Abstract
Objective
Moyamoya disease (MMD) is an uncommon cerebrovascular disorder, characterized by progressive occlusion at the terminal portion of the internal carotid artery. Incidence of the disease is high in East Asia and familial MMD accounts for about 15% of the disease. Although the pathogenesis is unknown, association of HLA class I or II alleles with MMD has been reported with conflicting results. We investigated whether there is a difference in HLA class II association between familial and non-familial forms of the disease.
Methods
A total of 70 Korean children with MMD, including 16 familial cases (10 probands), and 207 healthy controls were studied. Among familial cases, only 10 probands were used for the HLA frequency analysis. High resolution HLA-DRB1 and DQB1 genotyping was performed using polymerase chain reaction (PCR)-sequence specific oligonucleotide hybridization and PCR-single strand conformation polymorphism methods.
Results
The phenotype frequencies of HLA-DRB1*1302 (70.0%) and DQB1*0609 (40.0%) were significantly increased in familial MMD compared to both controls [vs. 15.5%, corrected p (pc) = 0.008, odds ratio (OR) = 12.76; vs. 4.3%, pc = 0.02, OR = 14.67] and non-familial MMD patients (vs. 14.8%, pc = 0.02, OR = 13.42; vs. 1.9%, pc = 0.02, OR = 35.33). The frequencies of DRB1 and DQB1 alleles in non-familial MMD patients were not significantly different from those in controls.
Conclusion
Our findings suggest that the genetic polymorphism of HLA class II genes or other closely linked disease relevant gene(s) could be a genetic predisposing factor for familial MMD.
Keywords: Moyamoya disease, Familial, HLA-DR, HLA-DQ
INTRODUCTION
Moyamoya disease (MMD) is an uncommon cerebrovascular disorder that is characterized by progressive stenoocclusive lesions of the terminal portion of the internal carotid artery and its main branches within the circle of Willis. This occlusion results in the formation of a fine vascular network of collateral circulation, the moyamoya vessels ("moyamoya" means "puff of smoke" in Japanese) at the base of the brain, as shown by cerebral angiography18). Definite cases of MMD are diagnosed in patients with bilateral lesions, whereas patients with unilateral lesions are diagnosed as probable cases5). The incidence of MMD is high in countries in East Asia, such as Korea and Japan, although worldwide distribution of the disease has been reported with much lower incidences7,18). The disease is distributed in all age groups, but the highest peak is in childhood at less than 10 years of age with a second low peak from 30s to 40s6,30).
The pathogenesis of MMD is still unclear18). Several pieces of evidence suggest the involvement of genetic factors in MMD : 1) over 10% of MMD patients have affected blood relatives, 2) concordance in the affection status has been proven in 80% of identical twins, and 3) there is an ethnic predisposition to MMD, the incidence of the disease being the highest in the Asian population9). Data from an epidemiological study of familial MMD have suggested that MMD is probably inherited in a polygenic or autosomal dominant mode with a low penetrance21). Microsatellite linkage analysis has identified genetic loci that are associated with MMD on chromosomes 3, 6, 8, and 1711,13,20,26,32). However, the relevant genes have not been identified so far18,20).
In relation to genetic factors associated with the disease, associations of various HLA class I or class II alleles with the disease have been reported in Japanese2,12,14,17) and Koreans8) with conflicting results. Although familial forms account 10-15% of MMD patients6,18,31), HLA association with the disease has not been studied separately in familial and non-familial (sporadic) MMD cases. Thus, it is not known whether there is a difference between these two forms of the disease with regard to HLA association. We have studied HLA association with the disease in 70 Korean children including 16 familial cases, and found a strong association of HLA-DR and -DQ genes with the disease in the familial form of the disease.
MATERIALS AND METHODS
Patients and controls
A total of 70 patients, who were diagnosed as having MMD after cerebral angiography at Seoul National University Children's Hospital between 2001 and 2004 were included in this study. The patients comprised of 29 boys and 41 girls. Fifty-four patients were sporadic (non-familial) cases and did not have other affected family members. Sixteen (22.9%) of the 70 patients had more than one family member diagnosed as MMD (familial MMD). They were from 10 family pedigrees (6 pairs of family members from 6 families and 4 single patients from 4 families) including 10 proband cases. Thirteen patients had first-degree (one in mother, 12 in siblings including 4 identical twins) and three had third-degree family members (cousins) affected. Age distribution was 7.1 ± 2.7 (SD) (range 7 months-12 years) in total patients; 7.5 ± 2.6 (7 months-12 years) in non-familial cases; and 5.9 ± 2.9 (13 months-10 years) in familial cases. Female to male ratio was 1.4 (41 : 29) in total patients; 1.2 (29 : 25) in non-familial cases; and 3.0 (12 : 4) in familial cases. Informed consent was obtained from each patient to acquire peripheral blood for genetic study. All the patients were of Korean ethnicity. Control group consisted of 207 healthy Koreans from our previous study, who were unrelated parents of 107 families recruited for family study of HLA polymorphism in this population28).
HLA-DR and -DQ typing
Genomic DNA was extracted from the peripheral blood using LaboPass™ Blood kit (CosmoGenetech, Seoul, Korea). HLA-DRB1 typing was performed in two steps. Intermediate-resolution HLA-DRB typing was carried out by the PCR-SSO (sequence specific oligonucleotide) hybridization method using the Dynal RELI™ SSO HLA-DRB Test (Dyanl Biotech Ltd., Wirral, UK). For high-resolution HLA-DRB1 typing, group-specific amplifications and PCR-SSCP (single strand conformation polymorphism) analysis was performed as described3) with minor modifications. HLA-DQB1 typing was also performed in two steps. Intermediate-resolution HLA-DQB1 typing was carried out by the PCR-SSO method using the Dynal RELI™ SSO HLA-DQB1 Test (Dyanl Biotech). For high-resolution HLA-DQB1 typing, group specific amplifications and PCR-SSCP was performed as described23).
Statistical analysis
The phenotype frequencies of HLA-DRB1 and -DQB1 alleles were compared using Fisher's exact test (2-tail). Among 16 familial cases, only 10 proband cases were included in the frequency analysis. The level of significance was set at p < 0.05, and odds ratios (OR) with 95% confidence intervals (CI) were calculated for those comparisons demonstrating significant p values. Where indicated as corrected p (pc), a Bonferroni correction was applied by multiplying the probability value by the number of comparisons made (27 for DRB1 and 15 for DQB1 alleles). SPSS for Windows version 12.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
RESULTS
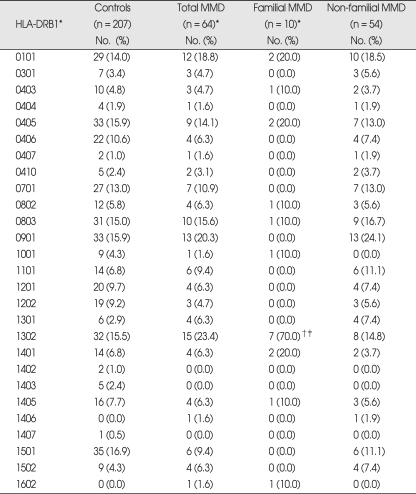
Phenotype frequencies of HLA-DRB1 alleles in MMD patients are presented in Table 1. Frequencies of HLA-DRB1 alleles were not significantly different between total MMD patients and controls. When the MMD patients of familial and non-familial cases were analyzed separately, familial MMD showed markedly increased frequency of DRB1*1302 (70%) compared to both controls (15.5%, p = 0.0003, pc = 0.008, OR = 12.76) and non-familial MMD (14.8%, p = 0.0008, pc = 0.02, OR = 13.42). However, the frequencies of DRB1 alleles in non-familial MMD were quite similar to those of controls, and no significant difference was observed.
Table 1.
Phenotype frequencies of HLA-DRB1 alleles in Korean patients with moyamoya disease (MMD)
*Among 16 familial cases, 10 proband cases were included in the analysis, †p = 0.0003, pc (corrected p) = 0.008. OR = 12.76 (3.13-51.95) vs. controls, ‡p = 0.0008, pc = 0.02, OR = 13.42 (2.86-63.02) vs. non-familial MMD
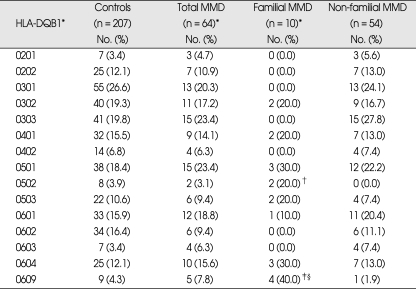
Frequencies of HLA-DQB1 alleles were not significantly different between total MMD patients and controls (Table 2). However, familial MMD patients showed significantly increased frequency of DQB1*0609 (40%) compared to both controls (4.3%, p = 0.001, pc = 0.02, OR = 14.67) and non-familial MMD (1.9%, p = 0.002, pc = 0.02, OR = 35.33). DQB1*0502 was increased in familial MMD when compared to non-familial MMD (20% vs. 0%, p = 0.02, OR = incalculable). The frequencies of DQB1 alleles in non-familial MMD were not significantly different from those in controls.
Table 2.
Phenotype frequencies of HLA-DQB1 alleles in Korean patients with moyamoya disease (MMD)
*Among 16 familial cases, 10 proband cases were included in the analysis, †p = 0.02 (OR = incalculable) vs. non-familial MMD, ‡p = 0.001, pc (corrected p) = 0.02, OR = 14.67 (3.51-61.33) vs. controls, §p = 0.002, pc = 0.02. OR = 35.33 (3.38-369.86) vs non-familial MMD
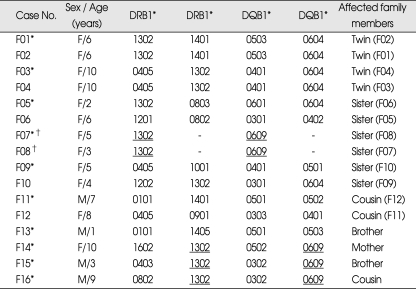
HLA-DRB1 and -DQB1 alleles and DRB1*1302-DQB1*0609 haplotypes in 16 familial MMD cases, including 10 proband cases are presented in Table 3. DRB1*1302 is exclusively associated with either DQB1*0604 or DQB1*0609 in Koreans28). Seven of the 10 proband cases carried DRB1*1302 allele, among which 3 and 4 were associated with DQB1*0604 and DQB1*0609, respectively. One patient was homozygous for the DRB1*1302-DQB1*0609 haplotype. Familial MMD patients showed significantly increased haplotype frequency of DRB1*1302-DQB1*0609 (25.0%) compared to both controls (2.4%, p = 0.0003, OR = 13.47) and non-familial MMD (0.9%, p = 0.0003, OR = 35.67). Although the haplotype frequency of DRB1*1302-DQB1*0604 was slightly increased in familial MMD patients compared to that of controls, it was not statistically significant (15% vs. 6.5%, p = 0.15).
Table 3.
HLA-DRB1 and -DQB1 alleles and DRB1*1302-DQB1*0609 haplotypes in 16 familial cases of moyamoya disease (MMD) including 10 proband cases
*Proband cases, †Homozygous for DRB1*1302-DQB1*0609 haplotype. DRB1*1302-DQB1*0609 haplotype (underlined) frequency in proband cases of familial MMD (25.0%, 5/20) : p = 0.0003, OR = 13.47 (4.09-44.30) vs. controls (2.4%, 10/414); p = 0.0003, OR = 35.67 (3.90-326.43) vs. non-familial MMD (0.9%, 1/108)
DISCUSSION
In our study, familial MMD patients showed markedly increased frequency of HLA-DRB1*1302 allele (phenotype frequency of 70%) compared to both controls (15.5%, pc = 0.008, OR = 12.76) and non-familial MMD patients (14.8%, pc = 0.02, OR = 13.42) (Table 1). Among the two HLA-DQB1 alleles (DQB1*0604 and *0609) tightly linked with DRB1*1302 in Koreans28), DQB1*0609 was significantly increased in familial MMD patients compared to both controls (pc = 0.02, OR = 14.67) and non-familial MMD patients (pc = 0.02, OR = 35.33) (Table 2). Thus, the haplotype frequency of DRB1*1302-DQB1*0609 was significantly increased in familial MMD patients compared to both controls (p = 0.0003, OR = 13.47) and non-familial MMD (p = 0.0003, OR = 35.67) (Table 3). In contrast, non-familial MMD patients did not show any significant change in the frequencies of HLA-DRB1 or DQB1 alleles compared to controls.
The strong association of HLA-DRB1* 1302 and DQB1*0609 alleles with familial MMD patients observed in the present study of Koreans is much stronger than those reported to date in Japanese or Korean MMD patients2,8,12,14,17). Previous studies included relatively small number of patients, and the investigators did not attempt to analyze the HLA association with familial and non-familial MMD, separately. Various HLA class I antigens were reported to be associated with MMD in three different studies of Japanese and Korean MMD patients. Increased frequencies of HLA-A24, B46, B51, B54, and B67 in Japanese2,17) and B35 in Korean MMD patients8) were reported. However, the associations were rather weak (p values in the range of < 0.05 - < 0.005; 18-32 patients studied) and none of the HLA class I antigens has been corroborated by more than one report.
There have been studies on the investigation of HLA class II association with the disease in Japanese, which also revealed non-concordant results. Aoyagi et al.2) reported weak association of DR1 [relative risk (RR) 9.1, p < 0.05] by serological typing. Inoue et al.14) studied HLA-DR and -DQ associations in MMD patients by molecular typing and found different alleles associated with early-onset (onset ≤ 10 years of age) and late-onset (onset > 10 years of age) MMD. HLA-DRB1*1501 (RR 2.3) and DQB1*0602 (RR 2.42) were increased and DRB1*0405 was decreased (RR 0.38) in early-onset, whereas DQB1*0502 was increased (RR 4.72) in late-onset MMD patients. The associations were weak (p values in the range of < 0.05 - < 0.025) and none of the associations was significant after Bonferroni correction. In our study, although majority (57/64, 89%) of the patients belonged to the early-onset MMD including 10 familial cases, we could not find those associations reported in the Japanese patients with early-onset MMD.
When particular HLA alleles are associated with disease susceptibility or resistance, there are two possible mechanisms of association. These HLA alleles may directly control the susceptibility to or resistance against the disease. Another possibility is that these HLA alleles are simply disease associated markers, showing linkage disequilibrium with other disease relevant genes located close to the HLA genes. The latter is more likely in MMD, because thus far reported HLA alleles associated with the disease quite vary among different studies. Then questions arise what would be the disease relevant gene(s) closely linked with DRB1*1302-DQB1*0609 haplotype in Korean patients with familial MMD. One of the possibilities is the tumor necrosis factor (TNF)-α gene considering the TNF-α promoter polymorphisms and extended HLA and TNF-α haplotypes in Koreans24). TNF-α high producer haplotype containing -308A allele in the promoter region of the TNF-α gene is strongly linked with HLA-A33, B58, and DRB1*1302 alleles24), which are in strong linkage disequilibrium with DQB1*0609 allele28). However, change in the TNF-α level or genetic polymorphism of TNF-α gene has not been investigated in MMD patients to date.
Pathological findings in the carotid terminations of cerebral arteries in MMD patients show fibrocellular thickening of the intima, irregular undulation (waving) of the internal elastic lamina, and attenuation of the media18). Although we have no definite explanation for the pathogenesis of MMD, the final common pathway seems to involve proliferation of smooth muscle cells and their migration from the media to the intima1,16). This process is regulated by various growth factors, and the results from previous studies have shown that concentrations of certain growth factors or cytokines are increased in the cerebrospinal fluid and/or their expression is increased in the intracranial and extracranial arteries of patients with MMD18). These include basic fibroblast growth factor10), soluble adhesion molecules29), cellular retinoic-acid-binding protein I (CRABP-I)16), and hepatocyte growth factor, an angiogenic protein22). In vascular injury and repair, besides aforementioned various growth factors, a balance between the activities of connective tissue degrading enzymes, matrix metalloproteinases (MMPs) and their endogenous inhibitors, tissue inhibitors of metalloproteinases (TIMPs) is important27), which might be deranged in MMD. In this context, increased expression of serum MMP-94) and genetic polymorphism of TIMP2 gene15) were reported to be associated with MMD. It is of interest that the study of TIMP gene suggested a difference in the genetic predisposition of familial and non-familial MMD, as we have found in the present study of HLA gene. Presence of a G/C heterozygous genotype at position -418 in TIMP2 promoter (located in chromosome 17q25), which was expected to reduce the transcription of TIMP2, was associated with familial, but not with non-familial MMD15).
We have found that HLA-DRB1*1302 and DQB1*0609 alleles were strongly associated with familial MMD in the present study, and DRB1*1302-DQB1*0609 haplotype in Koreans might be a predisposing factor for increased intimal fibrosis and occlusion of cerebral arteries observed in MMD patients. It is of interest that HLA-DRB1*1302 and DQB1*0609 alleles were significantly associated with liver fibrosis following Schistosoma japonicum infection in Southern Chinese, in whom these two HLA class II alleles are in very strong linkage disequilibrium, forming a common haplotype19). In contrast, this haplotype is very rare in Japanese25). We speculated that HLA-DRB1*1302 and DQB1*0609 alleles associated with familial MMD might be related with TNF-α gene polymorphism of high producer type. Whether genetic polymorphism of TNF-α gene is associated with the disease needs to be studied to get more insight into the pathogenesis of the disease.
CONCLUSION
We for the first time found a difference in the genetic predisposition of familial and non-familial MMD in terms of HLA association. HLA-DRB1*1302 and DQB1*0609 alleles were significantly associated with familial, but not with non-familial form of the disease in Koreans. Our findings suggest that the genetic polymorphism of HLA class II genes or other closely linked disease relevant gene(s) could be a genetic predisposing factor for familial MMD.
References
- 1.Aoyagi M, Fukai N, Yamamoto M, Nakagawa K, Matsushima Y, Yamamoto K. Early development of intimal thickening in superficial temporal arteries in patients with moyamoya disease. Stroke. 1996;27:1750–1754. doi: 10.1161/01.str.27.10.1750. [DOI] [PubMed] [Google Scholar]
- 2.Aoyagi M, Ogami K, Matsushima Y, Shikata M, Yamamoto M, Yamamoto K. Human leukocyte antigen in patients with moyamoya disease. Stroke. 1995;26:415–417. doi: 10.1161/01.str.26.3.415. [DOI] [PubMed] [Google Scholar]
- 3.Bannai M, Tokunaga K, Lin L, Kuwata S, Mazda T, Amaki I, et al. Discrimination of human HLA-DRB1 alleles by PCR-SSCP (single-strand conformation polymorphism) method. Eur J Immunogenet. 1994;21:1–9. doi: 10.1111/j.1744-313x.1994.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 4.Fujimura M, Watanabe M, Narisawa A, Shimizu H, Tominaga T. Surg Neurol. 2009. Increased expression of serum matrix metalloproteinase-9 in patients with moyamoya disease. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Fukui M. Guidelines for the diagnosis and treatment of spontaneous occlusion of the circle of willis ('moyamoya' diseases). Research Committee on Spontaneous Occlusion of the Circle of Willis (Moyamoya Disease) of the Ministry of Health and Welfare, Japan. Clin Neurol Neurosurg. 1997;99:S238–S240. [PubMed] [Google Scholar]
- 6.Fukui M, Kono S, Sueishi K, Ikezaki K. Moyamoya disease. Neuropathology. 2000;20:S61–S64. doi: 10.1046/j.1440-1789.2000.00300.x. [DOI] [PubMed] [Google Scholar]
- 7.Goto Y, Yonekawa Y. Worldwide distribution of moyamoya disease. Neurol Med Chir (Tokyo) 1992;32:883–886. doi: 10.2176/nmc.32.883. [DOI] [PubMed] [Google Scholar]
- 8.Han H, Pyo CW, Yoo DS, Huh PW, Cho KS, Kim DS. Associations of Moyamoya patients with HLA class I and class II alleles in the Korean population. J Korean Med Sci. 2003;18:876–880. doi: 10.3346/jkms.2003.18.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashitaka H, Liu W, Mineharu Y, Inoue K, Takenaka K, Ikeda H, et al. [Current knowledge on the genetic factors involved in moyamoya disease.] Brain Nerve. 2008;60:1261–1269. [PubMed] [Google Scholar]
- 10.Houkin K, Yoshimoto T, Abe H, Nagashima K, Nagashima M, Takeda M, et al. Role of basic fibroblast growth factor in the pathogenesis of moyamoya disease. Neurosurg Focus. 1998;5:1–5. doi: 10.3171/foc.1998.5.5.5. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda H, Sasaki T, Yoshimoto T, Fukui M, Arinami T. Mapping of a familial moyamoya disease gene to chromosome 3p24.2-p26. Am J Hum Genet. 1999;64:533–537. doi: 10.1086/302243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M. Analysis of class II genes of human leukocyte antigen in patients with moyamoya disease. Clinical Neurol Neurosurg. 1997;99:S234–S237. doi: 10.1016/s0303-8467(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 13.Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M. Linkage analysis of moyamoya disease on chromosome 6. J Child Neurol. 2000;15:179–182. doi: 10.1177/088307380001500307. [DOI] [PubMed] [Google Scholar]
- 14.Inoue TK, Ikezaki K, Sasazuki T, Ono T, Kamikawaji N, Matsushima T, et al. DNA typing of HLA in the patients with moyamoya disease. Jpn J Hum Genet. 1997;42:507–515. doi: 10.1007/BF02767027. [DOI] [PubMed] [Google Scholar]
- 15.Kang HS, Kim SK, Cho BK, Kim YY, Hwang YS, Wang KC. Single nucleotide polymorphisms of tissue inhibitor of metalloproteinase genes in familial moyamoya disease. Neurosurgery. 2006;58:1074–1080. doi: 10.1227/01.NEU.0000215854.66011.4F. [DOI] [PubMed] [Google Scholar]
- 16.Kim SK, Yoo JI, Cho BK, Hong SJ, Kim YK, Moon JA, et al. Elevation of CRABP-I in the cerebrospinal fluid of patients with moyamoya disease. Stroke. 2003;34:2835–2841. doi: 10.1161/01.STR.0000100159.43123.D7. [DOI] [PubMed] [Google Scholar]
- 17.Kitahara T, Okumura K, Semba A, Yamaura A, Makino H. Genetic and immunologic analysis on moya-moya. J Neurol Neurosurg Psychiatry. 1982;45:1048–1052. doi: 10.1136/jnnp.45.11.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda S, Houkin K. Moyamoya disease : current concepts and future perspectives. Lancet Neurol. 2008;7:1056–1066. doi: 10.1016/S1474-4422(08)70240-0. [DOI] [PubMed] [Google Scholar]
- 19.McManus DP, Ross AG, Williams GM, Sleigh AC, Wiest P, Erlich H, et al. HLA class II antigens positively and negatively associated with hepatosplenic schistosomiasis in a Chinese population. Int J Parasitol. 2001;31:674–680. doi: 10.1016/s0020-7519(01)00132-1. [DOI] [PubMed] [Google Scholar]
- 20.Mineharu Y, Liu W, Inoue K, Matsuura N, Inoue S, Takenaka K, et al. Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology. 2008;70:2357–2363. doi: 10.1212/01.wnl.0000291012.49986.f9. [DOI] [PubMed] [Google Scholar]
- 21.Mineharu Y, Takenaka K, Yamakawa H, Inoue K, Ikeda H, Kikuta KI, et al. Inheritance pattern of familial moyamoya disease : autosomal dominant mode and genomic imprinting. J Neurol Neurosurg Psychiatry. 2006;77:1025–1029. doi: 10.1136/jnnp.2006.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanba R, Kuroda S, Ishikawa T, Houkin K, Iwasaki Y. Increased expression of hepatocyte growth factor in cerebrospinal fluid and intracranial artery in moyamoya disease. Stroke. 2004;35:2837–2842. doi: 10.1161/01.STR.0000148237.13659.e6. [DOI] [PubMed] [Google Scholar]
- 23.Park MH, Whang DH, Kang SJ. High resolution HLA-DQB1 typing by combination of PCR-RFLP and PCR-SSCP. Hum Immunol. 1999;60:901–907. doi: 10.1016/s0198-8859(99)00069-5. [DOI] [PubMed] [Google Scholar]
- 24.Park YJ, Park H, Park MH. TNF-α promoter polymorphisms and extended HLA and TNF-α haplotypes in Koreans based on 100 families. Tissue Antigens. 2004;63:75–79. doi: 10.1111/j.1399-0039.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- 25.Saito S, Ota S, Yamada E, Inoko H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens. 2000;56:522–529. doi: 10.1034/j.1399-0039.2000.560606.x. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai K, Horiuchi Y, Ikeda H, Ikezaki K, Yoshimoto T, Fukui M, et al. A novel susceptibility locus for moyamoya disease on chromosome 8q23. J Hum Genet. 2004;49:278–281. doi: 10.1007/s10038-004-0143-6. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Patel S, Niculescu R, Chung W, Desrochers P, Zalewski A. Role of matrix metalloproteinases and their tissue inhibitors in the regulation of coronary cell migration. Arterioscler Thromb Vasc Biol. 1999;19:1150–1155. doi: 10.1161/01.atv.19.5.1150. [DOI] [PubMed] [Google Scholar]
- 28.Song EY, Park MH, Kang SJ, Park HJ, Kim BC, Tokunaga K, et al. HLA class II allele and haplotype frequencies in Koreans based on 107 families. Tissue Antigens. 2002;59:475–486. doi: 10.1034/j.1399-0039.2002.590604.x. [DOI] [PubMed] [Google Scholar]
- 29.Soriano SG, Cowan DB, Proctor MR, Scott RM. Levels of soluble adhesion molecules are elevated in the cerebrospinal fluid of children with moyamoya syndrome. Neurosurgery. 2002;50:544–549. doi: 10.1097/00006123-200203000-00022. [DOI] [PubMed] [Google Scholar]
- 30.Wakai K, Tamakoshi A, Ikezaki K, Fukui M, Kawamura T, Aoki R, et al. Epidemiological features of moyamoya disease in Japan : findings from a nationwide survey. Clin Neurol Neurosurg. 1997;99:S1–S5. doi: 10.1016/s0303-8467(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 31.Yamauchi T, Houkin K, Tada M, Abe H. Familial occurrence of moyamoya disease. Clin Neurol Neurosurg. 1997;99:S162–S167. doi: 10.1016/s0303-8467(97)00054-1. [DOI] [PubMed] [Google Scholar]
- 32.Yamauchi T, Tada M, Houkin K, Tanaka T, Nakamura Y, Kuroda S, et al. Linkage of familial moyamoya disease (spontaneous occlusion of the circle of Willis) to chromosome 17q25. Stroke. 2000;31:930–935. doi: 10.1161/01.str.31.4.930. [DOI] [PubMed] [Google Scholar]