Abstract
Pathogens can utilize DNA recombination to promote antigenic variation of surface structures to avoid immune detection. We identified a cis-acting DNA sequence near the antigenically variable pilin locus of the human pathogen, Neisseria gonorrhoeae. This 16 base pair G-rich sequence was required for pilin antigenic variation and formed a guanine quartet (G4) structure in vitro. Individual mutations that disrupted the structure also blocked pilin antigenic variation and prevented nicks required for recombination from occurring within the G4 region. A compound that binds and stabilizes G4 structures also inhibited pilin antigenic variation and prevented nicks from occurring on the G-rich strand. This site constitutes a recombination initiation sequence/structure that directs gene conversion to a specific chromosomal locus.
DNA recombination is a process shared by all DNA carrying organisms that is used for a variety of cellular processes including DNA repair, genetic exchange, and meiotic chromosome segregation (1). Additionally, recombination mediates many high frequency gene diversification systems including yeast mating type switches, immunoglobin diversity and pathogenesis-associated antigenic variation (Av) (2-4). Most recombination reactions occur at low frequency, but several diversity generating systems can enact programmed recombination reactions between specific loci at relatively high frequencies (2, 3, 5, 6).
Neisseria gonorrhoeae is the sole causative agent of gonorrhea and has evolved three high-frequency, diversity-generation systems to avoid immune surveillance (7). This antigenic variability of gonococcal populations is one reason that natural immunity to re-infection has never been demonstrated and has prevented development of an effective vaccine. One of these Av systems is mediated by high-frequency gene conversion events between one of many silent pilin loci and the single expressed pilin locus, pilE. This produces variant pilin proteins (2) that form antigenically divergent pili, which are the hair-like appendages expressed by many bacteria.
This system uses normal homologous recombination factors to mediate specialized gene conversion reactions (8-12), but no DNA element required for pilin Av has been described. Previous work showed that some transposon insertions in the intergenic region upstream of pilE block pilin Av without altering pilin expression (11, 13). To define the DNA element being disrupted by these transposons, the pilE upstream region was randomly mutagenized, and the mutations were introduced into the gonococcal chromosome by DNA transformation and linkage to a transposon insertion that does not affect pilin Av (Fig. S1A) (13). Two independent screens were performed. In both screens, mutants unable to undergo pilin Av were selected by screening for a stable, piliated colony morphology (Avd phenotype, Fig. S1B), while in the second screen both Av deficient (Avd) and Av transformants were analyzed (Table S1) (14). DNA sequence analysis of the transformants coupled with site directed mutagenesis revealed that alteration of any one of 12 guanine-cytosine (GC) bp within a 16bp region each inhibits pilin Av, while mutation of the other bps within or surrounding this region did not (Fig. 1A-B, S2, Table S2). The 12 GC bps defining this DNA element are conserved in 14 sequenced gonococcal isolates and three Neisseria meningitis genomes upstream of pilE but are not found in commensal Neisseria (Table S3). The ability of the single nucleotide mutations to replicate the genetic behavior of the pilin Av-disrupting transposon insertions was determined by introducing one of the GC bp mutations into a recG/ruvB double mutant. Previously, the RecG and RuvABC Holliday junction processing pathways, were both found to be required for pilin Av (10). RecA expression in a recG/ruvB double mutant is synthetically lethal, but can be overcome by a pilin Av-disrupting transposon insertion (10). Introduction of one of the GC bp mutations also rescued the RecA-dependent synthetically lethal phenotype showing they have the identical phenotypes as the neighboring transposon insertions that act upstream of homologous recombination factors (Fig. S3).
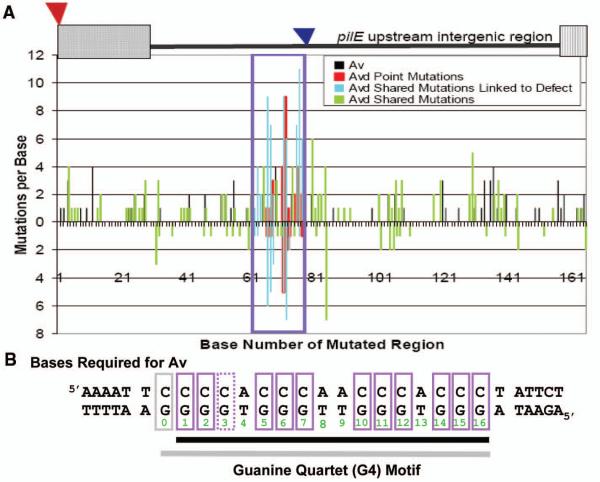
Figure 1. Identification of a DNA sequence in the intergenic region upstream of pilE required for gonococcal pilin Av.
A. Mutational Spectrum of Genetic Screens: A cartoon of the mutagenized region from the transposon insertion (red triangle) that has no affect on pilin Av (13) to 165bps downstream showing the location of the previously identified Av-disrupting transposons (blue triangle) (11, 13). X-axis indicates base number while y-axis indicates number of mutations isolated per base. Screen #1 (bottom axis) represents 103 Avd mutants (only mutants with mutations <9 are shown) (Table S1A). Screen #2 (top axis) represents 204 Av transformants and 106 Avd mutants (only mutants with mutations <17 are shown) (Table S1B). The region linked to the Avd phenotype is boxed. Bars indicate mutations that allow Av (black), single mutations that cause an Avd phenotype (red), and Avd mutants that have mutations within the boxed region (blue) but have mutations elsewhere (green). B. Bases required for pilin Av: Mutation of individual purple boxed bps result in an Avd phenotype. Solid purple boxes indicate a complete block of pilin Av while the G-3 mutant shows residual activity (Fig. S2). The DNA element on the bottom strand forms a guanine quartet (G4) motif (black underlined bases 1-16), when G-3 is mutated, the alternative G4 using G-0 (grey box) is shown underlined in grey.
The G-rich sequence defined by the genetic screens conforms to a guanine-quartet (G4) motif (Fig. 1B) (15, 16). G4 forming sequences have been implicated in many biological processes (17-20) but their existence within a cell has yet to be proven. To test whether the pilE-linked G4 motif forms a G4 structure we synthesized a series of oligonucleotides and evaluated their ability to form a G4 structure. CD spectrum analysis showed that the DNA sequence required for pilin Av adopts a parallel G4 structure and that mutation of two thymines in the loop regions have no effect on the structure (Fig. 2A-B). Methylase protection assay revealed two alternative G4 structures (Fig. 2C). Interestingly, mutation of the G-3 residue yielded a mutant having residual pilin Av suggesting this alternate G4 structure can form when the G-3 site is mutated (Fig. 1B, S2). Replication stop assays using three different DNA polymerases confirmed that the G4 structure forms and can halt replication (Fig. S4). Individual mutations that block pilin Av were found to block structure formation in all three assays (Fig. 2A, 2C, S4).
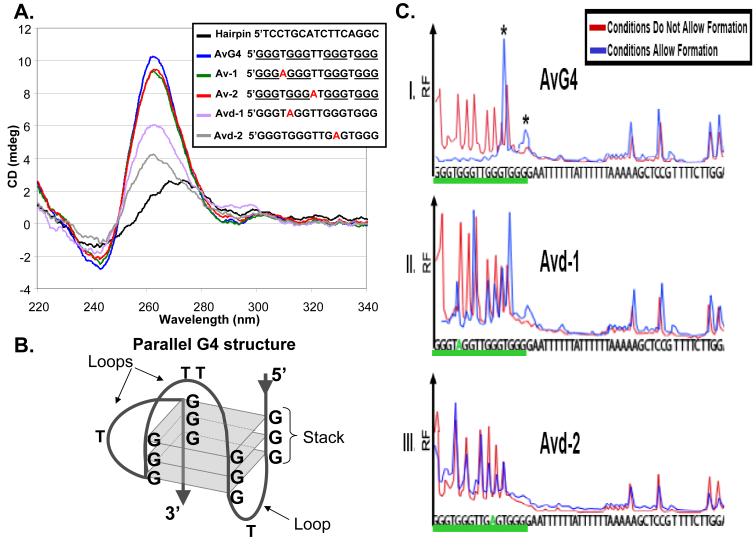
Figure 2. Analysis of G4 structure formation in vitro.
A. Circular dichroism (CD) spectrometry: The Hairpin oligonucleotide forms a double stranded DNA molecule producing a B-DNA CD spectrum (25). The AvG4 oligonucleotide has the pilE G4 sequence that allows pilin Av, Av-1 and Av-2 oligonucleotides have A-T mutations that do not alter Av, and all produce parallel G4 structure CD spectrums (25). Avd-1 and Avd-2 oligonucleotides contain G-A transversions that block pilin Av and show altered CD spectrums. B. Parallel G4 structure: Three G-quartets (squares) in a parallel G4 configuration. C. Dimethyl sulfate (DMS) methylase protection assay: Formed G4 guanines are protected from DMS attack (26). Shown are representative electropherograms of a FAM-labeled 52bp oligonucleotide containing: I. The wild-type G4 sequence (AvG4 underlined), or II and III, two mutations that block pilin Av (Avd-1 and Avd-2, underlined). N=3. X-axis indicates the base and y-axis shows relative fluorescence (RF). Blue and red traces indicate G4 forming and non-forming conditions, respectively. Under G4 forming conditions, AvG4 forms two alternative G4 structures (5′GGGTGGGTTGGGTGGGG or 5‘GGGTGGGTTGGGTGGGG, guanines forming the structure are underlined, asterisks) while mutant oligonucleotides do not form the structure.
There are 46 predicted G4 forming sequences in the gonococcal chromosome (15, 16), and none of these sequences are identical to the pilE-linked G4 nor are they located near any gene involved in pilin Av. Mutation of two irrelevant G4 forming sequences in the genome had no effect on pilus phase variation ruling out a nonspecific role for other G4 sequences in pilin Av (Fig. S5). When a copy of the pilE-linked G4 sequence was introduced upstream of the pilS7 silent pilin locus at a position similar to the pilE-linked copy in a strain where the pilE-linked copy was mutated (Fig. S6), there was no observed changes at pilS7 or pilE, even though 4-31 pilE variants carrying pilS7 sequences would normally be found in a pool of 374 progeny (Table S4). Moreover, when the pilE-linked G4 was replaced with four other G4 forming sequences or an inverted copy, pilin Av was lost (Fig. 3C). Thus, the specific G4 forming sequence, its position and orientation were all essential to allow pilin Av.
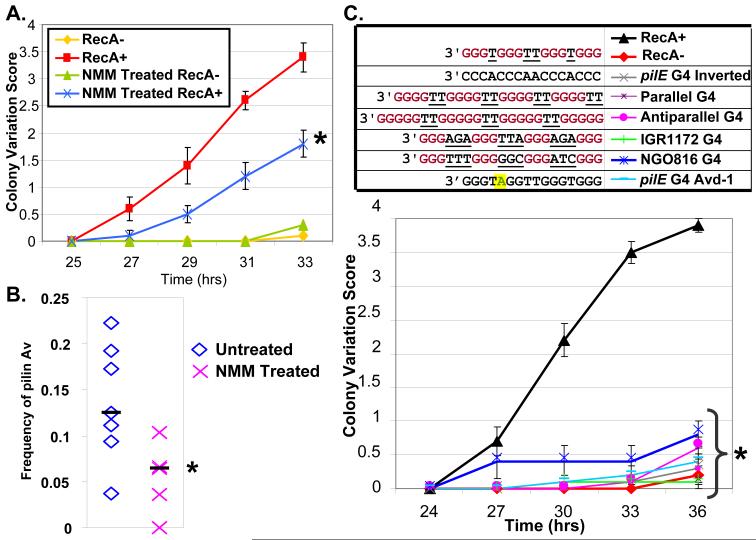
Figure 3. Stabilization, substitution or inversion of the pilE G4 structure inhibits Av.
A. Kinetic pilus-dependent colony phase variation assay: Shown is a representative assay (N=3) of NMM treated or untreated RecA+ and RecA− bacteria, (RecA is required for pilin Av), with mean phase variation score (11) and SEM of 10 colonies. The mean phase variation of NMM treated RecA+ is different from untreated RecA+ by Student’s T-test, P<0.05 (asterisk). B. DNA sequencing assay for pilin Av frequency (2): 28 piliated progeny from seven founder colonies of untreated and NMM treated bacteria were analyzed. ◇ and X show the mean frequency of untreated and NMM treated founders, respectively. Bars show the median of the means. The pilin Av frequency of NMM treated is less than untreated bacteria by Wilcoxon rank-sum test, P<0.05 (asterisk). C. G4 substitution colony phase variation assay: RecA+ and RecA− strains have the wild-type pilE G4 sequence. The pilE G4 inverted strain has the identical G4 sequence inverted in the same location with no other sequence changes. The pilE G4 Avd-1 mutant has a G-A transversion at G-5 that blocks pilin Av. The other mutants contain different G4 forming sequences: a parallel G4 from T. thermophila (27), an in vitro characterized antiparallel G4 (28), and two G4s (IGR1172 G4 and NGO816 G4) found in other locations in the gonococcal chromosome. Shown is a representative assay (N=3) with mean phase variation score (11) and SEM of 10 colonies. All mutants are different than RecA+ by Student’s T-test, P<0.05 (asterisk).
To directly probe for a role of the G4 structure in live gonococci, we used N-methyl mesoporphyrin IX, (NMM) (Fig. S7A-B), a compound which can enter live cells and specifically binds G4 DNA but not duplex DNA (21, 22). Gonococci grown on a concentration of NMM that did not alter growth (Fig. S7C) were assayed for pilin phase and Av. Bacterial growth on NMM showed significantly decreased pilus phase and antigenic variation (Fig. 3A-B). Surprisingly, ~70% of piliated antigenic variants arising on NMM were produced by recombination with pilS3 copy 1 which normally contributes to ~20% of variants produced from the same parental variant (Table S4) (2). These results are consistent with formation and further processing of the G4 structure being required for pilin Av.
Since homologous recombination is initiated by a nick or break, we used break site mapping (BSM) (23) to assay for G4-depedent nicks or breaks in this chromosomal region. We detected nicks throughout the pilE upstream region that were at a higher concentration per bp in the G4 forming sequence in Av bacteria (Table S5), and were in a higher density throughout the region as compared to the analogous region in pilS7 (Table S6). Occasionally, double strand breaks were also detected in the G4 forming sequence (Fig. 4A). No nicks or breaks were detected in the G-rich sequence in the Avd-1 G4 point mutant suggesting that nicks are the result of the G4 structure (Fig. 4A-B). NMM treated bacteria had a similar pattern of nicks as untreated bacteria on the top strand, but no nicks were detected on the bottom strand (Fig. 4F-G). To test whether specific proteins required for pilin Av process the pilE G4, BSM of selected mutants was performed. A recJ exonuclease mutant and recQ/rep helicase double mutant are both deficient in pilin Av. BSM of the recJ mutant showed an increase in nicks at the G4 forming sequence but all G4 localized nicks were lost in a recJ/Avd-1 mutant (Fig. 4D-E). A recQ/rep double mutant showed a slight increase in number of nicks (Fig. 4C). Thus, an intact G4 forming sequence is required to produce nicks in the G4 DNA and these nicks are processed by RecJ exonuclease and RecQ and Rep helicases. The involvement of RecQ in pilin Av (9, 11) is consistent with the established affinity of RecQ helicases for G4 structures (22, 24).
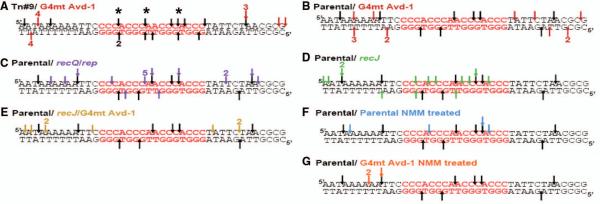
Figure 4. The pilE G4 is processed during pilin Av.
Shown is a 40bp region upstream of pilE containing the pilE G4 (red font). Arrows indicate location of nicks detected by Break Site Mapping (23) on top or bottom DNA strand with number detected in four replicate reactions from two separate cultures. Asterisks mark double strand breaks. A. Tn#9 is Av strain used in the genetic screen (black arrows) with an isogenic G4 mutant strain, Avd-1 (red arrows). B-G. Parental strain nicks (black arrows). B. G4 mutant strain Avd-1 nicks (red arrows). C. recQ/rep double mutant nicks (purple arrows). D. recJ mutant strain nicks (green arrows). E. G4 Avd-1/ recJ double mutant nicks (yellow arrows). F. NMM treated parental strain nicks (blue arrows). G. NMM treated G4 mutant strain Avd-1 nicks (orange arrows).
Our data suggest that formation of the pilE G4 structure is required for pilin Av. It is likely that the structure forms only when the DNA duplex is melted, possibly during DNA replication since the G-rich sequence is on the lagging strand. As only the pilE G4 forming sequence can mediate pilin Av, we propose that this G4 structure is a specialized recombination initiation sequence/structure (G4-RIS). The recognition that some G4 forming sequences are important for transcription (G4-TSC) and telomere maintenance (G4-TEL) suggests divergent evolution of these sequences/structures for diverse molecular processes.
Supplementary Material
Acknowledgments
We thank A. Chen and P. Schook for reading the manuscript, and the Seifert laboratory members and W. Anderson for input. This work was supported by NIH grants R37AI033493, R01AI055977, and R01AI044239 to HSS. LAC was partially supported by NIH grant T32GM08061.
References
- 1.Li X, Heyer WD. Cell Res. 2008;18:99. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Criss AK, Kline KA, Seifert HS. Mol Microbiol. 2005;58:510. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haber JE. Annu Rev Genet. 1998;32:561. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 4.Maizels N. Annu Rev Genet. 2005;39:23. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 5.Finlay BB, McFadden G. Cell. 2006;124:767. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 6.Stavnezer J, Guikema JE, Schrader CE. Annu Rev Immunol. 2008;26:261. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kline KA, Sechman EV, Skaar EP, Seifert HS. Mol Microbiol. 2003;50:3. doi: 10.1046/j.1365-2958.2003.03679.x. [DOI] [PubMed] [Google Scholar]
- 8.Kline KA, Seifert HS. J Bacteriol. 2005;187:2903. doi: 10.1128/JB.187.8.2903-2907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehr IJ, Seifert HS. Mol Microbiol. 1998;30:697. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 10.Sechman EV, Kline KA, Seifert HS. Mol Microbiol. 2006;61:185. doi: 10.1111/j.1365-2958.2006.05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sechman EV, Rohrer MS, Seifert HS. Mol Microbiol. 2005;57:468. doi: 10.1111/j.1365-2958.2005.04657.x. [DOI] [PubMed] [Google Scholar]
- 12.Skaar EP, Lazio MP, Seifert HS. J Bacteriol. 2002;184:919. doi: 10.1128/jb.184.4.919-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kline KA, Criss AK, Wallace A, Seifert HS. J Bacteriol. 2007;189:3462. doi: 10.1128/JB.01911-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanson J, Bergstrom S, Boslego J, Koomey M. Antonie Van Leeuwenhoek. 1987;53:441. doi: 10.1007/BF00415500. [DOI] [PubMed] [Google Scholar]
- 15.Kikin O, D’Antonio L, Bagga PS. Nucleic Acids Res. 2006;34:W676. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaria V, Hariharan M, Arora A, Maiti S. Nucleic Acids Res. 2006;34:W683. doi: 10.1093/nar/gkl299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muniyappa K, Anuradha S, Byers B. Mol Cell Biol. 2000;20:1361. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paeschke K, et al. Nat Struct Mol Biol. 2008;15:598. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 19.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc Natl Acad Sci U S A. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fry M. Front Biosci. 2007;12:4336. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 21.Ren J, Chaires JB. Biochemistry. 1999;38:16067. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Maizels N. Nucleic Acids Res. 2001;29:1765. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong Q, Maizels N. Nucleic Acids Res. 2001;29:E33. doi: 10.1093/nar/29.6.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber MD, Duquette ML, Shiels JC, Maizels N. J Mol Biol. 2006;358:1071. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 25.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Nucleic Acids Res. 2006;34:5402. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, et al. Nucleic Acids Res. 2008;36:1200. doi: 10.1093/nar/gkm1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schierer T, Henderson E. Biochemistry. 1994;33:2240. doi: 10.1021/bi00174a034. [DOI] [PubMed] [Google Scholar]
- 28.Rachwal PA, Brown T, Fox KR. Biochemistry. 2007;46:3036. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.