Abstract
LC/ESI-MS/MS has been previously demonstrated to be a powerful method to detect and quantify molecular species of glycerophospholipids including lysophospholipids. In this study, we provide an improved pre-mass spectrometry lipid extraction procedure that avoids the acid-catalyzed decomposition of plasmenyl phospholipids that is problematic with previously reported methods. We show that the use of lysophospholipid internal standards with perdeuterated fatty acyl chains avoids isobar problems associated with the use of internal standards containing odd carbon number fatty acyl chains. We also show that LC prior to MS is required to avoid numerous problems associated with isobars and with MS in-source decomposition of lysophosphatidylserine. The reported method of using normal phase chromatography/ESI-MS is used to quantify lysophospholipids in serum and to quantify lysophospholipids produced in mammalian cells by human group X secreted phospholipase A2. The latter shows that group X phospholipase A2 added exogenously to cells generates a different set of lysophospholipids compared with enzyme produced endogenously in cells, which supports earlier studies showing that this phospholipase A2 can act on cell membranes prior to externalization from cells.
Keywords: lysophosphatidic acid, plasmenyl, lipidomics
Detection and quantification of lysophospholipid species are important in lipid biology because such species mark the action of lipolytic enzymes such as phospholipases A2 (1) and are themselves ligands for numerous signaling receptors (2). Combined high pressure LC/ESI-MS/MS is a powerful method to detect and quantify a large number of glycerophospholipid molecular species in a single analysis (3). In this study we focus on the use of LC/ESI-MS/MS to detect and quantify lysophospholipid molecular species.
Prior to LC/ESI-MSMS, it is helpful to partially purify the lysophospholipids by some form of liquid-liquid or liquid-solid extraction. Although extraction of biological aqueous samples with chloroform/methanol is useful for many phospholipid and lysophospholipid species, extraction of lysophosphatidic acid and lysophosphatidylinositol are low yielding unless the aqueous phase is acidified (4). It is also possible to extract LPA using 1-butanol (5, 6), but high yield extraction also requires acidification, and 1-butanol is more cumbersome to remove than chloroform/methanol. The problem with acidification during extraction is that plasmalogens are rapidly degraded at low pH due to spontaneous cleavage of their sn-1-enol ether chain to give the corresponding sn-1 hydroxyl group (4). This leads to formation of sn-2-acyl-lysophosphatidylcholine and sn-2-acyl-lysophosphidylethanolamine and possible other species. These sn-2 lysophospholipids behave similarly to the corresponding sn-1-acyl lysophospholipids when examined by ESI-MS/MS because they are isobaric and often give rise to the same major fragment ions. This acid-catalyzed breakdown leads to an underestimation of the amount of enol ether-lysophospholipid species and an overestimation of the amount of lysophospholipid species. In this paper, we describe a simple multi-step extraction procedure that allows for the rapid analysis of lysophospholipid and enol ether lysophospholipid molecular species using LC/ESI-MS/MS and that avoids the decomposition problems described above. We also carried out an analysis of isobaric lysophospholipid species and show that the use of lysophospholipids with perdeuterated fatty acyl chains as internal standards is prefered over the use of those with fatty acyl chains with odd carbon numbers and that LC prior to ESI-MS/MS is required to avoid problems with isobars. We show that lysophosphatidylserine species break down in the mass spectrometer source to generate lysophosphatidic acid species, which underscores the need for LC separation of these species prior to mass spectrometry. We apply the new method to quantify lysophospholipids in serum and in cells expressing or treated with group X secreted phospholipase A2.
METHODS
Materials
CHCl3 and CH3OH were from Fisher Scientific (HPLC grade), and n-hexane and isopropanol from Fisher (Optima grade). Water was purified with a Milli-Q system (Millipore). Phosphate buffer saline without Ca2+ and Mg2+ was from Gibco (Cat. 14190-144). The following lipids were purchased from Avanti Polar Lipids: 14:0-lysophosphatidic acid (LPA), 17:0-LPA, 18:1-LPA, 12:0-lysophosphatidylcholine (LPC), 17:0-LPC, egg-LPC, 24:0-LPC, 19:2-enyl-LPC, 14:0-lysophosphatidylethanolamine (LPE), 16:0-LPE, 17:1-LPE, 18:0-LPE, 19:2-enyl-LPE, 14:0-lysophosphatidylglycerol (LPG), 16:0-LPG, 17:1-LPG, 18:0-LPG, liver lysophosphatidylinositol (LPI), 17:1-LPI, 17:1-lysophosphatidylserine (LPS), brain-LPS, 1-O-1′-(Z)-octadecenyl-2-arachidonyl-sn-glycero-3- phosphocholine, d31-16:0-LPC and 1-O-1′-(Z)-octadecenyl-2-arachidonyl-sn-glycero-3-phosphoethanolamine. 16:0-alkyl-LPC was from BioMol, and d4-16:0-alkyl-LPC was from Cayman Chemicals.
Deuterated lysophospholipid internal standards were prepared as follows. Into a 1.5 ml polypropylene microfuge tube was added 10 μg of each of the following deuterated phospholipids (all from Avanti Polar Lipids) in CHCl3: 1-d31-palmitoyl-2-palmitoyl-glycero-sn-3-phosphoinositol, 1-d31-palmitoyl-2-palmitoyl-glycero- sn-3-phospho(rac)glycerol; 1-d31-palmitoyl-2-palmitoyl-glycero- sn-3-phosphoethanolamine,
1-d31-palmitoyl-2-palmitoyl-glycero-sn-3-phosphoserine, and 1-d31-palmitoyl-2-palmitoyl-glycero-sn-3-phosphate. CHCl3 was removed with a stream of N2. To the residue was added 50 μl of 10 mM MOPS, pH 7.2, 0.5 mM EGTA, 100 mM KCl, 4 mM CaCl2, and the capped tube was placed in a high-powered bath sonicator for ∼30 s (a microtip probe sonicator can also be used). A stock solution of bee venom phospholipase A2 (Sigma Cat. P9279, 1020 Units per mg of solid) was prepared at 17.9 Units/μl in the above buffer, and a 2 μl aliquot was added to the phospholipid mixture. The sample was left at room temperature for 3 h and then placed in a centrifugal evaporator (Speed-Vac) to remove all liquid. The residue was taken up in ethanol and stored at −80°C in a capped vial. d31-16:0-LPC and d4-alkyl-16:0-LPC were added to the internal standard stock solution to give a mixture of all six internal standards. It was determined by MS that bee venom phospholipase A2 hydrolyzed phosphatidic acid poorly. Thus, we generated d31-16:0-LPA by treating commercial d31-16:0-LPC with cabbage phospholipase D as described (7) and added it to the internal standard stock solution. To determine the concentration of each deuterated lysophospholipid in the stock solution, an aliquot was spiked with known amounts of 16:0-LPA, 16:0-LPE, 16:0-LPG, 16:0-LPI, 16:0-LPS, 16:0-LPC, and alkyl-16:0-LPC (stock solutions calibrated by inorganic phosphorous assay as described above), and the mixture was analyzed by LC/MS/MS as described below. By comparing the peak area for each internal standard to that from the corresponding non-deutrated lysophospholipid peak, the amount of each internal standard was obtained. We assumed that the deuterated and the non-deuterated lysophospholipid species give the same mass spectrometry peak area per mole, which is not true because of likely slight differences in the intrinsic ionization efficiencies of these species and because the deuterated species contains a significant amount of d30 and d29 species (discussed further below), and we selected only the d31 species in the first quadrupole mass analyzer. However, the factor by which the deuterated and non-deuterated samples differ is not needed because it cancels out of all calculations to derive the absolute moles of lysophospholipid analytes in test samples.
Sample preparation
Stock solutions of phospholipids were made in CHCl3/CH3OH (2/1, v/v) and stored at −80°C under argon in Teflon septum-capped glass vials. Concentrations were determined by total phosphorous assay (8). Aqueous samples for lysophospholipid analysis were extracted in 16 ×125 mm disposable glass culture tubes and extraction solvent was transferred with glass Pasteur pipets. Aqueous samples with volumes >1 ml were lyophilized overnight and reconstituted in 1 ml PBS prior to extraction. Cell pellets were thawed on ice in 1 ml PBS. Prior to extraction, each sample was spiked with 10 μl of the internal standard mix. Each 1 ml of PBS was first extracted under neutral conditions with 4 ml of CHCl3/CH3OH (2/1, v/v) followed by 2 ml of PBS-saturated CHCl3/CH3OH (2/1, v/v). Each extraction step was followed by a 60 s vortex and a 1 min centrifugation step in a bench-top clinical centrifuge in a cold room at 4°C at ∼3,000 rpm to resolve the organic and aqueous phases. The lower organic phases from both neutral extraction steps were pooled and dried under a gentle stream of N2. The remaining aqueous phase was chilled on ice for 10 min and then acidified with ice-cold 0.3 M citric acid solution to pH ∼3–4 (small aliquot spotted on pH paper, checked on the first two samples to determine the volume of citric acid needed per sample thereafter). The sample was then extracted twice with ice-cold PBS-saturated CHCl3/CH3OH (2/1, v/v). The sample was vortexed as above, and the organic phases were again resolved by centrifugation. The lower organic phases were pooled and neutralized to pH ∼6–7 (checked with pH paper as above) with 10% triethylamine in CHCl3/CH3OH (2/1, v/v) on ice. The neutralized organic phase from the acidic extraction was combined with the intial neutral fraction and dried under a stream of N2. The resulting residue was taken up in 0.5 ml of CHCl3/CH3OH (2/1, v/v), and after brief vortex mixing, the solution was transferred to the autosampler vial (Microsolv Technology Corporationo, MRQ Max Recovery Vials, 1.2 ml, 12 × 32 mm, Cat. 9512C-0CV-T). The tube was rinsed with 0.25 ml of the same solvent and this was combined with the solution in the autosampler vial. Solvent was removed with a stream of N2 and the residue was taken up in 0.1 ml of initial LC solvent (see below). The vial was capped with a Teflon-Silicone cap (Agilent 5185-5838). The samples were analyzed by LC/ESI-MSMS or stored at −20°C under argon.
Analysis of lysophospholipids in serum was carried out with mouse serum from Atlantic Biologicals. A total of 100 μl of serum was extracted as above and analyzed by LC/ESI-MS/MS. The analysis was repeated with three additional 100 μl portions of serum. The growth of HEK293 cells and establishment of cells stably transfected with human group X secreted phospholipase A2 were carried out as described (9). Four million cells were seeded in a 10 cm dish, which was placed in the CO2 incubator for 48 h at 37°C. Cells were washed with PBS and then washed twice with RPMI-1640 medium without serum. Washed cells were covered with serum-free RPMI-1640 medium with or without recombinant human group X secreted phospholipase A2 (200 ng/ml) and placed for 4 h at 37°C in the CO2 incubator. Cells were washed once with PBS and scraped into 1 ml of PBS on ice. The cell suspension was stored at −80°C prior to lipid extraction. Cells stably transfected with human group X secreted phospholipase A2 were processed in the same way except that exogenous phospholipase A2 was not added.
Sample analysis
LC/ESI-MS/MS was carried out on a Waters Quattro triple quadrupole mass spectrometer, a 2795 Alliance HT LC/autosampler system, and the QuanLynx software package. Chromatography was carried out with a normal phase column (Phenomenex Luna, 5 μ, silica, 100 Ang., 250 × 2.0 mm, Cat 00G-4274-BO) equipped with a guard column (Phenomenex Security Guard Column Cat KJ0-4282) with a column temperature of 30°C. Solvent A is 30/40 (v/v) n-hexane/isopropanol, and solvent B is 30/60/15 (v/v/v) n-hexane/isopropanol/5 mM ammonium acetate in water. The solvent program is: 0–18 min, 50–86% linear B gradient (0.2 ml/min); 18–22 min, 86% B (0.2 ml/min); 22–25 min, 86–100% linear B gradient (0.2 ml/min); 25–42 min, 100% B (0.2 ml/min). At 42 min, solvent B was stepped to 50% and the flow rate was stepped to 0.3 ml/min. At 52 min, the flow rate was stepped to 0.2 ml/min. The vials were held at 15°C in the autosampler chamber and 20 μl injection volumes were used. Other injector parameters are given in supplementary Table I.
The ESI-MS/MS parameters are given in supplementary Table I. Parent and fragment ion m/z values, cone voltages, and collision energies for each analyte are given in supplementary Table II.
RESULTS and DISCUSSION
Extraction of lysophospholipids
The method that we developed involves extraction of the pH neutral aqueous biological sample with CHCl3/CH3OH (2/1), which is expected to well extract all lysophospholipids except LPA and LPI. The use of neutral pH conditions avoids spontaneous loss of species that contain the highly acid sensitive sn-1 enol ether. To extract LPA and LPI species, the remaining aqueous phase was acidified on ice and the mixture was extracted again with CHCl3/CH3OH (2/1). Ice was used to minimize acid decomposition of any enol ether species that were not extracted in the first round. The organic layer was neutralized with triethylamine before it was combined with the first organic phase, again, to avoid loss of enol ether species. The use of triethylamine avoids any possible transacylation reactions that might occur had a primary amine been used. The pH varies from 7.4 (phosphate buffered saline) to 3–4 (after citrate addition) to 6–7 (after triethylamine addition). A previous study of spontaneous migration of fatty acyl chains between the sn-1- and sn-2 position of LPC showed that the rate is pH dependent with a half-life at pH 7.4 of ∼6 h and at pH 3 of ∼90 h at room temperature (10). Thus, during the above extraction procedure, acyl chain migration should be minimal.
By applying the above extraction procedure to a standard mix of lysophospholipids and comparing the ESI-MS/MS peak areas of the species detected in the extracted sample versus those from a standard stock solution of lysophospholipids that were directly injected on to the LC column without extraction, we determined the percent recovery values for several lysophospholipid species (supplementary Table II). The yields were acceptable, the lowest being 55% for 16:0-LPI. In previous work, 1-butanol has been used together with acidification of the aqueous sample medium to ensure high yield recovery of LPA species (5, 6). If we extracted the sample with 1-butanol without acidification of the aqueous phase, yields of LPA species were very low (∼10%) as were yields of LPI (∼20%). If CHCl3/CH3OH (2/1, v/v) was used in the absence of acidification, yields of LPA species were ∼5%. In another study, CHCl3/CH3OH was used as the extraction solvent in the absence of acidification (11) but we could not reproduce the yields of LPA that were reported in that study. We prefer the use of CHCl3/CH3OH versus 1-butanol as the former is easier to remove by evaporation. We also the stress the need to acidify the aqueous phase to ensure high yield extraction of LPA and LPI species.
To examine whether the new extraction procedure leaves plasmenyl phospholipids intact, we subjected a mixture of plasmenyl-phosphatidylcholine and plasmenyl-phosphatidylethanolamine to the extraction procedure and analyzed the sample by LC/ESI-MS/MS. When 100 pmole of 1-O-1′-(Z)-octadecenyl-2-arachidonyl-sn-glycero-3-phosphocholine and 100 pmole of 1-O-1′-(Z)-octadecenyl-2-arachidonyl-sn-glycero-3-phosphoethanolamine were added to PBS and submitted to the extraction procedure given in Methods, the amount of 20:4-LPC and 20:4-LPE detected by LC/ESI-MS/MS corresponded to 1.6% and 0.6%, respectively, of the plasmenyl phospholipids present. When 100 pmoles of plasmenyl phospholipids were analyzed by LC/ESI-MS/MS without sample processing, the amount of 20:4-LPC and 20:4-LPE were 0.02% and 0.1%, respectively, of the plasmenyl species. Thus, the extraction procedure leads to a small amount of plasmenyl phospholipid breakdown. However, the amount of breakdown is very small compared with that which occurs if the aqueous sample of plasmenyl phospholipids is acidified with citric acid and then extracted with CHCl3/CH3OH. This latter procedure results in breakdown of 38% of the plasmenyl phosphatidylcholine and 10% of the plasmenyl phosphatidylethanolamine. We also tested the extraction procedure given by the Lipid Maps consortium (12), which involves quenching the lipid residue with ice-cold 1:1 CH3OH/0.1 N aqueous HCl (0.8 ml) plus CHCl3 (0.4 ml). This resulted in 4% and 2% breakdown of the plasmenyl phosphatidylcholine and plasmenyl phosphatidylethanolamine, respectively. Thus, acidification leaves the plasmenyl species mainly intact when the extraction is done with cold solvents, but use of the procedure described in Methods is superior if lysophospholipids are to be analyzed. The use of acidification with 1-butanol extraction (5, 6) is problematic for analysis of lysophospholipid species. If LPA and LPI species are the only ones analyzed, acid catalyzed breakdown would be a problem if the sample contains enol ether phosphatidic acid and phosphatidylinositol species. For the analysis of LPC and LPE species, the acid-catalyzed breakdown of the enol ethers could potentially pose a significant problem since many cell membranes contain a relatively large amount of plasmenyl phosphatidylcholine and plasmenyl phosphatidylethanolamine species.
General features of the LC/ESI-MS/MS analysis
All lysophospholipids except LPC, enyl-LPC, and alkyl-LPC species were analyzed in negative ion mode. In the case of LPE, LPG, and LPI species, the major anionic fragment detected after collision induced dissociation is the fatty acid carboxylate chain (3), and this was used for detection. For LPA species, the major fragment is due to loss of the sn-1 fatty acyl chain followed by attack of the sn-1 hydroxyl to form the the cyclic-phosphate mono anion (m/z = −153) (3), and this was used for analyte detection. For LPS species, serine is lost from the phosphate to give the LPA intermediate, which then converts to the same cyclic phosphate (m/z = −153). For enyl-LPE species, we detected the fragment at m/z = −196, presumably due to cleavage of the enol ether and formation of the phosphate diester mono anion [analogous to Fig. 7 of (13)]. For enyl-LPC species, we detected the m/z = +181 fragment ion due to formation of a phosphate diester mono cation [see Fig. 7 of (13)].
We developed a simple procedure to prepare all of the deuterated internal standard lysophospholipids using commercially available reagents. We used a separate internal standard for each lysophospholipid head group class. For example, we used d31-16:0-LPC as the internal standard to quantify all fatty acyl LPC species, and we assumed that all fatty acyl LPC species ionize with the same efficiency in the MS source (however, see below). It is simply not practical to have an internal standard for each of the 116 lysophospholipid species analyzed by LC/ESI-MS/MS in this study. We obtained standard curves for all of the commercially available lysophospholipids used in this study (see supplementary Fig. I). A linear response was obtained for all species in the 50–1000 fmole range. This suggests that aggregation of lysophospholipids during LC does not occur. In the case of commercial LPC species, we obtained data for 12:0-LPC, 16:0-LPC, 18:1-LPC, and 24:0-LPC. The relative peak areas of the ion trace peaks are as follows: 12:0-LPC (1.0), 16:0-LPC (1.0), 18:1-LPC (1.0), and 24:0-LPC (3.0). For LPG, LPI, LPE, and LPS, the relative areas vary by less than 1.3-fold in going from 14:0 to 18:1. For LPA species, the relative areas are: 14:0-LPA (1.0), 16:0-LPA (0.37), and 18:1-LPA (0.27). Because we did not correct the ESI-MS/MS responses for the variation of ionization efficiency with fatty acyl chain length, the absolute values of the amounts of lysophospholipids reported in this study may be off by as much as ∼3-fold. However, in our study of group X secreted phospholipase A2-induced lysophospholipid generation described below, it is the relative change in analyte levels that we are most interested in, i.e., the fold-increase in lysophospholipid levels when the phospholipase A2 is added to cells or comparing nontransfected to phospholipase A2-transfected cells. Relative quantification of lysophospholipids is not influenced by fatty acyl chain dependence on ESI-MS/MS ionization efficiencies.
We assumed that each fatty acyl LPC species fragmented to the same extent, which should be valid because the fragment ion chosen is the major ion after collision-induced dissociation. For quantification of enol ether LPC species, we injected a standard amount of 19:2-enyl-LPC and 16:0-LPC to obtain the relative integrals of the fragment ion traces after LC/ESI-MS/MS analysis. This factor was then used to quantify all enol ether LPC species based on the signal for d31-16:0-LPC. We used d4-16:0-alkyl-LPC as the internal standard to quantify alkyl ether LPC species. Likewise, for LPE, we used d31-16:0-LPE to quantify all fatty acyl LPE species, and we analyzed a standard amount of 19:2-enyl-LPE and 16:0-LPE to obtain the relative detection signals. We made no attempt to quantify alkyl ether LPE species or enol ether and alkyl ether species of LPA, LPG, LPI, and LPS. Accurate quantification of these species would require ESI-MS/MS analysis of stock solutions of appropriate lysophospholipid standards of known concentration.
Supplementary Table II also gives the optimized ESI-MS/MS conditions (cone voltage and collision energy) for several lysophospholipid species. Cone voltages and collision energies were optimized for one to two lysophospholipid species representative of each head group class and we used the same conditions for each member of each class. According to the User Manual for the Waters Quattro mass spectrometer, the optimal cone voltage is generally higher as the molecular weight of the analyte increases. In the case of LPC species, the optimal cone voltage for 12:0-LPC was found to be +30 V and it was +35 V for 24:0-LPC. The analyte signal versus cone voltage peaks are fairly broad; the use of +30 V for all LPC species reduces detection sensitivity only by ∼5% in the worst case. The same trends are seen for other lysophospholipid head group species. This justifies our use of a single cone voltage for each lysophospholipid head group class.
We found that LPS species undergo in-source conversion to the corresponding LPA species (data not shown). The amount of this decomposition depends on the ESI source conditions. As noted above, the pathway for collision-induced fragmentation of LPS involves, first, conversion to LPA as an intermediate. This in-source LPS to LPA conversion underscores the need to resolve LPA and LPS species by LC prior to ESI-MS/MS.
To look for isobaric species, we carefully considered the masses of a general set of lysophospholipids that contain either a fatty acyl sn-1 chain or an ether linked sn-1 chain (both enol ether and alkyl ether). Polar head groups included were phosphate, phosphocholine, phosphoethanolamine, phosphoglycerol, phosphoinositol, and phosphoserine. We considered acyl and ether sn-1 chains 12:0, 14:0, 16:0, 16:1, 18:0, 18:1, 18:2, 18:3, 20:0, 20:1, 20:2, 20:3, 20:4, 22:4, 22:5, and 22:6 (and 24:0 for LPC only). We also considered fatty chains with an odd number of carbons (17:0, 17:1, and 19:2) because commercially available lysosphospholipids with these chains are often used as internal standards in earlier studies. The full analysis is given in the supplementary material. Supplementary Table III lists all isobars along with a statement about whether an unambiguous identification of the lysophospholipid species can be made. Several general points emerge. Within each polar head group lysophospholipid class, there are no isobars other than double bond positional isomers as long as the deuterated internal standards described in this study are used. On the other hand, use of the commercially available internal standards with an sn-1 chain containing an odd number of carbons (6) is not advised. For example, 17:0-LPC is isobaric with 18:0-alkyl-LPC, and 17:1-LPC is isobaric with 18:1-alkyl-LPC. It is not known a priori whether all of these isobaric species resolve by LC. Likewise, 19:2-enyl-LPC and 19:2-enyl-LPE were not used as internal standards.
The isobaric problem with the use of odd carbon fatty acyl lysophospholipids is eliminated by the use of the d31-lysophospholipid internal standards. A potential problem with the latter is that they are not completely deuterated and thus, they give rise to an envelope of isotopic species. ESI-MS analysis of the d31-internal standards used in this study showed that the the d30/d31 ratio is ∼1, the d29/d31 ratio is ∼0.3, and the d28/d31 ratio is ∼0.05. For this reason, we also included in our isobar analysis the d30 and d29 species for all of the internal standards. No additional isobar problems arise from these species (supplementary Table III). It should be noted that if the amount of internal standard used is in vast excess over the analyte being quantified that isobar problems from partially deuterated species of d28 or less may arise if the two lysophospholipids in question are not resolved by LC. Thus, caution is advised, and it is recommended to use amounts of internal standards that are not orders of magnitude greater than the amounts of analytes being quantified. All things considered, the use of perdeuterated internal standards is preferred over those with odd carbon fatty acyl chains.
Several other points emerge. Many of the isobaric issues are resolved by the use of LC preceding MS. Thus, lysophospholipid analysis by direct infusion of the sample into the mass spectrometer is not recommended (also because of the LPS to LPA conversion as noted above). The use of MS/MS rather than MS helps to resolve isobaric species but the number of such resolutions is small compared with those resolved by LC. The main advantage of MS/MS over MS is to improve signal-to-noise and to distinguish lysophospholipids from nonlysophospholipid isobaric species that may be present in the sample.
Some trends in the LC retention time of lysophospholipids were noted. For fatty acyl lysophospholipids, long chain species elute earlier than short chain species, and the the presence of double bonds tends to delay the chromatographic elution. For LPE and LPC species, the fatty enol ether lysophospholipid elutes prior to the fatty acyl lysophospholipid with the same carbon chain number and number of double bonds (i.e., 18:2-enyl-LPC elutes before 18:2-LPC). Comparable data for other head group lysophospholipids was not obtained. On the other hand, the fatty alkyl ether LPC species tend to comigrate with the fatty acyl LPC species with the same carbon chain number. Although we did not obtain comparable data for fatty alkyl ether versus fatty acyl lyosphospholipids with noncholine head groups, we did observed that 16:0-alkyl-LPA comigrated with 16:0-LPA. We used these LC trends to make predictions on which isobaric species can be resolved.
Of the 360 lysophospholipid masses considered, there are 182 isobars (see supplementary data), 10 of which are unlikely to be resolved by LC or by MS/MS. Thus, the vast majority of isobaric issues are resolved by the methods described in this paper.
As already noted, the above analysis treats double bond regioisomers as a single species. The most convincing way to distinguish an isobaric lysophospholipid pair in which one species has a double bond as an enol ether from the other species that does not have an enol ether is to treat the sample under acidic conditions and to look for the loss of the LC/ESI-MS/MS signal compared with the sample that was processed in the absence of acid. The other method, which is shown in this study to be reliable for LPE and LPC species, is to rely on the earlier LC retention time for the enol ether species (as noted above). In the case of other double bond regioisomers, for example, a lysophospholipid containing a γ-linolenoyl chain (6,9,12-18:3) versus an α linolenoyl chain (9,12,15-18:3), the methods presented here will likely not allow a distinction.
LC/ESI-MS/MS analysis of lysophospholipid mixtures
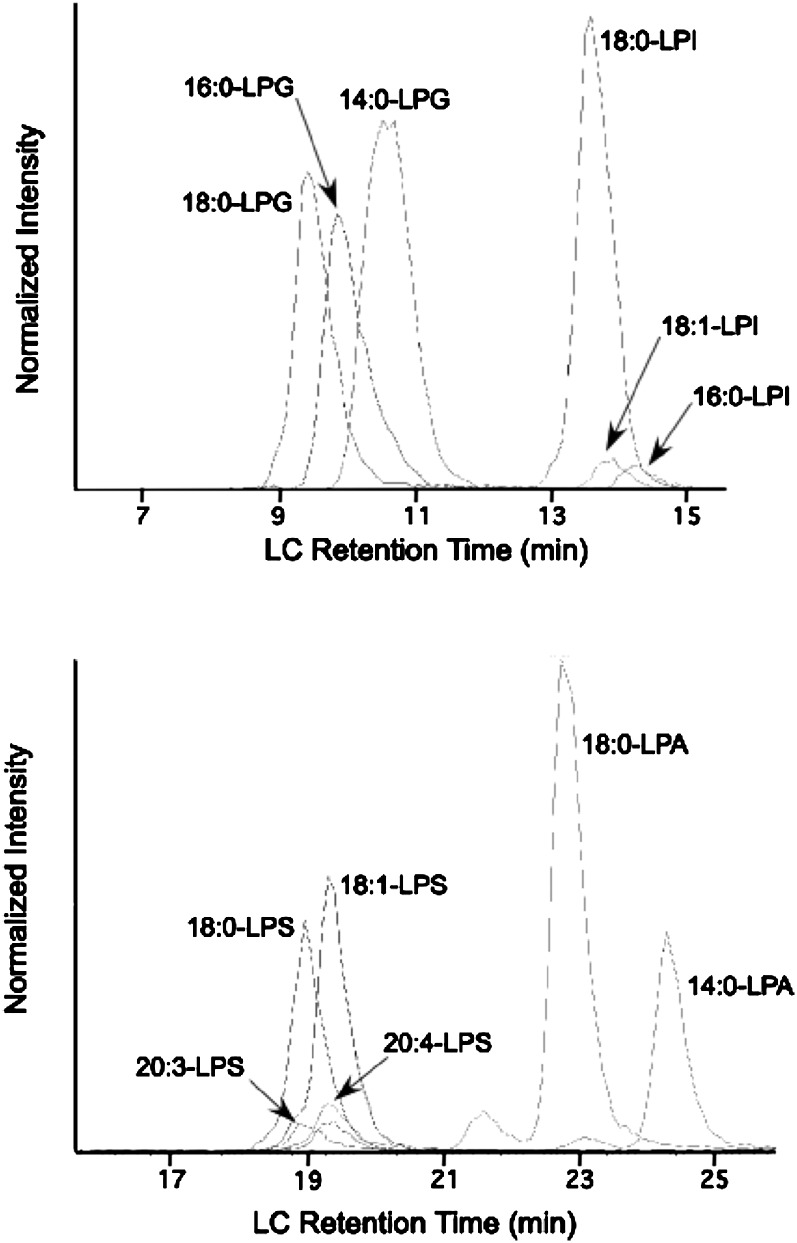
Fig. 1 shows an LC/ESI-MS/MS analysis of the standard test mixture of LPG, LPI, LPS, and LPA species (traces for LPE and LPC species are given in supplementary Fig. II). Limit of quantification (defined as a signal peak height-to-noise height of ∼10/1) values are given in supplementary Table II and are in the range of ∼20 pmole for LPC species and ∼1000 fmole for enyl-LPE species. Ion traces for quantities of lysophospholipids near their limit of quantification are shown in supplementary Fig. III. We found that it is important to include 5 mM ammonium acetate in the LC mobile phase as the use of lower concentrations leads to significant LC peak tailing for LPA and LPS species (not shown). Note that the LPA and LPS peaks have similar peak shapes to those for other lysophospholipid species (Fig. 1). Results with 10 mM ammonium acetate were similar to those with 5 mM (not shown).
Fig. 1.
LC/ESI-MS/MS detection of a standard mixture of lysophospholipids. The top trace shows negative mode detection of 14:0-LPG (9.4 pmole), 16:0-LPG (6.1 pmole), 18:0-LPG (6.4 pmole), 16:0-LPI (0.4 pmole), 18:1-LPI (0.55 pmole), and 18:0-LPI (7.9 pmole). The bottom panel shows negative mode detection of 18:0-LPA (7.9 pmole), 14:0-LPA (3.2 pmole), 20:3-LPS (0.5 pmole), 20:4-LPA (0.5 pmole), 18:0-LPS (3.7 pmole), and 18:1-LPS (4.4 pmole). The X-axis is the LC retention time (min) and the Y-axis is the ion intensities normalized to the largest peak in the group. Ion traces for LPC and LPE species are shown in supplementary Fig II.
In the analysis of lysophospholipid species reported here, we cycled through 89 single reaction monitoring events during the entire period of time in which the instrument was in negative mode. We compared the lysophospholipid detection sensitivity under these conditions to that observed under conditions where we monitored a smaller set of single ion reactions in time windows appropriate for the elution of each lysophospholipid head group class. Results were comparable under both conditions (not shown). This is expected as each cycle of 89 ion reactions takes ∼5 s, and given the LC peak widths of about 1 min (Fig. 1), there are about 12 cycles per LC peak, which is presumably sufficient to well define the peak.
We analyzed the lysophospholipid species present in mouse serum mainly to determine statistics for the quantification of lysophospholipids using our LC/ESI-MS/MS method. Among the full set of lysophospholipids analyzed by LC/ESI-MS/MS (116 species; see supplementary Table II), 80 gave discernable peaks in the selective ion trace chromatograms (by visual inspection) and were quantified with the aid of the perdeuterated internal standards. Data for all 80 species is given as supplementary Table IV. The method was carried out on four 100 μl aliquots of the same batch of mouse serum. The coefficient of variation (100 × (standard deviation/mean) were all <20% except five species were in the 20–25% range, one was 35%, and one was 43%. The distribution of coefficients of variation is shown as supplementary Fig. IV. Thus, the LC/ESI-MS/MS method is highly reproducible. Also highly reproducible among the four runs are the LC retention times for the 80 detected lysophospholipid species (supplementary Table IV). The concentration of the various lysophospholipid classes in mouse serum are as follows: total LPG, 0.05 μM; total LPI, 2.5 μM; total LPE, 1.1 μM; total LPS, 0.13 μM; total LPA, 5.7 μM; total LPC, 66 μM; total enyl-LPE, 0.33 μM; total enyl-LPC, 0.50 μM; total alkyl-LPC, 3.2 μM. Previously reported values for serum lysophospholipid concentrations include: 2 μM for LPA (14) and 200 μM for LPC (15).
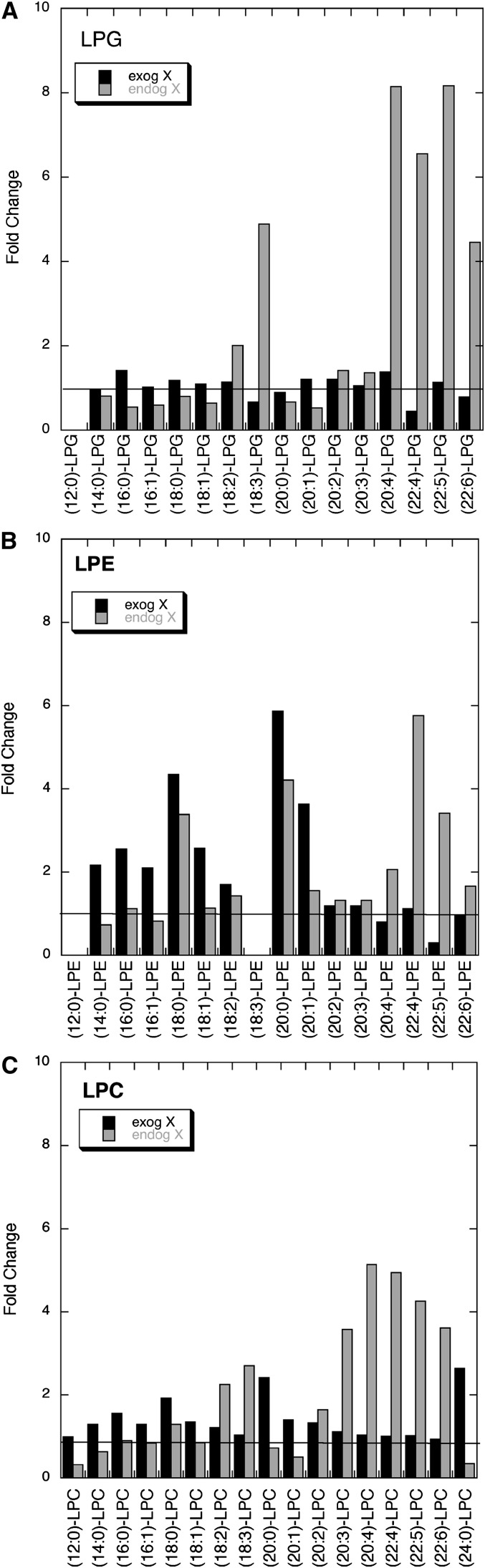
We analyzed the changes in lysophospholipid levels in HEK293 cells in response to exogenous addition of mature human group X secreted phospholipase A2 or in cells stably transfected with the human group X secreted phospholipase A2 gene (endogenously expressed enzyme). Relatively large changes were seen in the levels of LPG, LPE, and LPC species and these are shown in Fig. 2. Smaller changes in lysophospholipid levels were seen for the other head group species, and these changes are listed in in supplementary Table V. In the case of LPG, only small changes were seen after treating cells with exogenously added phospholipase A2, but transfected cells displayed relatively large changes in a subset of LPG species (18:3-, 20:4-, 22:4-, 22:5-, and 22:6-LPG) (Fig. 2A). Both exogenous and endogenous phospholipase A2 led to significant increases in specific LPE species, but the species produced were different depending on the presentation of the enzyme (Fig. 2B). In particular, 22:4-, 22:5-, and 22:6-LPE species were produced only by endogenous phospholipase A2. LPC levels showed a similar trend as that for LPE levels (Fig. 2C). A significant increase in 24:0-LPC was seen only with exogenous phospholipase A2 (Fig. 2C). These data clearly show that exogenous and endogenous human group X secreted phospholipase A2 act at different membrane sites. This in turn is consistent with a wealth of data from our earlier study showing that endogenously produced group X enzyme acts on membranes of the secretory compartment prior to externalization from cells, whereas exogenously added enzyme likely acts on the extracellular face of the plasma membrane (9).
Fig. 2.
Lysophospholipid molecular species changes in HEK293 cells in response to group X secreted phospholipase A2 action. Results are shown for LPG (A), LPE (B), and LPC (C) species. Data for all lysophospholipid species are given in supplementary Table V. Black bars give the ratio of lysophospholipid species (indicated on the X-axis) measured in HEK293 cells treated exogenously for 4 h at 37°C with 200 ng/ml group X secreted phospholipase A2 to the amount of lysophospholipid measured in HEK293 cells cultured for 4 h at 37°C in the absence of the phospholipase A2. Gray bars give the ratio of lysophospholipid species measured in HEK293 cells stably expressing group X secreted phospholipase A2 after culturing for 4 at 37°C to the amount of lysophospholipid measured in nontransfected HEK293 cells after culturing for 4 h at 37°C. Each LC/ESI-MS/MS analysis was carried out using one 10 cm dish of cells.
Summary
We developed an improved sample extraction procedure that best prevents the acid-catalyzed breakdown of enol ether containing phospholipids and lysophospholipids compared with previously reported methods. The new method also results in high recovery of the major types of lysophospholipid species including LPA and LPI, which are generally more difficult to isolate than the more hydrophobic species. Breakdown of even 5–10% of the plasmenyl phospholipids is a significant problem if one is trying to assess the presence of lysophospholipids in a sample, as lysophospholipids are in relatively low abundance compared with plasmenyl phospholipids. It should also be pointed out that the new extraction method does result in a low level of plasmenyl phospholipid breakdown, thus, lysophospholipid analysis is best carried out by looking at a relative change in molecular species level as a result of a biological stimulus rather than looking at absolute levels in a single sample. Still, relative changes in lysophospholipids will be hard to spot if there is significant breakdown of plasmenyl phospholipid species; thus, the new extraction procedure is recommended in all studies of lysophospholipid abundance when using biological membranes that contain plasmenyl phospholipids. Analysis of lysophospholipids by the LC/ESI-MS/MS method described here allows for the detection of 20–1000 fmoles of molecular species using an instrument manufactured about a decade ago. With use of more updated instruments, it should be possible to extend the limit of quantification by a factor of ∼5- to 10-fold. Finally, we analyzed isobaric species of sn-1 ester and ether lysophospholipids and showed that the use of internal standards with fatty chains with odd carbon number, i.e., 17:0 or 17:1 chains is problematic and the use of d31-16:0 lysophospholipid internal standards is preferred. Also, LC is needed along with ESI-MS/MS to detect all the sn-1 ester and ether lysophospholipid with a high degree of confidence of species identification. The new method was used to quantify lysophospholipids present in mouse serum with acceptable inter-sample reproducibility. Finally, the method was used to illustrate that human X secreted phospholipase A2 added exogenously to HEK293 cells acts on a different membrane site than cells expressing endogenous phospholipase A2 within the cell.
Supplementary Material
Footnotes
Abbreviations:
- LPA
- lysophosphatidic acid
- LPC
- lysophosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- LPG
- lysophosphatidylglycerol
- LPI
- lysophosphatidylinositol
- LPS
- lysophosphatidylserine
- alkyl-LPC
- 1-O-alkyl-LPC
- enyl-LPC and enyl-LPE
- LPC and LPE with the sn-1 ester replaced with a 1-O-1′-(Z)-enol ether
This work was supported by the National Institutes of Health Grants HL36235 and HL50040. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five tables and four figures.
REFERENCES
- 1.Surrel F., Ikram J., Boilard E., Bollinger J. G., Payre C., Mounier C. M., Talvinen K. A., Laine V. J. O., Nevalainen T., Gelb M. H., et al. 2009. Group X phospholipase A2 stimulates the proliferation of colon cancer cells by producing various lipid mediators. Mol. Pharmacol. 76: 778–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrill A. L. 2008. Lysophospholipid interactions with protein targets. Biochim. Biophys. Acta. 1781: 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulfer M., Murphy R. C. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 4.Christie W. W. 1982. Lipid Analysis. Pergamon, New York [Google Scholar]
- 5.Baker D. L., Desiderio D. M., Miller D. D., Tolley B., Tigyi G. J. 2001. Direct quantitative analysis of lysophosphatidic acid molecular species by stable isotope dilution electrospray ionization liquid chromatography-mass spectrometry. Anal. Biochem. 292: 287–295. [DOI] [PubMed] [Google Scholar]
- 6.Murph M., Tanaka T., Pang J., Felix E., Liu S., Trost R., Godwin A. K., Newman R., Mills G. 2007. Liquid chromatography mass spectrometry for quantifying plasma lysophospholipids: potential biomarkers for cancer diagnosis. Methods Enzymol. 433: 1–25. [DOI] [PubMed] [Google Scholar]
- 7.Bezzine S., Koduri R. S., Valentin E., Murakami M., Kudo I., Ghomashchi F., Sadilek M., Lambeau G., Gelb M. H. 2000. Exogenously added human group X secreted phospholipase A2 but not the group IB, IIA, and V enzymes efficiently release arachidonic acid from adherent mammalian cells. J. Biol. Chem. 275: 3179–3191. [DOI] [PubMed] [Google Scholar]
- 8.Eaton B. R., Dennis E. A. 1976. Analysis of phospholipase C (Bacillus cereus) action toward mixed micelles of phospholipid and surfactant. Arch. Biochem. Biophys. 176: 604–609. [DOI] [PubMed] [Google Scholar]
- 9.Mounier C. M., Ghomashchi F., Lindsay M. R., James S., Singer A. G., Parton R. G., Gelb M. H. 2004. Arachidonic acid release from mammalian cells transfected with human groups IIA and X secreted phospholipase A2 occurs predominantly during the secretory process and with the involvement of cytosolic phospholipase A2-alpha. J. Biol. Chem. 279: 25024–25038. [DOI] [PubMed] [Google Scholar]
- 10.Pluckthun A., Dennis E. A. 1982. Acyl and phosphoryl migration in lysophospholipids: importance in phospholipid synthesis and phospholipase specificity. Biochemistry. 21: 1743–1750. [DOI] [PubMed] [Google Scholar]
- 11.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipid Maps [Internet]. Cited 16 May 2009 at http://www.lipidmaps.org.
- 13.Khaselev N., Murphy R. C. 2000. Electrospray ionization mass spectrometry of lysoglycerophosphocholine lipid subclasses. J. Am. Soc. Mass Spectrom. 11: 283–291. [DOI] [PubMed] [Google Scholar]
- 14.Aoki J., Taira A., Takanezawa Y., Kishi Y., Hama K., Kishimoto T., Mizuno K., Saku K., Taguchi R., Arai H. 2002. Electrospray ionization mass spectrometry of lysoglycerophosphocholine lipid subclasses. J. Biol. Chem. 277: 48737–48744. [DOI] [PubMed] [Google Scholar]
- 15.Kishimoto T., Soda Y., Matsuyama Y., Mizuno K. 2002. An enzymatic assay for lysophosphatidylcholine concentration in human serum and plasma. Clin. Biochem. 35: 411–416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.