Abstract
Group IVA cytosolic phospholipase A2α (cPLA2α) plays a role in the microbicidal machinery of immune cells by translocating to phagosomes to initiate the production of antimicrobial eicosanoids. In this work, we have studied the involvement of the cationic cluster of cPLA2α (Lys488/Lys541/Lys543/Lys544) in the translocation of the enzyme to the phagosomal cup in human macrophages responding to opsonized zymosan. Phagocytosis was accompanied by an increased mobilization of free arachidonic acid, which was strongly inhibited by pyrrophenone. In transfected cells, a catalytically active enhanced green fluorescent protein-cPLA2α translocated to the phagocytic cup, which was corroborated by frustrated phagocytosis experiments using immunoglobulin G-coated plates. However, a cPLA2α mutant in the polybasic cluster that cannot bind the anionic phospholipid phosphatidylinositol 4, 5-bisphosphate (PIP2) did not translocate to the phagocytic cup. Moreover, an enhanced yellow fluorescent protein (EYFP)-cPLA2α and an enhanced cyan fluorescent protein-pleckstrin homology (PH) domain of the phospholipase Cδ1 (PLCδ1) construct that specifically recognizes endogenous PIP2 in the cells both localized at the same sites on the phagosome. High cellular expression of the PH domain inhibited EYFP-cPLA2α translocation. On the other hand, group V-secreted phospholipase A2 and group VIA calcium-independent phospholipase A2 were also studied, but the results indicated that neither of these translocated to the phagosome. Collectively, these data indicate that the polybasic cluster of cPLA2α (Lys488/Lys541/Lys543/Lys544) regulates the subcellular localization of the enzyme in intact cells under physiologically relevant conditions.
Keywords: lipid mediators, monocytes/macrophages, phagocytosis, phospholipase A2, inflammation
Phospholipase A2 (PLA2) enzymes cleave membrane phospholipids at the sn-2 position of the glycerol backbone, releasing a free fatty acid and a lysophospholipid (1). One of the better studied roles of PLA2 enzymes is their involvement in inflammatory processes due to their ability to liberate free arachidonic acid (AA) from membrane phospholipids (2–5). Free AA will in turn be oxygenated by specific enzymes to generate a wide variety of compounds with potent pro- and anti-inflammatory actions, collectively called the eicosanoids (6, 7). Some of these compounds have also been described as important bactericidal agents (8).
Of the many existing PLA2 enzymes, only one, the group IVA PLA2 [also known as cytosolic phospholipase A2α (cPLA2α)], manifests specificity for AA-containing phospholipids (9). Today, it is widely accepted that cPLA2α is the key enzyme in AA release leading to physiological and/or pathophysiological eicosanoid production (9–11). Structurally, cPLA2α possesses a C2 domain containing the binding site for Ca2+ [and a binding site for ceramide-1 phosphate as well (12)] and a catalytic domain where the residues involved in enzymatic activity are located (9). The catalytic domain also contains a cluster of cationic amino acids (Lys488, Lys541, Lys543, and Lys544) that may mediate enzyme binding to anionic phospholipids in membranes (13). In fact, in vitro, anionic phospholipids such as phosphatidylinositol 4,5-bisphosphate (PIP2), PI(3,4,5)P3, and PI(3,4)P2 strongly increase the cPLA2α specific activity when incorporated into the vesicle substrate (14, 15). Mutation of the aforementioned polybasic 4-Lys cluster eliminates the activating effect of PIP2 on cPLA2α activity (13). Also, by plasmon resonance experiments, it has been demonstrated that PIP2 activates cPLA2α by increasing the catalytic efficiency of the enzyme as a result of increasing membrane penetration (16). In live cells, exogenous PIP2 and PI(3,4)P2 induce the translocation of cPLA2α to the intracellular membranes (17). More recently, it has been described that mutation of the Lys488, Lys543, and Lys544, decreases AA release in cells activated by serum, pointing to an important role for that cluster in cPLA2 activation under physiological stimulation (18).
Phagocytosis is a specialized process of cells of the innate immune system to engulf invading microorganisms, foreign particles, and apoptotic cells and debris. Phagocytosis is initiated at the site of particle attachment, producing a polarized region within the membrane that enlarges as a pseudopod to engulf the particle and drive it into the cytoplasm where it will be degraded (19). It has been previously shown that cPLA2α translocates to the phagosomal cup during the ingestion of particles, and such a translocation appears to be important for in situ eicosanoid generation and efficient elimination of microbes (20–23). However, the mechanism regulating this translocation remains largely unknown. Using human macrophages responding to opsonized zymosan, we demonstrate in this work that translocation of cPLA2α to the phagocytic cup depends on an intact cationic cluster Lys488/Lys541/Lys543/Lys544) in the catalytic domain of the enzyme. These results reveal a novel role for the cluster and provide new insights into the complex cellular regulation of the cPLA2α.
MATERIALS AND METHODS
Materials
[5,6,8,9,11,12,14,15-3H]AA 200 Ci/mmol was purchased from Amersham Ibérica (Madrid, Spain). Zymosan labeled with Alexa Fluor 594 and 1,2-bis-(4,4-difluoro-5,7-dimethyl-4-bora-3a, 4a-diaza-sindecane-3-undecanoyl)-sn-glycero-3-phosphocholine (bis-BODIPY FL C11-PC) were from Molecular Probes (Carlsbad, CA). Ficoll-PaqueTM Plus was from GE Healthcare (Uppsala, Sweden). Gentamicin was purchased from Biowhittaker (Walkersville, MD). Human macrophage Nucleofection solution was from Amaxa (Gaithersburg, MD). Macrophage serum free medium and RPMI 1640 were purchased from GIBCO (Carlsbad, CA). The enhanced green fluorescent protein (EGFP)-PLCδ1-PH construct with the pleckstrin domain from the PLCδ1 was kindly provided by Dr. Tobias Meyer (Stanford University Medical Center, Stanford, CA) (24). The EGFP-cPLA2α plasmid, the mutant for the PIP2 binding site EGFP-4KE/A-cPLA2α, the iPLA2-VIA- enhanced yellow fluorescent protein (EYFP) plasmid, and the sPLA2-V-EGFP plasmid have been described elsewhere (17, 25, 26). pDsRed-Monomer-C1 was obtained from Clontech (Mountain View, CA). All other reagents were from Sigma.
Cells
Human macrophages were obtained from Buffy coats of healthy volunteer donors obtained from the Centro de Hemoterapia y Hemodonación de Castilla y León (Valladolid, Spain). Briefly, blood cells were diluted 1:1 with PBS, layered over a cushion of Ficoll-Paque and centrifuged at 750 g for 30 min. The mononuclear cellular layer was then recovered and washed three times with PBS, resuspended in RPMI supplemented with 2 mM L-glutamine, 40 μg/ml of gentamicin, and allowed to adhere to plastic in sterile dishes for 2 h. Nonadherent cells were then removed by extensively washing with PBS. Macrophage differentiation was achieved by incubating the adhered monocytes in RPMI supplemented with 2 mM L-glutamine, 40 μg/ml of gentamicin, and 5% human serum for 2 weeks in the absence of exogenous cytokine mixtures.
Plasmid transfection
Human macrophages were transfected by the Nucleofection technique (Amaxa), following the kit specifications for human macrophages. Briefly, cells were harvested by treatment with trypsin for 90 min and then by gentle scraping. After washing, the cells were resuspended in 100 μl Human Macrophage Nucleofector solution plus 5 μg of plasmid. Nucleofection was carried out using the program Y-010 and the cells were resuspended in 400 μl of macrophage serum free medium (GIBCO) plus 5% heat inactivated human serum.
Synchronized phagocytosis
These experiments were carried out as described elsewhere with minor modifications (27). Macrophages were seeded over glass coverslips, allowed to adhere, and then washed with RPMI and resuspended in this medium. Cells were then kept at 4°C for 5 min and opsonized zymosan was then added. After 15 min incubation, coverslips were transferred to plates with RPMI at 37°C and the phagocytosis was allowed to proceed for different periods of time. Reactions were stopped by fixation with 3% paraformaldehyde and 3% sacarose for 15 min if they were to be analyzed by microscopy. After three washes with PBS, the coverslips were mounted in glass slides with antifade medium. Samples were then analyzed by epifluorescent or confocal microscopy.
Confocal microscopy
Transfected cells were seeded in MatTek dishes and allowed to adhere for 24 h in RPMI supplemented with 2 mM L-glutamine, 40 μg/ml of gentamicin, and 5% human serum. Medium was then changed by HBSS with 10 mM HEPES and 1.3 mM CaCl2 and fluorescence was monitored by confocal a Bio-Rad Radiance 2100 laser-scanning system coupled to a Nikon TE-2000U with a thermostatized chamber (Warner Instruments). The objective was CFI Plan Apo 60×, 1.4 numerical aperture, oil immersion. The fluorescence of enhanced cyan fluorescent protein (ECFP) was monitored at 457 nm argon excitation using the combination of a long pass barrier filter HQ470LP and a short pass filter HQ520SP. The fluorescence of EGFP was monitored at 488 nm Argon excitation using the combination of a long pass filter HQ500LP and a short pass filter HQ560SP. The EYFP was monitored at 514 nm Argon excitation and the filters HQ520LP and HQ560 SP. The Alexa Fluor 594 fluorescence was monitored at 543 nm HeNe excitation using a long band pass filter HQ570LP.
AA release
Cells were labeled with 0.5 μCi/ml of [3H]AA overnight. Afterward, the cells were washed three times with PBS supplemented with 0.5 mg/ml fatty acid free-BSA and incubated with RPMI supplemented with 0.5 mg/ml fatty acid-free BSA and 1% ITS. Cells were then stimulated and supernatants were removed at different time periods. Cells monolayers were overlaid with ice-cold phosphate buffer containing 0.05% Triton X-100 and scraped. Radioactivity was quantified by liquid scintillation counting and AA release in the supernatants was referred to total radioactivity for each condition.
Frustrated phagocytosis
This was carried out as described by Marshall et al. (28). Briefly, human macrophages were dislodged from the culture plates by trypsin treatment, and were resuspended in RPMI containing 10 mM HEPES and 2 mM EDTA. Cells were gently stirred for 2–3 h to allow for the reexpression of membrane receptors. Cells were then washed, resuspended in RPMI containing 10 mM HEPES and 2 mM MgCl2, and plated over MatTek dishes treated or not with 10 mg/ml pure IgG. After 30 min at 37°C, the cells were monitored by confocal microscopy. Some pictures were obtained in the XZ axis to have a better view of the cell membranes attached to the glass. In some experiments, the cells were labeled with 5 μM bis-BODIPY FL C11-PC for 30 min, washed twice, and processed for frustrated phagocytosis. Fluorescence was monitored by confocal microscopy using 488 argon excitation and the combination of a HQ500 long band pass filter and HQ560 short band pass filter.
RESULTS
Translocation of cPLA2alpha to the phagosome in zymosan-stimulated human macrophages
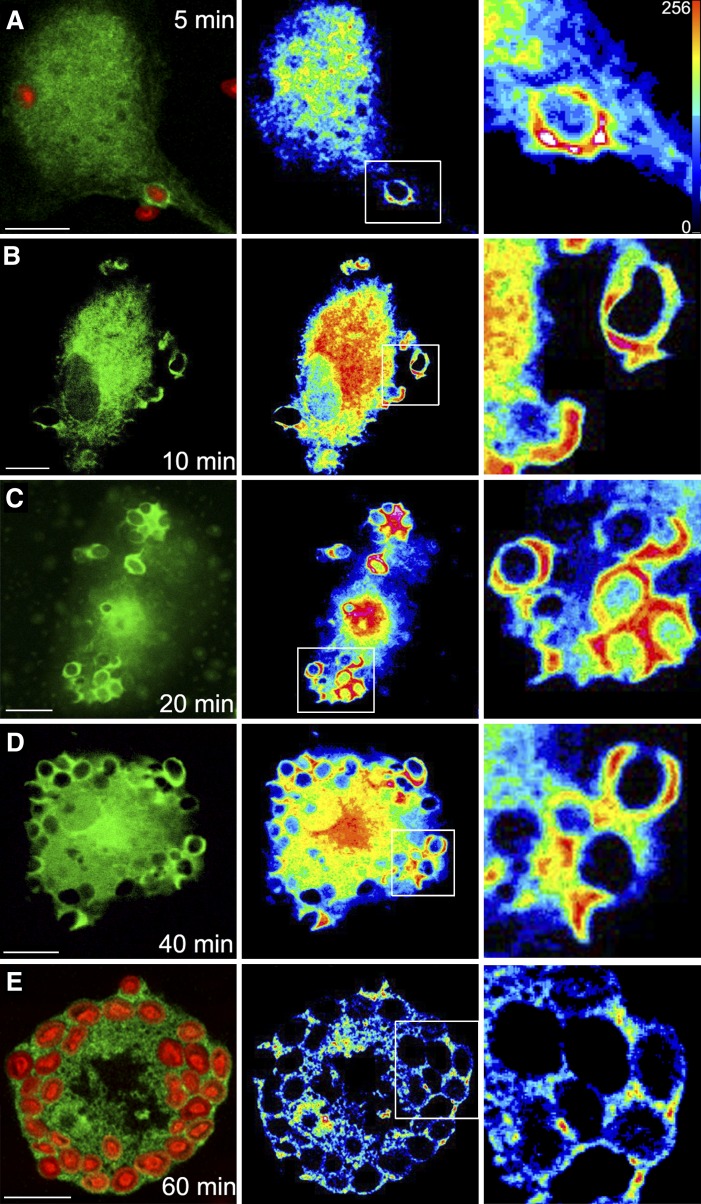
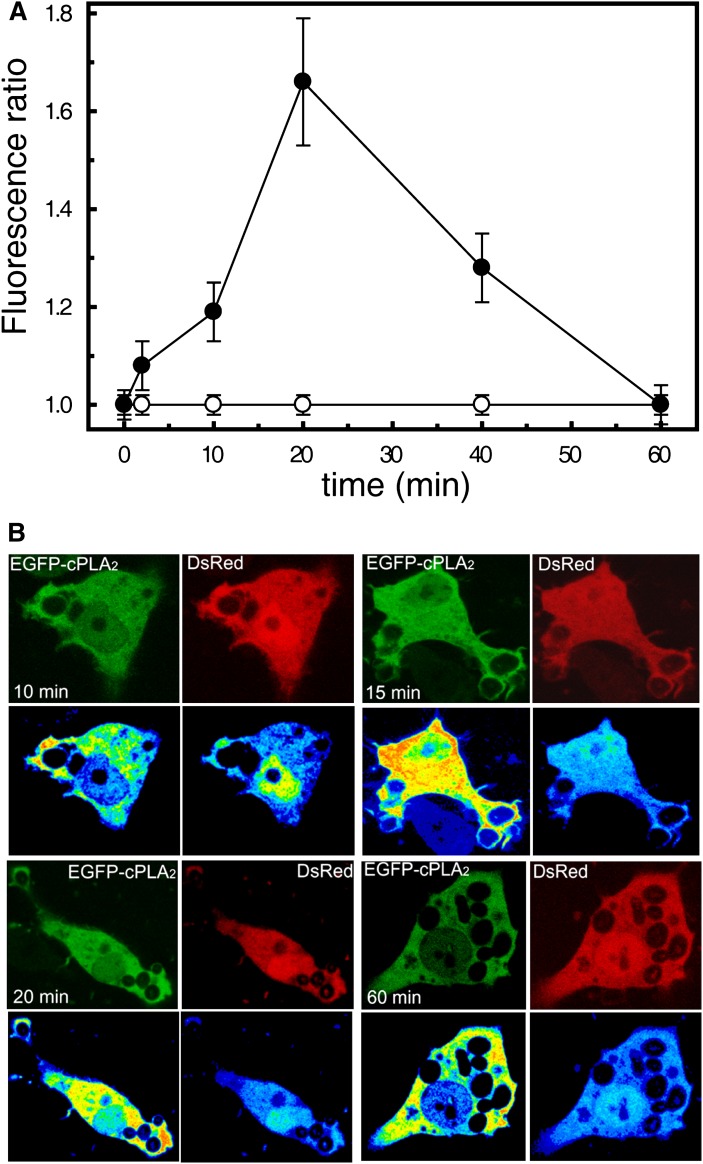
Human macrophages are capable of recognizing yeast-derived zymosan particles and engulf them. By using transfected cells, we detected the translocation of a chimeric construct EGFP-cPLA2α from the cytosol to the phagocytic cups (Fig. 1). Translocation of the enzyme was particularly prominent in nonsealed phagocytic cups (Fig. 1, 10–15 min). Once the phagosome was sealed and internalized, the EGFP-cPLA2α separated from it (Fig. 1, 60 min). Experiments were performed next to rule out the possibility that the increased fluorescence arising from EGFP-cPLA2α in the forming phagosome was due to an increased volume of cytoplasm imaged in the plane. This was addressed by imaging in the same cell the construct EGFP-cPLA2α and a monomeric form of DsRed (to correct for local variations in cytoplasmic volume). Figure 2 shows a clear increase in EGFP-cPLA2α fluorescence in the phagosomes that does not correspond with an increase in cytoplasmic volume (ratio EGFP-cPLA2α/DsRed) at any time, thus indicating that enzyme translocation actually occurs.
Fig. 1.
Translocation of cPLA2α to the phagosome in zymosan-treated human macrophages. Human macrophages were transfected with the construct EGFP-cPLA2α, treated with 1 mg/ml opsonized zymosan for 5 min (A), 10 min (B), 20 min (C), 40 min (D), and 60 min (E), and analyzed by confocal microscopy. In (A) and (E), Alexa Fluor 594-labeled zymosan was used to better visualize the particles. Fluorescence arising from the EGFP-cPLA2α (green) or zymosan (red) is shown on the left column. Middle-column panels show a pseudocolored fluorescence intensity from the EGFP-cPLA2α. Panels on the right column show a detailed amplification of the phagosomes framed in the middle panels. Scale bar = 10 μm.
Fig. 2.
Analysis of the translocation of cPLA2α and the monomeric DsRed to phagosomal membranes. Human macrophages were cotransfected with the construct EGFP-cPLA2α or the soluble fluorescent protein DsRed (monomeric form). Cells were stimulated with opsonized zymosan, fixed, and fluorescence was analyzed by confocal microscopy at different times. A: The figure represents the fluorescence ratio EGFP-cPLA2α / DsRed in phagosomes versus cytosol at each time point. At least four phagosomes per cell were analyzed from many cells. B: Pictures of the cellular fluorescence of the EGFP-cPLA2α and the DsRed at different times of phagocytosis are shown. Pseudocolor analysis of the fluorescence is shown in the lower panels at each time point.
Phagocytosis of opsonized zymosan activates cPLA2α in human macrophages
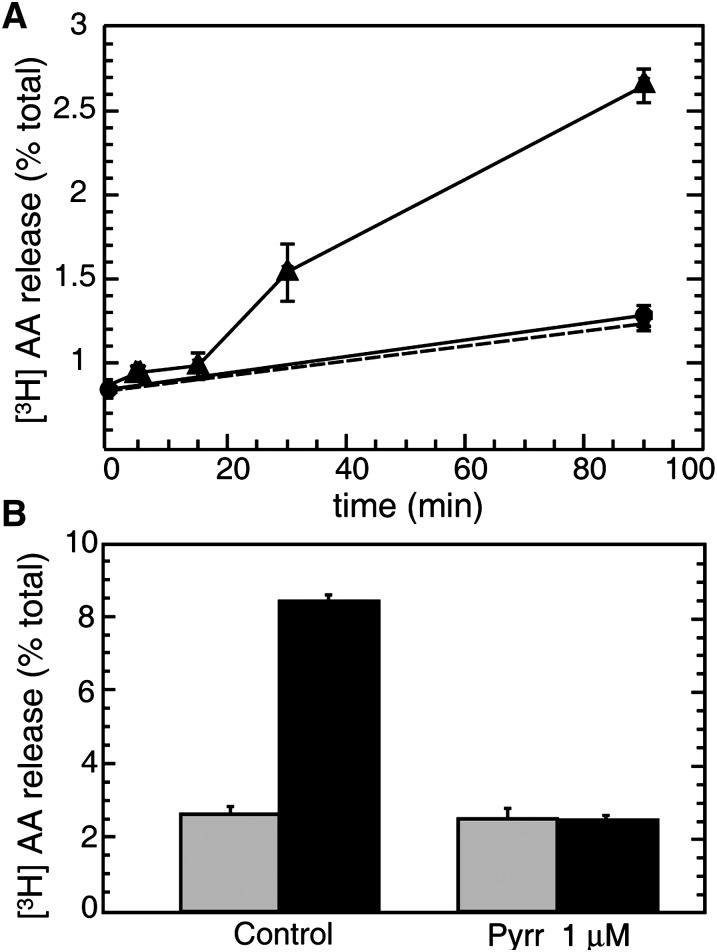
Zymosan induced a significant release of AA (and metabolites) to the extracellular medium, suggesting the activation of a PLA2 (Fig. 3A). This PLA2 was identified as cPLA2α on the basis of complete inhibition of the response by a low concentration of pyrrophenone, a cPLA2α inhibitor (29) (Fig. 3B).
Fig. 3.
AA release in human macrophages challenged with opsonized zymosan. A: Human macrophages labeled with [3H]AA were treated with 1 mg/ml opsonized zymosan (black triangles) or vehicle (black circles). AA release was assessed at different times as described under Materials and Methods. B: Cells labeled with [3H]AA were pretreated with 1 μM pyrrophenone (pyrr) or vehicle (control) for 30 min and then treated with 1 mg/ml opsonized zymosan (black bars) or vehicle (gray bars) for 1 h.
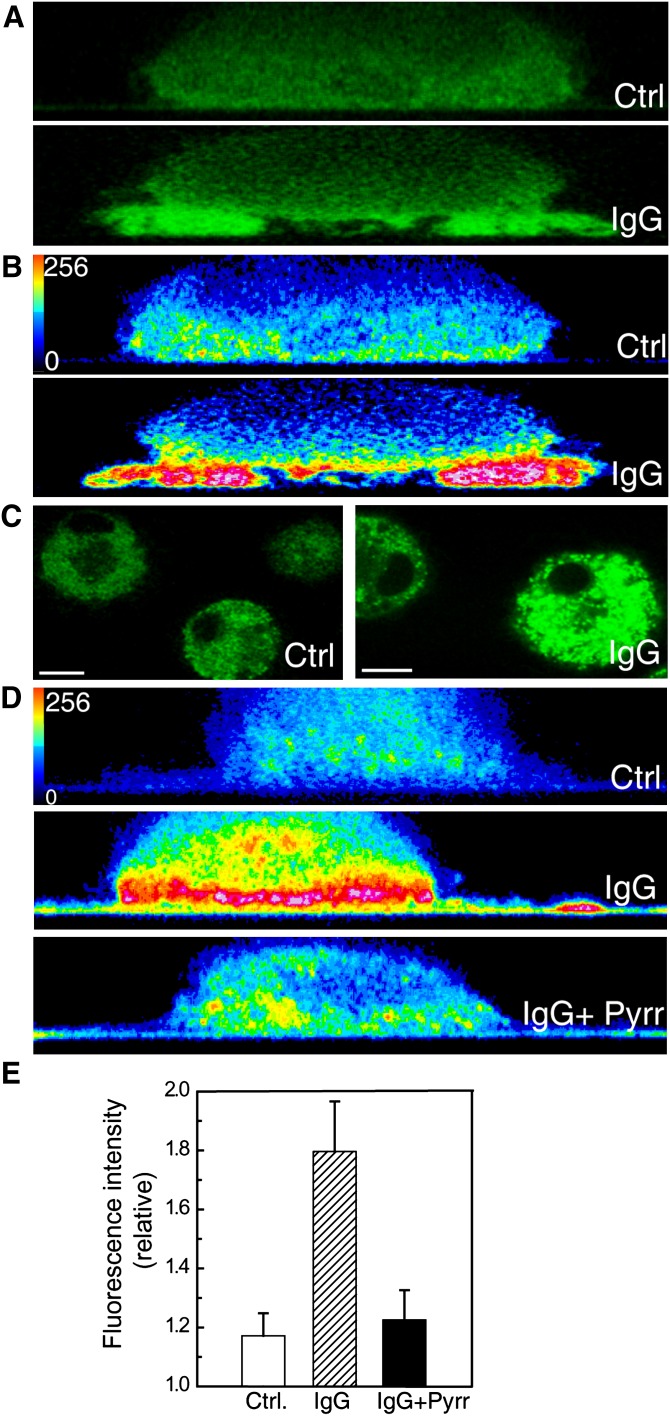
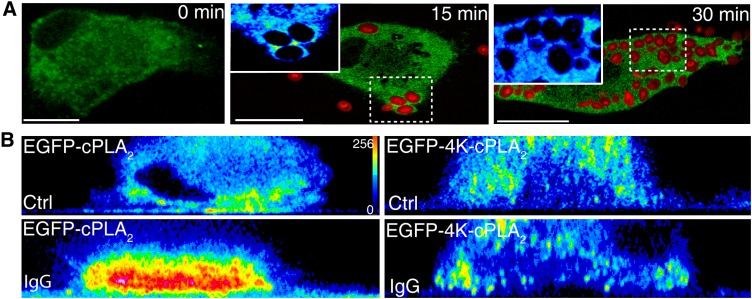
To confirm that the cPLA2α that translocates to the phagocytic cup is functionally active, an experiment of frustrated phagocytosis was performed, using glass plates coated with IgG (28). Macrophages exposed to IgG-coated glass surfaces responded by translocating the EGFP-cPLA2α to the membranes more proximal to the glass surface but not to other cellular membranes (Fig. 4A, B, E). In this experiment, the IgG-coated glass would represent the phagocytosable particle (28). Next, we loaded the cells with the fluorogenic phospholipase substrate bis-BODIPY FL C11-PC (30), and subjected them to the frustrated phagocytosis assay. A dramatic increase in fluorescence was observed, especially in the proximity of the IgG-coated glass (Fig. 4C, D and F), indicating that an A-type phospholipase is acting at that place (where the “phagosome” is being initiated). To confirm that such a phospholipase is actually cPLA2α, we conducted experiments in the presence of pyrrophenone (Fig. 4D and F). As expected, pyrrophenone at doses as low as 1 µM strongly blocked the fluorescence increase in the cells exposed to IgG-coated glass, indicating that such a fluorescence increase is mainly due to cPLA2α activation.
Fig. 4.
cPLA2α is functionally active in the phagosomal cup in human macrophages. A: Cells transfected with EGFP-cPLA2 were subjected to a frustrated phagocytosis assay on noncoated glass (Ctrl) or IgG-coated glass (IgG), as indicated, and analyzed by confocal microscopy. Pictures of the XZ axis of the cells were taken. B: The intensity of the fluorescence obtained in (A) was analyzed in pseudocolor. C: Cells, labeled with bis-BODIPY FL C11-PC, were plated on noncoated glass (Ctrl) or on IgG-coated glass (IgG) for 30 min, and fluorescence was analyzed by confocal microscopy. The mean of fluorescence intensity in Ctrl was 66 and in IgG was 128. D: The cells were labeled with bis-BODIPY FL C11-PC and subjected to a frustrated phagocytosis assay. Pictures of the XZ axis of the cells were taken and the intensity of the fluorescence was analyzed in pseudocolor. In the picture on the bottom, the cells were treated with 1 μM pyrrophenone (Pyrr). White bar = 10 μm. E: Statistical analysis of the fluorescence intensity in cells assayed for frustrated phagocytosis as in D. Data are represented as relative fluorescence intensities (mean fluorescence intensity in membranes closer to the glass/mean fluorescence intensity in the cytosol). At least 15 different cells were analyzed.
Mutation of a four-lysine cluster in cPLA2α that is involved in binding of anionic phospholipids suppresses EGFP-cPLA2α translocation to the phagocytic cup
We have previously described that mutations on Lys488, Lys541, Lys543, and Lys544 of cPLA2α result in a defective translocation of cPLA2α to intracellular membranes in response to exogenous PIP2 (17). Because it is well described that PIP2 increases in the phagosome (31), we next studied the behavior of the 4-Lys mutant (EGFP-4KE/A-cPLA2α) in macrophages exposed to opsonized zymosan. The results, as shown in Fig. 5A, clearly indicated that this mutant does not translocate to the phagocytic cup in activated cells at any time tested, suggesting that a functional binding site in cPLA2α for anionic phospholipids is necessary for such a translocation to be observed. To further substantiate this observation, we performed experiments of frustrated phagocytosis, similar to those shown in Fig. 4, using cells transfected with the EGFP-4KE/A-cPLA2α mutant. Fig. 5B shows that, unlike the wild-type enzyme, the mutant did not translocate to the membranes that are closer to the IgG-coated plate. These data highlight the importance of the 4-Lys cluster for proper binding of cPLA2α to the phagosomal membranes.
Fig. 5.
The mutant EGFP-4KE/A-cPLA2α does not translocate to the phagosome in human macrophages. A: Human macrophages transfected with the mutant EGFP-4KE/A-cPLA2α were subjected to synchronized phagocytosis with Alexa Fluor 594-labeled opsonized zymosan for the indicted times, fixed and analyzed by confocal microscopy. Framed phagosomes have been enlarged and the intensity of the fluorescence analyzed by pseudocolor (left upper inserts at 15 and 30 min). B: Cells transfected with the construct EGFP-cPLA2α or EGFP-4KE/A-cPLA2α were subjected to a frustrated phagocytosis assay and plated over noncoated glasses (Ctrl) or IgG- coated glasses (IgG) as indicated, and analyzed by confocal microscopy. Pictures of the XZ axis of the cells were taken and the intensity of the fluorescence obtained was analyzed in pseudocolor. The figure is representative of more than 40 cells that were analyzed per experiment and the experiment was repeated four times. White bar = 10 μm.
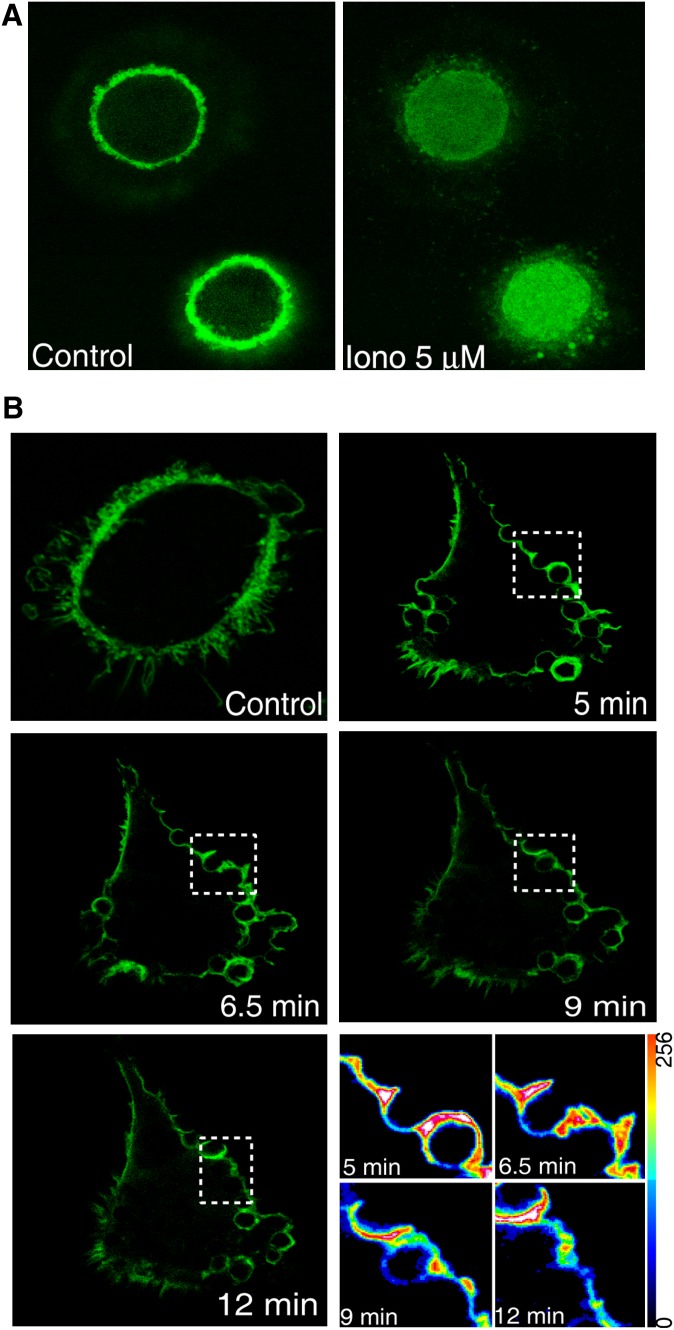
Experiments were next conducted to study a possible role for anionic phospholipids in cPLA2α translocation to the phagocytic cup. To this end, we took advantage of the PIP2-binding properties of a fluorescent chimeric protein of EGFP with the pleckstrin homology domain of the PLCδ1 (EGF-PLCδ1-PH) (28). Figure 6A shows that, when transfected into human macrophages, the chimera labels the main cellular reservoir of PIP2, i.e., the plasma membrane (24). Immediately after promoting PIP2 hydrolysis by activating the cells with a calcium ionophore, the fluorescence disappears from the plasma membrane and accumulates in the cytoplasm as would be expected from a functional PH domain (Fig. 6A). By using this chimera, we confirmed in human macrophages previous observations by Botelho et al. (31), indicating that PIP2 levels increase at the phagocytic cup while the phagosome is being formed and decrease when it seals (Fig. 6B).
Fig. 6.
PIP2 accumulation in the forming phagosomes during zymosan phagocytosis in human macrophages. A: Cells were transfected with the construct EGFP-PLCδ1-PH and analyzed in vivo by confocal microscopy. Pictures were taken before (control) and after 5 min of treatment with 5 μM ionomycin (Iono 5 μM). B: Cells were transfected with the construct EGFP-PLCδ1-PH, treated with opsonized zymosan, and analyzed in vivo by confocal microscopy. Pictures were taken at different time points after exposure to zymosan, as indicated. The pictures on the bottom right correspond to the phagosomes selected with a dotted white box at each time point, and the fluorescence intensity was analyzed in pseudocolor.
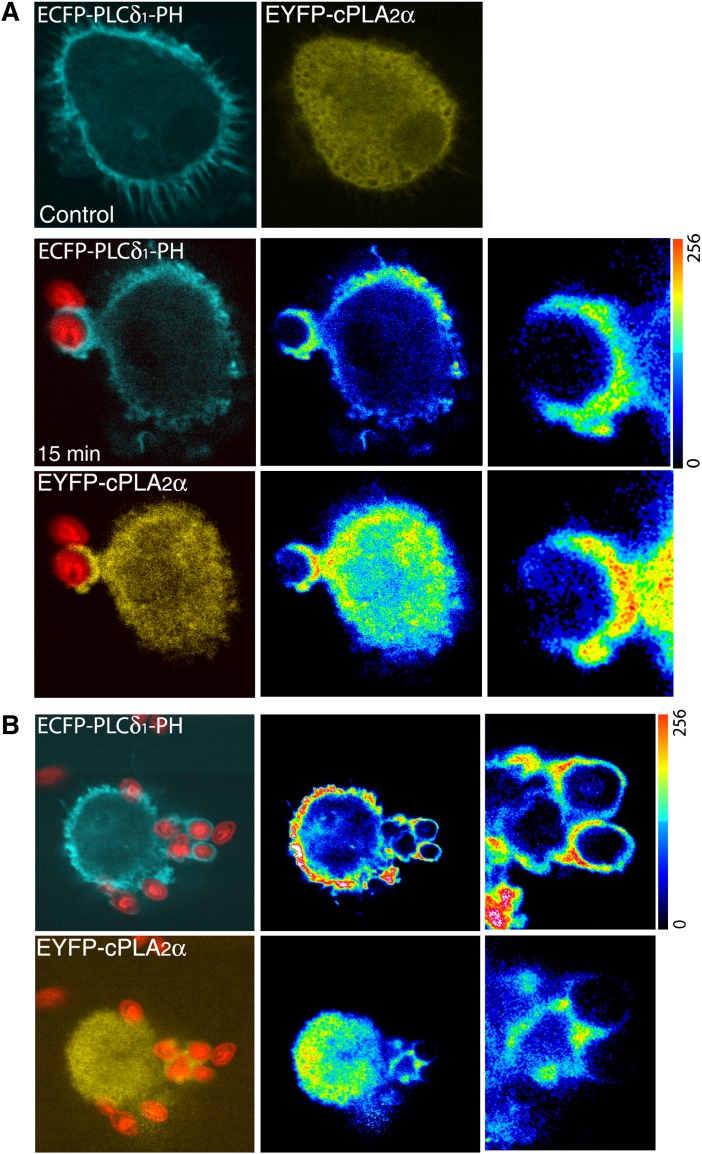
Subsequently, the cells were cotransfected with both constructs, namely ECFP-PLCδ1-PH and EYFP-cPLA2α. In resting cells, ECFP-PLCδ1-PH was present only in the plasma membrane whereas EYFP-cPLA2α was found primarily in the cytoplasm (Fig. 7). However, after exposure of the cells to opsonized zymosan, both chimeric proteins localized at the forming phagocytic cups (Fig. 7B). There was no colocalization in the cytoplasm of resting or stimulated cells. We noticed also that in those cells where ECFP-PLCδ1-PH construct was expressed at higher levels the translocation of the cPLA2α to the phagosomes was inhibited (Fig. 7 C).
Fig. 7.
Localization of ECFP-PLCδ1-PH and EYFP-cPLA2α during phagocytosis in human macrophages. Human macrophages, cotransfected with the ECFP-PLCδ1-PH and the EYFP-cPLA2α constructs, were subjected to synchronized phagocytosis using Alexa Fluor 594-labeled opsonized zymosan, fixed at 0 (Control) and 15 min, and analyzed by confocal microscopy (A). B: A cell with high expression of the construct ECFP-PLCδ1-PH is shown. In A and B, middle panels, fluorescence intensities are shown in pseudocolor, and detailed fluorescence in forming phagosomes is shown in panels to the right.
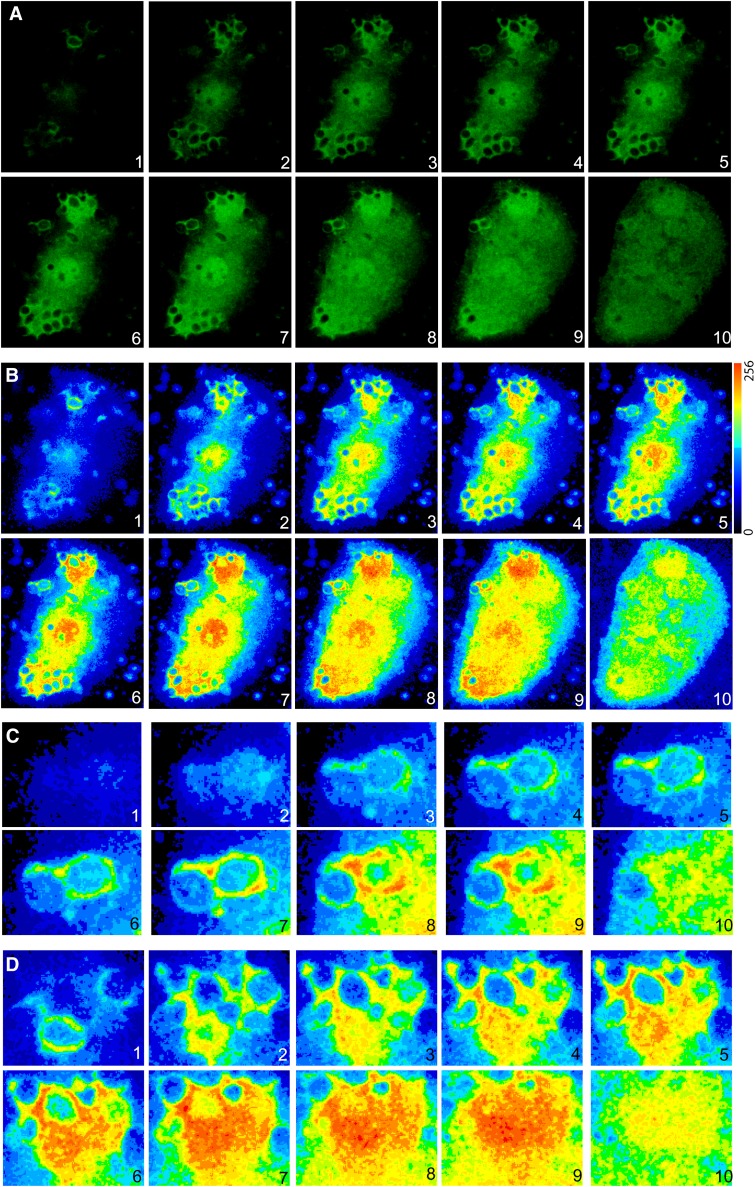
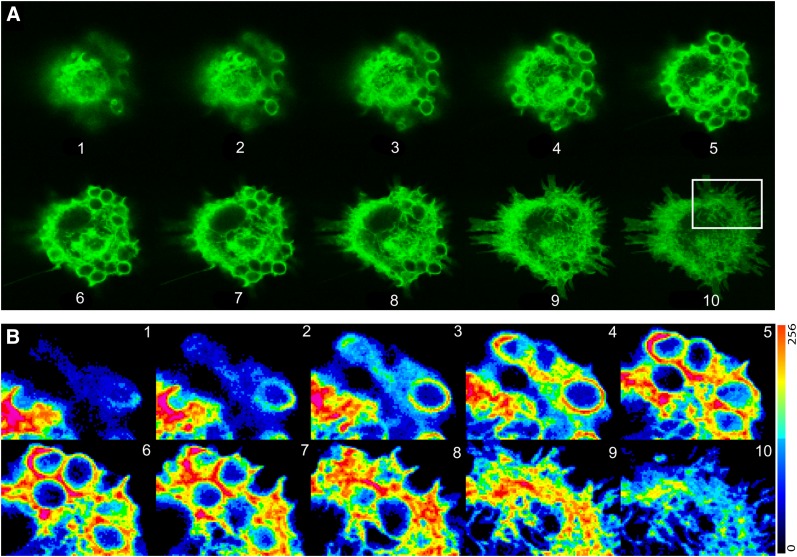
The behavior of EGFP-PLCδ1-PH and EGFP-cPLA2α were analyzed in more detail by confocal analysis of z-stack series from cells that were engulfing particles at similar stages of phagocytosis (Figs. 8, 9). We observed that both constructs were enriched in the same sites of the phagosomes, especially along the particles and in the base. It is worth noting that translocation of the EGFP-cPLA2α to the base of some of the phagosome was more prominent than that of EGFP-PLCδ1-PH. It is possible that in the base of the phagosome other factors, in addition to or independently of PIP2, contribute to the translocation of cPLA2α.
Fig. 8.
Analysis of human macrophages transfected with EGFP-cPLA2α during phagocytosis of zymosan. Human macrophages transfected with the construction EGFP-cPLA2α were treated with opsonized zymosan for 20 min, fixed, and fluorescence analyzed by confocal microscopy. A: Ten different z-stacks of the same cell are shown (from top to bottom). B: Pseudocolor analysis of the intensity of fluorescence of the cell in A is shown. C and D: Detailed phagosomes from B.
Fig. 9.
Analysis of a human macrophage transfected with EGFP-PLCδ1-PH during phagocytosis of zymosan. Human macrophages transfected with the construction EGFP- PLCδ1-PH were treated with zymosan for 20 min, fixed, and fluorescence analyzed by confocal microscopy. A: Ten different z-stacks of the same cell are shown (from top to bottom). B: Pseudocolor analysis of the intensity of fluorescence from some phagosomes is shown (white square in A).
Secreted group V and cytosolic calcium-independent group VIA PLA2s do not translocate to the phagocytic cup
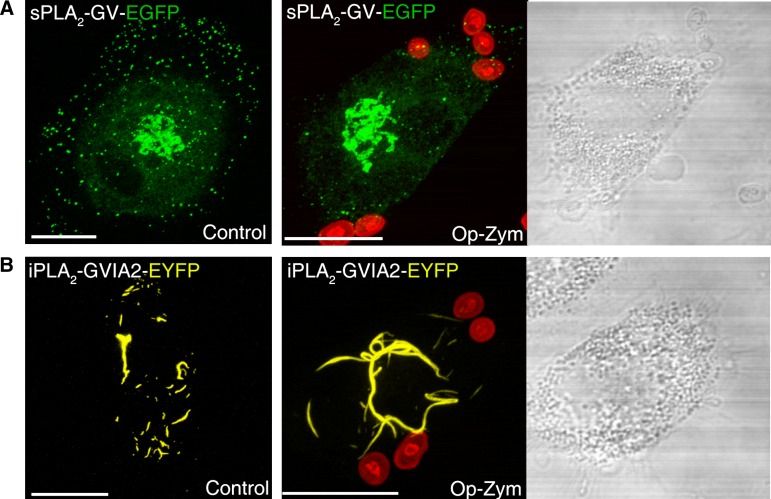
To address whether other PLA2s in addition to cPLA2α could also translocate to the phagocytic cup during phagocytosis of opsonized zymosan, we used human macrophages transfected with either sPLA2-V-EGFP or iPLA2-VIA-EYFP. In agreement with our previous studies in murine macrophages (25, 32), sPLA2-V-EGFP was found in resting cells associated with secretory granules and Golgi-like structures (Fig. 10A). iPLA2-VIA-EYFP had a mitochondrial localization in unstimulated cells (Fig. 10B), which is in accordance with previous estimates (33). After the cells where challenged with opsonized zymosan, localization was studied between the chimeric proteins and the fluorescent particles. We failed to detect association of the fluorescence arising from either sPLA2-V-EGFP or iPLA2-VIA-EYFP to the phagocytic cups under any condition (Fig. 10). Thus, of the three major PLA2 classes potentially capable of generating lipid mediators during inflammation (2–5, 34), only the cPLA2α translocates to the phagocytic cup in human macrophages.
Fig. 10.
sPLA2-V and iPLA2-VIA do not translocate to the phagocytic cup in human macrophages. Cells transfected with the fluorescent constructs sPLA2-V-EGFP (A) or iPLA2-VIA-EYFP (B) were subjected to synchronized phagocytosis using Alexa Fluor 594-labeled opsonized zymosan (op-zym) or vehicle (control) for 15 min, fixed, and analyzed by confocal microscopy. Images are the projection to the Z axis of more than 13 stacks (0.25 μm each). Transmission images are shown in the right panel. White bar = 10 μm.
DISCUSSION
A major regulatory mechanism of cPLA2α activity in cells is the Ca2+-dependent control of the physical state of the enzyme. In resting cells, the enzyme resides in the cytosol and hence, having no access to its substrate in the membrane, has no activity. In stimulated cells, cPLA2α translocates to the membrane in a Ca2+-dependent process resulting in phospholipid hydrolysis and free AA release (9). Most of the research carried out to date has thoroughly documented the translocation of cPLA2α to intracellular membranes such as those of the nuclear envelope, Golgi complex, or endoplasmic reticulum (9). A few instances have been reported of translocation of cPLA2α to cellular membranes different to those indicated above (20, 35, 36). This study adds to these studies by showing that the phagosomal membrane of human macrophages is also a site for cPLA2α translocation during activation conditions. Particularly relevant to our report is the work by Girotti et. al. (20) showing translocation of cPLA2α to forming phagosomes in murine macrophages. Although the molecular mechanism was not investigated, the authors noted that chelation of extracellular calcium decreased the total number of phagocytic events, but cPLA2α still remained associated to the lasting phagosomes at intracellular calcium levels equaling those of resting cells (20). In previous work from our laboratory, we showed that introducing exogenous short-chain PIP2 into cells promotes the translocation of cPLA2α from cytosol to perinuclear membranes at basal levels of intracellular Ca2+ (17). Moreover, other studies have demonstrated that PIP2 transiently increases in the forming phagosome and disappears after phagosome sealing, being undetectable in the sealed phagosome (31). Intrigued by these observations, we speculated that translocation of cPLA2α to the forming phagosome could require the cationic cluster of four Lys (Lys488/Lys541/Lys543/Lys544) that is present in cPLA2α and has been shown to bind anionic phospholipids such as PIP2 (13). Using confocal microscopy techniques, we document the localization of cPLA2α and a PIP2-binding construct, ECFP-PLCδ1-PH, in forming phagosomes. Also, high level expression of the ECFP-PLCδ1-PH inhibits the translocation of the cPLA2α to phagosomes. Moreover, mutation of the cationic cluster of Lys488, Lys541, Lys543, and Lys544 that serves as a PIP2-binding site eliminates the ability of the enzyme to translocate to the phagosomal membrane. Collectively, these results provide the first example of a physiologically-relevant condition where the cationic cluster of cPLA2α participates in regulating enzyme association to membranes and hence, its activity in intact cells.
The presence of cPLA2α at the membrane of the nascent phagosome may serve important pathophysiological roles because during phagocytosis large quantities of eicosanoids are produced, which could be involved in the killing of the ingested microorganism at the phagosome (8, 21–23). In the mouse model, other PLA2s in addition of cPLA2α have been suggested to be related to phagocytic events. Particularly relevant to this work, it has been shown that group V secreted PLA2 also translocates to the phagosome in zymosan-stimulated murine peritoneal macrophages (23). Intrigued by this report, we also studied the possible movement of sPLA2-V to phagosomes in our human macrophage cell system. We failed to detect translocation of a chimeric sPLA2-V-EGFP protein to phagosomes in our systems. We had previously demonstrated that this chimeric sPLA2-V behaves the same as the native sPLA2-V protein in terms of biochemical properties, enzymatic activity, and subcellular localization (25, 32). There are many differences between the murine and human macrophage models of zymosan phagocytosis that may explain these different results. For instance, in murine macrophages, zymosan induces abundant AA release (37–43) and may be internalized primarily via dectin-1 receptors (44). However, human macrophages do not respond readily to zymosan by releasing AA, and opsonization of the particle appears to be required for full responses, which may occur primarily via Fc receptors. Thus, it appears likely that the different mechanisms of internalization of phagocytosable particles in mouse versus human may account, at least in part, for the remarkable differences in the translocation ability of sPLA2-V to the phagosomes.
The one other PLA2 that we investigated is iPLA2-VIA, a calcium-independent enzyme. Unlike cPLA2α and sPLA2 enzymes, the involvement of iPLA2-VIA in receptor-mediated AA mobilization appears not to be a general one but to depend on cell type and stimulation conditions (2, 45–48). Using a chimeric construct, iPLA2-VIA-EYFP, we detected no appreciable change in the subcellular localization of this enzyme during opsonized zymosan challenge; the enzyme always remained associated to mitochondria, which is consistent with previous data (33). Thus, from the three major PLA2 families potentially capable of effecting AA release for eicosanoid production, only one, cPLA2α, translocates to the phagosome in human cells.
Acknowledgments
The authors thank Montse Duque and Yolanda Sáez for expert technical assistance, and the personnel from Centro de Hemoterapia y Hemodonación de Castilla y León for supplying Buffy coats from healthy volunteers.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- BEL
- bromoenol lactone
- bis-BODIPY FL C11-PC
- 1,2-bis-(4,4-difluoro-5,7-dimethyl-4-bora-3a, 4a-diaza-sindecane-3-undecanoyl)-sn-glycero-3-phosphocholine
- cPLA2α
- group IVA cytosolic phospholipase A2
- ECFP
- enhanced cyan fluorescent protein
- EGFP
- enhanced green fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- iPLA2
- calcium-independent phospholipase A2
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PLA2
- phospholipase A2
- PLCδ1
- phospholipase Cdelta1
- PLCδ1-PH
- pleckstrin homology domain of PLCδ1
- sPLA2
- secreted phospholipase A2
- sPLA2-V
- group V secreted phospholipase A2
This work was supported by the Spanish Ministry of Science and Innovation (Grants SAF2007-60055 and BFU2007-67154) and the Regional Government of Castile and León (Grant CSI09-A08). CIBERDEM is an initiative of Instituto de Salud Carlos III.
REFERENCES
- 1.Schaloske R. H., Dennis E. A. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 1761: 1246–1259. [DOI] [PubMed] [Google Scholar]
- 2.Balsinde J., Balboa M. A., Insel P. A., Dennis E. A. 1999. Regulation and inhibition of phospholipase A2. Annu. Rev. Pharmacol. Toxicol. 39: 175–189. [DOI] [PubMed] [Google Scholar]
- 3.Balsinde J., Winstead M. V., Dennis E. A. 2002. Phospholipase A2 regulation of arachidonic acid mobilization. FEBS Lett. 531: 2–6. [DOI] [PubMed] [Google Scholar]
- 4.Balsinde J., Balboa M. A. 2005. Cellular regulation and proposed biological functions of group VIA calcium-independent phospholipase A2 in activated cells. Cell. Signal. 17: 1052–1062. [DOI] [PubMed] [Google Scholar]
- 5.Balboa M. A., Balsinde J. 2006. Oxidative stress and arachidonic acid mobilization. Biochim. Biophys. Acta. 1761: 385–391. [DOI] [PubMed] [Google Scholar]
- 6.Funk C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294: 1871–1875. [DOI] [PubMed] [Google Scholar]
- 7.Serhan C. N., Chiang N., Van Dyke T. E. 2008. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 8: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailie M. B., Standiford T. J., Laichalk L. L., Coffery M. J., Strieter R., Peters-Golden M. 1996. Leukotriene-deficient mice manifest enhanced lethality from Klebsiella pneumonia in association with decreased alveolar macrophage phagocytic and bactericidal activities. J. Immunol. 157: 5221–5224. [PubMed] [Google Scholar]
- 9.Ghosh M., Tucker D. E., Burchett S. A., Leslie C. C. 2006. Properties of the Group IV phospholipase A2 family. Prog. Lipid Res. 45: 487–510. [DOI] [PubMed] [Google Scholar]
- 10.Bonventre J. V., Huang Z., Taheri M. R., O'Leary E., Li E., Moskowitz M. A., Sapirstein A. 1997. Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2. Nature. 390: 622–625. [DOI] [PubMed] [Google Scholar]
- 11.Uozumi N., Kume K., Nagase T., Nakatani N., Ishii S., Tashiro F., Komagata Y., Maki K., Ikuta K., Ouchi Y., et al. 1997. Role of cytosolic phospholipase A2 in allergic response and parturition. Nature. 390: 618–622. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian P., Stahelin R. V., Szulc Z., Bielawska A., Cho W., Chalfant C. E. 2005. Ceramide 1-phosphate acts as a positive allosteric activator of group IVA cytosolic phospholipase A2α and enhances the interaction of the enzyme with phosphatidylcholine. J. Biol. Chem. 280: 17601–17607. [DOI] [PubMed] [Google Scholar]
- 13.Das S., Cho W. 2002. Roles of catalytic domain residues in interfacial binding and activation of group IV cytosolic phospholipase A2. J. Biol. Chem. 277: 23838–23846. [DOI] [PubMed] [Google Scholar]
- 14.Mosior M., Six D. A., Dennis E. A. 1998. Group IV cytosolic phospholipase A2 binds with high affinity and specificity to phosphatidylinositol 4,5-bisphosphate resulting in dramatic increases in activity. J. Biol. Chem. 273: 2184–2191. [DOI] [PubMed] [Google Scholar]
- 15.Six D. A., Dennis E. A. 2003. Essential Ca2+-independent role of the group IVA cytosolic phospholipase A2 C2 domain for interfacial activity. J. Biol. Chem. 278: 23842–23850. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian P., Vora M., Gentile L. B., Stahelin R. V., Chalfant C. E. 2007. Anionic lipids activate group IVA cytosolic phospholipase A2 via distinct and separate mechanisms. J. Lipid Res. 48: 2701–2708. [DOI] [PubMed] [Google Scholar]
- 17.Casas J., Gijón M. A., Vigo A. G., Crespo M. S., Balsinde J., Balboa M. A. 2006. Phosphatidylinositol 4,5-bisphosphate anchors cytosolic group IVA phospholipase A2 to perinuclear membranes and decreases its calcium requirement for translocation in live cells. Mol. Biol. Cell. 17: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker D. E., Ghosh M., Ghomashchi F., Loper R., Suram S., John B. S., Girotti M., Bollinger J. G., Gelb M. H., Leslie C. C. 2009. Role of phosphorylation and basic residues in the catalytic domain of cytosolic phospholipase A2α in regulating interfacial kinetics and binding and cellular function. J. Biol. Chem. 284: 9596–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart L. M., Ezekowitz A. B. 2005. Phagocytosis: elegant complexity. Immunity. 22: 539–550. [DOI] [PubMed] [Google Scholar]
- 20.Girotti M., Evans J. H., Burke D., Leslie C. C. 2004. Cytosolic phospholipase A2 translocates to forming phagosomes during phagocytosis of zymosan in macrophages. J. Biol. Chem. 279: 19113–19121. [DOI] [PubMed] [Google Scholar]
- 21.Tomioka H., Sano C., Sato K., Ogasawara K., Akaki T., Sano K., Cai S. S., Shimizu T. 2005. Combined effects of ATP on the therapeutic efficacy of antimicrobial drug regimens against Mycobacterium avium complex infection in mice and roles of cytosolic phospholipase A2-dependent mechanisms in the ATP-mediated potentiation of antimycobacterial host resistance. J. Immunol. 175: 6741–6749. [DOI] [PubMed] [Google Scholar]
- 22.Rubin B. B., Downey G. P., Koh A., Degousee N., Ghomashchi F., Nallan L., Stefanski E., Harkin D. W., Sun C., Smart B. P., et al. 2005. Cytosolic phospholipase A2α is necessary for platelet-activating factor biosynthesis, efficient neutrophil mediated bacterial killing, and the innate immune response to pulmonary infection: cPLA2α does not regulate neutrophil NADPH oxidase activity. J. Biol. Chem. 280: 7519–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balestrieri B., Hsu V. W., Gilbert H., Leslie C. C., Han W. K., Bonventre J. V., Arm J. P. 2006. Group V secretory phospholipase A2 translocates to the phagosome after zymosan stimulation of mouse peritoneal macrophages and regulates phagocytosis. J. Biol. Chem. 281: 6691–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stauffer T. P., Ahn S., Meyer T. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8: 343–346. [DOI] [PubMed] [Google Scholar]
- 25.Shirai Y., Balsinde J., Dennis E. A. 2005. Localization and functional interrelationships among cytosolic group IV, secreted group V, and Ca2+-independent group VI phospholipase A2s in P388D1 macrophages using GFP/RFP constructs. Biochim. Biophys. Acta. 1735: 119–129. [DOI] [PubMed] [Google Scholar]
- 26.Casas J., Gijón M. A., Vigo A. G., Crespo M. S., Balsinde J., Balboa M. A. 2006. Overexpression of cytosolic group IVA phospholipase A2 protects cells from calcium-dependent death. J. Biol. Chem. 281: 6106–6116. [DOI] [PubMed] [Google Scholar]
- 27.Larsen E. C., DiGennaro J. A., Saito N., Mehta S., Loegering D. J., Mazurkiewicz J. E., Lennartz M. R. 2000. Differential requirement for classic and novel PKC isoforms in respiratory burst and phagocytosis in RAW 264.7 cells. J. Immunol. 165: 2809–2817. [DOI] [PubMed] [Google Scholar]
- 28.Marshall J. G., Booth J. W., Stambolic V., Mak T., Balla T., Schreiber A. D., Meyer T., Grinstein S. 2001. Restricted accumulation of phosphatidylinositol 3-kinase products in a plasmalemmal subdomain during Fc gamma receptor-mediated phagocytosis. J. Cell Biol. 153: 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono T., Yamada K., Chikazawa Y., Ueno M., Nakamoto S., Okuno T., Seno K. 2002. Characterization of a novel inhibitor of cytosolic phospholipase A2α, pyrrophenone. Biochem. J. 363: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manna D., Cho W. 2007. Real-time cell assays of phospholipase A2s using fluorogenic phospholipids. Methods Enzymol. 434: 15–27. [DOI] [PubMed] [Google Scholar]
- 31.Botelho R. J., Teruel M., Dierckman R., Anderson R., Wells A., York J. D., Meyer Y., Grinstein S. 2000. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 151: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balboa M. A., Shirai Y., Gaietta G., Ellisman M. E., Balsinde J., Dennis E. A. 2003. Localization of group V phospholipase A2 in caveolin-enriched granules in activated P388D1 macrophage-like cells. J. Biol. Chem. 278: 48059–48065. [DOI] [PubMed] [Google Scholar]
- 33.Seleznev K., Zhao C., Zhang X. H., Song K., Ma Z. A. 2006. Calcium-independent phospholipase A2 localizes in and protects mitochondria during apoptotic induction by staurosporine. J. Biol. Chem. 281: 22275–22288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzpatrick F. A., Soberman R. 2001. Regulated formation of eicosanoids. J. Clin. Invest. 107: 1347–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shmelzer Z., Haddad N., Admon E., Pessach I., Leto T. L., Eitan-Hazan Z., Hershfinkel M., Levy R. 2003. Unique targeting of cytosolic phospholipase A2 to plasma membranes mediated by the NADPH oxidase in phagocytes. J. Cell Biol. 162: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wooten R. E., Willingham M. C., Daniel L. W., Leslie C. C., Rogers L. C., Sergeant S., O'Flaherty J. T. 2008. Novel translocation responses of cytosolic phospholipase A2alpha fluorescent proteins. Biochim. Biophys. Acta. 1783: 1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balsinde J., Fernandez B., Diez E. 1990. Regulation of arachidonic acid release in mouse peritoneal macrophages. The role of extracellular calcium and protein kinase C. J. Immunol. 144: 4298–4304. [PubMed] [Google Scholar]
- 38.Balsinde J., Fernández B., Solís-Herruzo J. A., Diez E. 1992. Pathways for arachidonic acid mobilization in zymosan-stimulated mouse peritoneal macrophages. Biochim. Biophys. Acta. 1136: 75–82. [DOI] [PubMed] [Google Scholar]
- 39.Qiu Z. H., de Carvalho M. S., Leslie C. C. 1993. Regulation of phospholipase A2 activation by phosphorylation in mouse peritoneal macrophages. J. Biol. Chem. 268: 24506–24513. [PubMed] [Google Scholar]
- 40.Balsinde J., Balboa M. A., Insel P. A., Dennis E. A. 1997. Differential regulation of phospholipase D and phospholipase A2 by protein kinase C in P388D1 macrophages. Biochem. J. 321: 805–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balsinde J., Balboa M. A., Yedgar S., Dennis E. A. 2000. Group V phospholipase A2-mediated oleic acid mobilization in lipopolysaccharide-stimulated P388D1 macrophages. J. Biol. Chem. 275: 4783–4786. [DOI] [PubMed] [Google Scholar]
- 42.Gijón M. A., Spencer D. M., Siddiqui A. R., Bonventre J. V., Leslie C. C. 2000. Cytosolic phospholipase A2 is required for macrophage arachidonic acid release by agonists that do and do not mobilize calcium. Novel role of mitogen-activated protein kinase pathways in cytosolic phospholipase A2 regulation. J. Biol. Chem. 275: 20146–20156. [DOI] [PubMed] [Google Scholar]
- 43.Balsinde J., Balboa M. A., Dennis E. A. 2000. Identification of a third pathway for arachidonic acid mobilization and prostaglandin production in activated P388D1 macrophage-like cells. J. Biol. Chem. 275: 22544–22549. [DOI] [PubMed] [Google Scholar]
- 44.Suram S., Brown G. D., Ghosh M., Gordon S., Loper R., Taylor P. R., Akira S., Uematsu S., Williams D. L., Leslie C. C. 2006. Regulation of cytosolic phospholipase A2 activation and cyclooxygenase 2 expression in macrophages by the beta-glucan receptor. J. Biol. Chem. 281: 5506–5514. [DOI] [PubMed] [Google Scholar]
- 45.Balsinde J. 2002. Roles of various phospholipases A2 in providing lysophospholipid acceptors for fatty acid phospholipid incorporation and remodelling. Biochem. J. 364: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balboa M. A., Sáez Y., Balsinde J. 2003. Calcium-independent phospholipase A2 is required for lysozyme secretion in U937 promonocytes. J. Immunol. 170: 5276–5280. [DOI] [PubMed] [Google Scholar]
- 47.Balboa M. A., Balsinde J. 2002. Involvement of calcium-independent phospholipase A2 in hydrogen peroxide-induced accumulation of free fatty acids in human U937 cells. J. Biol. Chem. 277: 40384–40389. [DOI] [PubMed] [Google Scholar]
- 48.Pérez R., Melero R., Balboa M. A., Balsinde J. 2004. Role of group VIA calcium-independent phospholipase A2 in arachidonic acid release, phospholipid fatty acid incorporation, and apoptosis in U937 cells responding to hydrogen peroxide. J. Biol. Chem. 279: 40385–40391. [DOI] [PubMed] [Google Scholar]