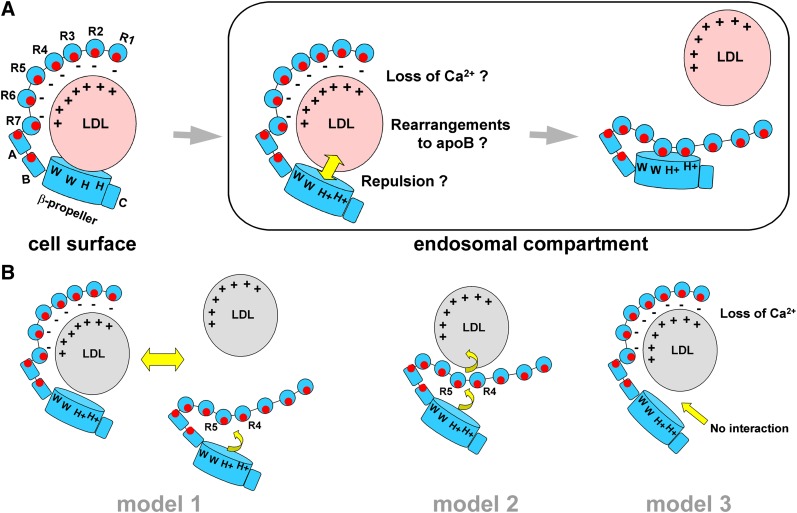
A number of models have recently been proposed describing the release mechanism of LDL by LDL-R. Model 1: At acidic pH, R4 and R5 preferentially interact with the β-propeller using the region surrounding their Ca2+-binding sites and promote LDL release (12, 13). Model 2: At acidic pH, the β-propeller binds R4 and R5 using the regions surrounding the Ca2+-binding sites, which triggers allosteric release of LDL that is bound elsewhere (for example, on the backsides of R4 and R5 away from the Ca2+-binding sites) (14). Model 3: An acidic pH causes cysteine-rich repeats of LDL-R to lose their Ca2+ ions and triggers release of LDL (25). Models 1 and 3 require that LDL binds residues near or at the Ca2+-binding sites of cysteine-rich repeats in LDL-R, held in a binding compatible conformation by Ca2+ ions. Model 2, in contrast, requires that LDL binds residues on LDL-R that do not participate in the R4,R5,β-propeller interface, thus excluding the residues surrounding the Ca2+-binding sites of R4 and R5 and the β-propeller residues buried in the interface. Furthermore, models 1 and 2 require that the β-propeller interacts with R4 and R5 to promote LDL release, while model 3 does not.
Our data are incompatible with aspects of all three models. Our results argue against model 1, whereby formation of the R4, R5, and the β-propeller interface is the sole driving force behind LDL release. If this were true, W144A, which packs against W515 and W541, should have the same negative effect on release as W515A and W541A from a structural perspective. In fact, W144A shows no effect on LDL release at all. Our results argue against model 2 because a number of residues buried in the R4,R5,β-propeller interface affect the affinity for LDL, suggesting that they contact LDL, precluding the proposed allosteric action of the β-propeller. Finally, our results also argue against model 3 as the primary mode of LDL release. In a study examining the Ca2+-binding affinity of R5 at both acidic and neutral pH, R5 has a Kd for Ca2+ of ∼13 μM at pH 5 and a Kd of <0.5 μM at pH 7.4 (33). The Ca2+ concentration in endocytotic vesicles spans a wide range (3–40 μM) (34, 35), so it is unclear at which pH significant loss of Ca2+ ions could occur. In our study, however, most LDL dissociates from LDL-R at pH 6–7 where Ca2+-binding sites are likely still occupied. Furthermore, the β-propeller mutations disrupting LDL release suggest that while Ca2+ ion depletion in LDL-R might contribute to ligand release at highly acidic pH, it cannot be a primary or sole release mechanism in the range pH 6–7 where most of the LDL is released.
We propose a fourth model of LDL release (Fig. 8). R4 and R5 mediate binding to LDL using their Ca2+-binding sites and surrounding residues, along with other modules, including the β-propeller. However at acidic pH, R4, R5, and the β-propeller work together to mediate release. For example, the tryptophan residues of the β-propeller could recruit interactions with LDL at neutral pH, that position His562 and His586 with respect to key residues in apolipoprotein B; at acidic pH, the LDL-R:LDL interactions would be incompatible due to positive charges acquired by H562 and H586, and any pH-induced rearrangements that apoB might undergo, and LDL-R and LDL would repel each other. The binding properties of the triple mutant H190A/H562A/H586A support this idea; this mutant binds an apoE-derived ligand three times better than WT at pH 6, but not at pH 7 (15), compatible with the idea that this mutant receptor has lost an LDL repulsive structural element that exists at pH 6 but not pH 7. In our model, efficient release at pH 6–7 would require intact β-propellers, whereas at highly acidic pH (pH 5.0–5.5), the ligand binding domain would contribute to LDL release, aided, for example, by loss of Ca2+ ions. Upon LDL release, the closed form of LDL-R is adopted and buries key residues for binding and release in the R4,R5,β-propeller interface, preventing untimely association with ligands inside the cell as the receptor cycles back to the cell surface. New mutants specifically testing this model and complementary studies using biophysical methods and cell surfaces studies will help further delineate the mechanism of LDL-R for LDL binding and release.
Fig. 8.
Proposed model for LDL binding and release by LDL-R. A: At neutral plasma pH, LDL binds to the extracellular domain of LDL-R on the cell surface via at least R4, R5, and the β-propeller, but likely other modules as well. Complex formation includes interactions between positively charged regions on LDL (depicted with + signs), and negatively charged regions on the cysteine-rich repeats of LDL-R (− signs). At acidic pH in endosomal compartments, His562 and His586 on the β-propeller acquire positive charges that are no longer compatible with the binding surface on LDL, leading to electrostatic repulsion of LDL; conformational rearrangements to apoB as well as loss of Ca2+ ions to LDL-R may further enhance LDL release. Upon LDL release, the LDL-R adopts a closed form that obscures residues involved in LDL binding and release and that may aid in its transport back to the cell surface. B: Key differences between our model and those previously proposed. Model 1 requires competition between the β-propeller and LDL to take place for interaction with R4 and R5. At acidic pH, the interaction of the β-propeller with R4 and R5 is favored, replacing LDL bound to LDL-R. Model 2 requires that LDL bind to a surface on R4 and R5 that does not overlap with the binding site for the β-propeller at acidic pH. At acidic pH as the β-propeller docks onto R4 and R5, allosteric changes within the receptor occur altering the LDL binding site, triggering LDL release. Model 3 does not require the β-propeller for either LDL binding or release; LDL release is mediated by the loss of Ca2+ ions from the cysteine-rich repeats in LDL-R at acidic pH.
Supplementary Material
Acknowledgments
We thank Beverly Murray and John Gaubatz for preliminary studies and Faith Bjork for radiolabeling LDL. Dr. Johann Deisenhofer is most gratefully acknowledged for initial support and together with Dr. John Traynor thanked for helpful discussions.
Footnotes
Abbreviations:
- apo
- apolipoprotein
- EGF
- epidermal growth factor
- FH
- familial hypercholesterolemia
- LDL-R
- low density lipoprotein receptor
- RAP
- receptor-associated protein
- WT
- wild type
This work has been supported by the American Heart Association through a scientist development award to G.R.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and one table.
REFERENCES
- 1.Goldstein J. L., Brown M. S. 2001. Molecular medicine. The cholesterol quartet. Science. 292: 1310–1312. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J. L., Hobbs H. H., Brown M. S. 2001. Familial hypercholesterolemia. the Metabolic and Molecular Bases of Inherited Disease. 8th edition Vol. 2 Valle D., editor McGraw-Hill, New York: 2863–2914. [Google Scholar]
- 3.Davis C. G., Goldstein J. L., Südhof T. C., Anderson R. G., Russell D. W., Brown M. S. 1987. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 326: 760–765. [DOI] [PubMed] [Google Scholar]
- 4.Miyake Y., Tajima S., Funahashi T., Yamamoto A. 1989. Analysis of a recycling-impaired mutant of low density lipoprotein receptor in familial hypercholesterolemia. J. Biol. Chem. 264: 16584–16590. [PubMed] [Google Scholar]
- 5.van der Westhuyzen D. R., Stein M. L., Henderson H. E., Marais A. D., Fourie A. M., Coetzee G. A. 1991. Deletion of two growth-factor repeats from the low-density-lipoprotein receptor accelerates its degradation. Biochem. J. 278: 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Hoof D., Rodenburg K. W., Van der Horst D. J. 2005. Intracellular fate of LDL receptor family members depends on the cooperation between their ligand-binding and EGF domains. J. Cell Sci. 118: 1309–1320. [DOI] [PubMed] [Google Scholar]
- 7.Brown M. S., Goldstein J. L. 2006. Biomedicine. Lowering LDL–not only how low, but how long? Science. 311: 1721–1723. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russell D. W. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 39: 27–38. [DOI] [PubMed] [Google Scholar]
- 9.Jeon H., Meng W., Takagi J., Eck M., Springer T. A., Blacklow S. C. 2001. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat. Struct. Biol. 8: 499–504. [DOI] [PubMed] [Google Scholar]
- 10.Defesche J. C. 2004. Low-density lipoprotein receptor–its structure, function, and mutations. Semin. Vasc. Med. 4: 5–11. [DOI] [PubMed] [Google Scholar]
- 11.Fass D., Blacklow S., Kim P. S., Berger J. M. 1997. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 388: 691–693. [DOI] [PubMed] [Google Scholar]
- 12.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. 2002. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 298: 2353–2358. [DOI] [PubMed] [Google Scholar]
- 13.Rudenko G., Deisenhofer J. 2003. The low-density lipoprotein receptor: ligands, debates and lore. Curr. Opin. Struct. Biol. 13: 683–689. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z., Michaely P. 2008. The epidermal growth factor homology domain of the LDL receptor drives lipoprotein release through an allosteric mechanism involving H190, H562, and H586. J. Biol. Chem. 283: 26528–26537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto T., Chen H. C., Guigard E., Kay C. M., Ryan R. O. 2008. Molecular studies of pH-dependent ligand interactions with the low-density lipoprotein receptor. Biochemistry. 47: 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. 2008. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 105: 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Driel I. R., Brown M. S., Goldstein J. L. 1989. Stoichiometric binding of low density lipoprotein (LDL) and monoclonal antibodies to LDL receptors in a solid phase assay. J. Biol. Chem. 264: 9533–9538. [PubMed] [Google Scholar]
- 18.Esser V., Limbird L. E., Brown M. S., Goldstein J. L., Russell D. W. 1988. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 263: 13282–13290. [PubMed] [Google Scholar]
- 19.Russell D. W., Brown M. S., Goldstein J. L. 1989. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 264: 21682–21688. [PubMed] [Google Scholar]
- 20.Nguyen A. T., Hirama T., Chauhan V., Mackenzie R., Milne R. 2006. Binding characteristics of a panel of monoclonal antibodies against the ligand binding domain of the human LDLr. J. Lipid Res. 47: 1399–1405. [DOI] [PubMed] [Google Scholar]
- 21.Knott T. J., Rall S. C., Jr., Innerarity T. L., Jacobson S. F., Urdea M. S., Levy-Wilson B., Powell L. M., Pease R. J., Eddy R., Nakai H., et al. 1985. Human apolipoprotein B: structure of carboxyl-terminal domains, sites of gene expression, and chromosomal localization. Science. 230: 37–43. [DOI] [PubMed] [Google Scholar]
- 22.Mahley R. W. 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 240: 622–630. [DOI] [PubMed] [Google Scholar]
- 23.Boren J., Lee I., Zhu W., Arnold K., Taylor S., Innerarity T. L. 1998. Identification of the low density lipoprotein receptor-binding site in apolipoprotein B100 and the modulation of its binding activity by the carboxyl terminus in familial defective apo-B100. J. Clin. Invest. 101: 1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beglova N., Jeon H., Fisher C., Blacklow S. C. 2004. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol. Cell. 16: 281–292. [DOI] [PubMed] [Google Scholar]
- 25.Arias-Moreno X., Velazquez-Campoy A., Rodríguez J. C., Pocoví M., Sancho J. 2008. Mechanism of low density lipoprotein (LDL) release in the endosome: implications of the stability and Ca2+ affinity of the fifth binding module of the LDL receptor. J. Biol. Chem. 283: 22670–22679. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein J. L., Basu S. K., Brown M. S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260. [DOI] [PubMed] [Google Scholar]
- 27.Villéger L., Abifadel M., Allard D., Rabès J. P., Thiart R., Kotze M. J., Béroud C., Junien C., Boileau C., Varret M. 2002. The UMD-LDLR database: additions to the software and 490 new entries to the database. Hum. Mutat. 20: 81–87. [DOI] [PubMed] [Google Scholar]
- 28.Leigh S. E., Foster A. H., Whittall R. A., Hubbart C. S., Humphries S. E. 2008. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann. Hum. Genet. 72: 485–498. [DOI] [PubMed] [Google Scholar]
- 29.Fisher C., Beglova N., Blacklow S. C. 2006. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell. 22: 277–283. [DOI] [PubMed] [Google Scholar]
- 30.Jensen G. A., Andersen O. M., Bonvin A. M. J. J., Bjerrun-Bohr I., Etzerodt M., Thogersen H. C., O'Shea C., Poulsen F. M., Kragelund B. B. 2006. Binding site structure of one LRP- RAP complex: implications for a common ligand-receptor binding motif. J. Mol. Biol. 362: 700–716. [DOI] [PubMed] [Google Scholar]
- 31.Brown M. S., Goldstein J. L. 1974. Familial hypercholesterolemia: defective binding of lipoproteins to cultured fibroblasts associated with impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity. Proc. Natl. Acad. Sci. USA. 71: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdul-Aziz D., Fisher C., Beglova N., Blacklow S. C. 2005. Folding and binding integrity of variants of a prototype ligand-binding module from the LDL receptor possessing multiple alanine substitutions. Biochemistry. 44: 5075–5085. [DOI] [PubMed] [Google Scholar]
- 33.Simonovic M., Dolmer K., Huang W., Strickland D. K., Volz K., Gettins P. G. 2001. Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry. 40: 15127–15134. [DOI] [PubMed] [Google Scholar]
- 34.Gerasimenko J. V., Tepikin A. V., Petersen O. H., Gerasimenko O. V. 1998. Calcium uptake via endocytosis with rapid release from acidifying endosomes. Curr. Biol. 8: 1335–1338. [DOI] [PubMed] [Google Scholar]
- 35.Christensen K. A., Myers J. T., Swanson J. A. 2002. pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 115: 599–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.