Abstract
The International HapMap Consortium has determined the linkage disequilibrium (LD) patterns of four major human populations. The aim of our investigation was to compare the LD patterns of the HapMap CEPH (Centre d’Etude du Polymorphisme Humain) samples with a family-based cohort of similar ancestry to determine its usefulness as a reference population for disease association studies. We examined four genomic regions on chromosomes 7q, 12p, and 14q totaling 14.3 Mb, initially identified in our linkage study of obesity and the metabolic syndrome. Near identical patterns of LD were detected in both populations. Furthermore, tagSNPs selected based on the HapMap CEPH cohort data capture over 98% of the variants at an r2 > 0.8 in the disease cohort. This confirms the usefulness of the CEPH cohort of the HapMap as a reference sample for further investigations into the genomic variation of populations of Northern European descent.
Keywords: CEPH, HapMap, haplotype, tagSNP
Single-nucleotide polymorphisms (SNPs) are the most common form of genetic variation, with approximately 7 million at a minor allele frequency (MAF) above 5% estimated to be present in the human genome [1]. Accordingly, they are the marker of choice for disease association studies due to their prevalence and ease of genotyping. The availability of SNPs has generated a great deal of optimism for the detection of genes underlying complex human diseases. However, the identification of a quantitative trait locus (QTL) is merely the first step down a long road of genetic discovery and can yield any number of candidate genes (not counting potential regulatory elements located in the QTL) as putative sites of causal variants. It is this identification of the causal variant(s) that remains difficult.
The realization that the genome is split into discrete blocks of high and low linkage disequilibrium (LD) [2–4] and that the majority of common genetic variation can be captured by typing a subset of SNPs [5] has improved our ability to detect potential causal variants or the haplotypes they are associated with. The HapMap project has mapped the patterns of LD across the human genome by genotyping almost 4 million SNPs in 269 DNA samples from four distinct populations: 30 trios (60 independent individuals) from the Yoruba in Nigeria, a further 30 trios (60 independent individuals) from the Centre d’Etude du Polymorphisme Humain (CEPH) collection (Utah, USA), 45 independent Han Chinese individuals, and 44 independent Japanese individuals (http://www.hapmap.org [6]). The goal now is to use this information to help determine sequence variants that affect disease and ultimately develop diagnostic tools and identify targets for therapeutic intervention [6,7]. However, caution must be exercised in the widespread application of HapMap data without first ensuring its applicability to other populations. Indeed this need to confirm patterns of LD and tagSNPs is pointed out in the original HapMap publication [7], though it need be done only in a subset of 30–40 unrelated individuals as greater numbers do not result in a significant improvement in performance [8].
There are a number of reports comparing LD patterns between populations, though these are generally restricted to a comparison of different ethnic groups in which variation would be expected due to altered population histories and geographical restrictions. De La Vega et al. [9] compared the LD patterns of four major human populations across chromosomes 6, 21, and 22. They detected over one-third more LD in out-of-Africa populations than in African Americans, while overall patterns of LD remained similar. However, the authors also indicated that a significant proportion of haplotypes within LD blocks was not shared between populations, suggesting that markers for association studies may need to be selected from individual populations. Using a denser set of SNPs to compare a 10 Mb region of chromosome 20, Evans and Cardon [10] were able to demonstrate population-specific variations suggesting an altered recombination history or other historical factors affecting recombination. However, in the same study [10] a comparison of CEPH and UK unrelated samples generated very similar results suggesting (at least for this region of chromosome 20) that populations of close ancestry will demonstrate similar patterns of recombination. This is supported by studies comparing LD patterns of individual genes between European populations [8,11]. Nejentsev et al. [11] compared the LD patterns of the vitamin D receptor gene region between four European populations and an African population. Patterns for the European populations were identical, with greater evidence for historical recombination and the breakup of LD blocks in the African sample. Mueller et al. [8] compared eight populations with the CEPH cohort over four gene regions on four chromosomes and demonstrated that while there was good general agreement in LD patterns between populations of similar ancestry, those populations potentially isolated by language and geography may have an altered recombination history. In the most recent analysis published to date, Ribas et al. [12] detected very similar patterns of LD between the HapMap CEPH cohort and a sample of the Spanish population when they analyzed LD patterns and tagSNPs across 175 cancer-related genes. Despite all these studies, it remains to be seen if the close correspondence in LD patterns observed within gene regions, between the CEPH and other populations of similar ancestry, holds true for extended regions of the genome.
A number of reports have been published of late investigating the applicability and suitability of the HapMap data as a resource for interrogating LD patterns and selecting tagSNPs [12–14]. A study of samples from cases of multiple sclerosis in Australia demonstrated that the CEPH population can be used to predict patterns of LD accurately and select tagSNPs [13]. Other studies of worldwide populations have also indicated that tagSNPs are transferable to other populations [15,16]. A 13-Mb region of chromosome 1q was investigated, and while the HapMap proved effective in selecting tagSNPs that allow the capture of the majority of common (MAF >5%) SNPs when applied to other cohorts, rarer variants were missed in the analysis. This result was also supported by Taylor et al. [17], who indicated that common haplotypes within up to 50% of genes may be missed by the use of HapMap SNP data alone. However, for single-gene studies, it is up to individual researchers to determine whether the use (and/or detection) of rare SNPs in fine-mapping efforts is warranted [14].
It was the goal of our study to examine the usefulness of the HapMap data for the detailed analysis of QTL regions in a disease study cohort. For this, we compared the genotyping data from four QTL regions identified in a cohort of northern European descent with the HapMap data for the CEPH cohort. We compared a 5-Mb region on chromosome 7, in addition to a further 9.3 Mb distributed over three regions of two chromosomes. Our data suggest that the CEPH LD data from the HapMap project do provide a good reference for studying other cohorts of similar ancestry. Near identical patterns of LD in the two cohorts were observed, and tagSNPs selected based on HapMap CEPH data were able to capture 98% of SNPs at an r2 > 0.8 in the disease study cohort.
Results
The two cohorts analyzed in this investigation were the well-studied CEPH population of the HapMap Consortium study (http://www.hapmap.org) and a subset of the Metabolic Risk Complications of Obesity Genes (MRC-OB) project cohort [18,19]. The CEPH cohort consists of 30 family trio samples (two parents and a child) collected from residents of Utah, USA, with Northern and Western European ancestry. The MRC-OB cohort was recruited from the TOPS (Take Off Pounds Sensibly, Inc.) membership and consisted of 22 family quads (two parents and two children, one child of each sex when available). These provided 120 (CEPH) and 88 (MRC-OB) independent founder chromosomes for analysis and comparison.
A total of four genomic regions were investigated and compared between the two cohorts. All regions were selected from our initial linkage analysis of the MRC-OB cohort study. A 5 Mb interval on chromosome 7q36.1–q36.3 (Table 1), split into three regions (hereafter referred to as regions A, B, and C) by two gaps in the genome sequence (153,708,283–153,808,282 and 154,544,615–154,624,614 bp: UCSC May 2004 assembly), harbors a QTL for plasma triglyceride and low-density lipoprotein (LDL) concentrations [19]. Two additional regions on chromosome 12p13–p11 are located within a QTL for high-density lipoprotein (HDL)-cholesterol [19] and are 3.7 and 2.9 Mb in size. The fourth region investigated was a 2.7 Mb area of chromosome 14q12–q13 within a QTL for plasma adiponectin levels (unpublished data).
Table 1.
Genotyped single-nucleotide polymorphism (SNP) density along the 2.5, 0.7, and 1.8 Mb regions of chromosome 7 studied in the CEPH and MRC-OB cohorts and the density of SNPs genotyped in both cohorts
| Region | Size (Mb) | CEPH |
MRC-OB |
Shared |
|||
|---|---|---|---|---|---|---|---|
| No. of SNPs | SNP density (kb/SNP) | No. of SNPs | SNP density (kb/SNP) | No. of SNPs | SNP density (kb/SNP) | ||
| A | 2.5 | 810 | 3.1 | 439 | 5.6 | 355 | 7.0 |
| B | 0.7 | 258 | 2.9 | 194 | 3.8 | 162 | 4.6 |
| C | 1.8 | 597 | 3.1 | 415 | 4.4 | 371 | 5.0 |
Different strategies were used to select SNPs for genotyping in the four regions. SNPs were selected for fine-mapping of the QTL regions, and it is the ultimate goal to compare the success of these different approaches for QTL fine-mapping. For the QTL interval on chromosome 7, SNPs were selected across the entire region regardless of the location of genes with the aim of genotyping one SNP every 5 kb. First, individual tagSNPs were selected for genotyping based on LD by applying the method of Carlson et al. [20] with an r2 cutoff of 0.8 to the data for the CEPH population of the HapMap. For regions not contained in an LD block, additional SNPs were chosen approximately every 5 kb, irrespective of measures of LD. Additionally, all reported nonsynonymous SNPs for the genes in the interval were included in the genotyping.
For the other QTL intervals investigated in our study, SNPs were selected using a gene-centric approach with an emphasis on putative functional SNPs. SNPs were selected only within the coding region of all known genes and within 5 kb of the start and stop codons. For the interval on chromosome 14, approximately five SNPs per gene were selected from the following categories (in order of priority): nonsynonymous SNPs, promoter SNPs, putative splice site-altering SNPs, and synonymous SNPs in the coding sequence. For the two regions on chromosome 12, synonymous SNPs were not included, and additional tagSNPs based on LD were selected, again using the method of Carlson et al. [20] within the genic regions. Future investigations of the results obtained in our studies will allow the examination of QTL fine-mapping approaches; however, these analyses are outside the scope of the present investigation.
For the region on chromosome 7, a total of 1048 SNPs were genotyped for each sample of the MRC-OB panel. This consisted of 439 SNPs in region A (2.5 Mb), 194 SNPs in region B (0.7 Mb), and 415 SNPs in region C (1.8 Mb). Of these, 888 had corresponding genotypes in the CEPH cohort of the HapMap: 355, 162, and 371 in regions A, B, and C, respectively (Table 1). This resulted in an overall shared SNP density of 1 SNP per 5.5 kb for the entire region. When split into regions A, B, and C the shared SNP density was 1 SNP per 7.0 kb, 4.6 kb, and 5.0 kb, respectively (Table 1).
On chromosome 12 a total of 1055 SNPs were genotyped in the MRC-OB cohort; 603 in the 3.7 Mb region of chromosome 12p13 and 452 in the 2.9 Mb region of chromosome 12p12–p11. Of these, 1023 (584 and 439 on chromosomes 12p13 and 12p12–p11, respectively) had corresponding genotypes in the CEPH cohort (Table 2).
Table 2.
Genotyped single-nucleotide polymorphism (SNP) density in the CEPH and MRC-OB cohorts and those genotyped in both cohorts, in the 3.7, 2.9, and 2.7 Mb regions of chromosomes 12p13, 12p12–p11, and 14q12–q13, respectively
| Chromosome | Population | Region size (Mb) | No. of SNPs | No. of genes | SNPs/gene |
|
|---|---|---|---|---|---|---|
| Mean (range) | Median | |||||
| 12p13 | CEPH | 3.7 | 1697 | 37 | 45.9 (4–275) | 19 |
| MRC-OB | 3.7 | 603 | 37 | 16.3 (3–95) | 9 | |
| Shared | 3.7 | 584 | 37 | 15.8 (3–92) | 8 | |
| 12p12–p11 | CEPH | 2.9 | 1229 | 26 | 47.3 (1–165) | 40 |
| MRC-OB | 2.9 | 452 | 26 | 17.4 (1–68) | 13.5 | |
| Shared | 2.9 | 439 | 26 | 16.9 (1–68) | 13 | |
| 14q12–q13 | CEPH | 2.7 | 1076 | 13 | 82.8 (1–378) | 45 |
| MRC-OB | 2.7 | 354 | 13 | 27.2 (1–181) | 6 | |
| Shared | 2.7 | 348 | 13 | 26.8 (1–180) | 5 | |
Finally, on chromosome 14, 354 SNPs were genotyped in the MRC-OB cohort; 348 of these had corresponding CEPH genotypes (Table 2).
Linkage disequilibrium maps were constructed and tagSNPs determined for all data, based on phased parental haplotypes and for datasets restricted to SNPs with a MAF greater than 5%. Average minor allele frequencies for both populations were consistent over all chromosomes investigated (average MAF 0.25); and the value increased slightly for the restricted dataset, due to the removal of SNPs with a low MAF. A MAF cutoff level of 5% reduced the number of SNPs in chromosome 7 regions A, B, and C by 54, 16, and 33 (15.2, 9.9, and 8.9%), respectively. The number of SNPs in the other regions studied were reduced by 6.8% (12p13), 3.6% (12p12–p11), and 5.5% (14q12–q13).
LD comparison
Linkage disequilibrium in the two cohorts was compared visually by contrasting the patterns generated from the genotype data of SNPs typed in both cohorts (Fig. 1). This comparison of shared SNPs between cohorts resulted in a total of 144,511 pairwise comparisons across chromosome 7 (62,835, 13,041, and 68,635 in regions A, B, and C, respectively). There were 266,377 pair-wise comparisons along chromosomes 12p13 and 12p12–p11 each. On chromosome 14, the number of pair-wise comparisons was 60,378.
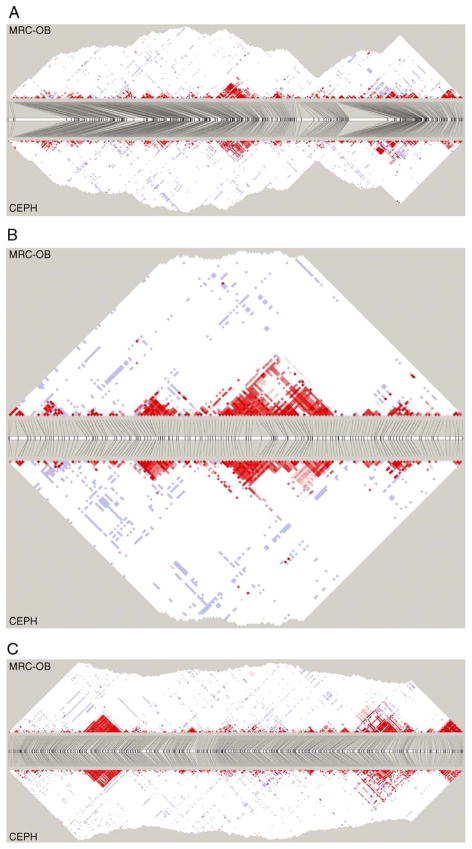
Fig. 1.
Patterns of linkage disequilibrium (LD) detected in the CEPH and MRC-OB cohorts along (A) a 2.5 Mb, (B) a 0.7 Mb, and (C) a 1.8 Mb stretch of chromosome 7q36. A, B, and C depict the regions studied around physical gaps in the genome. The horizontal white bar represents the genome, with tick marks indicating the approximate positions of genotyped SNPs. The angled lines connect the SNP positions in the genome to their location in the LD plot. Red squares indicate complete LD; shades of pink/red, some but not complete LD; blue squares, complete LD with a reduced lod score (and so confidence); and white squares, no apparent LD.
Almost identical patterns of LD were observed, with blocks of high LD interspersed by regions of little or no LD in the same areas of the genome in both groups (Supplementary Figs. S1–S3). The correspondence in patterns is striking and is a clear indicator of the genetic similarity of the cohorts.
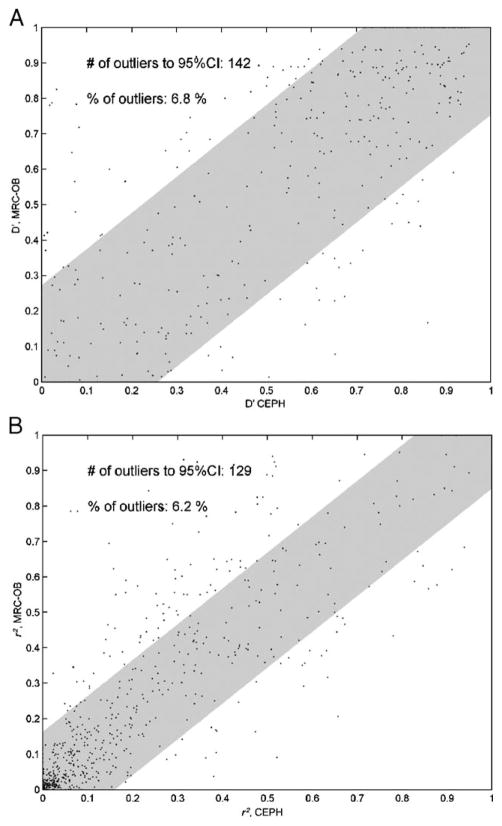
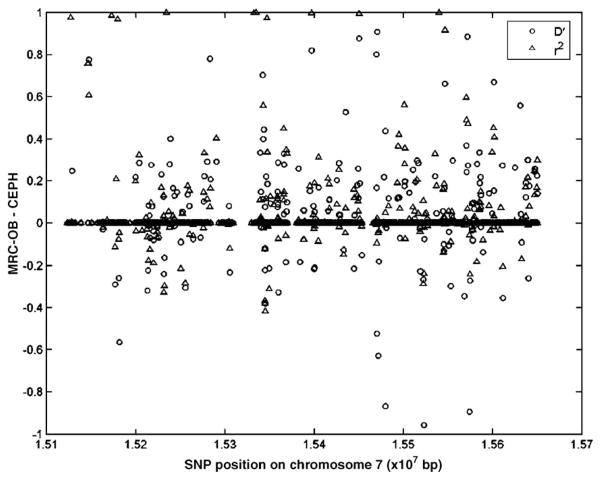
When the pair-wise measures of LD (|D′| and r2) between adjacent SNPs are compared between the two cohorts across all regions studied 6.8% (|D′|) and 6.2% (r2) fall outside the 95% confidence intervals (Fig. 2). This indicates that few SNPs compared in our study differ significantly between the two groups with respect to extent of LD. This is further supported by the high Spearman rank correlation values of r2 obtained between the two cohorts (Table 3), indicating almost identical LD patterns between the two groups. For the regions investigated on chromosomes 12 and 14 the values are greater than 0.95, whereas for regions A, B, and C on chromosome 7 the values are 0.88, 0.91, and 0.95, respectively. This observed variation in the correlation may be evidence for a (albeit small) population difference between the two groups and could be caused by population stratification. However, this result may also be influenced by the reduced shared SNP density in region A of chromosome 7 relative to the other regions. Furthermore, analysis of the variation in measures of LD (|D′| and r2) between the two groups relative to location within the regions studied (Fig. 3; Supplementary Figs. S4 and S5) demonstrates no clear pattern of localized variation, with differences distributed seemingly randomly throughout the regions investigated.
Fig. 2.
Correlation of (A) |D′| and (B) r2 between adjacent SNPs with a minor allele frequency >5%, present in all genomic regions studied, in the CEPH and MRC-OB cohorts. Each data point represents a single pair-wise comparison between adjacent SNPs; the shaded area indicates the extent of the 95% confidence interval of the mean.
Table 3.
Spearman rank correlations for r2 (|D′|) between the CEPH and the MRC-OB cohorts for the chromosomal regions indicated
| Region | 7A | 7B | 7C | 12p13 | 12p12–p11 | 14q12–q13 |
|---|---|---|---|---|---|---|
| 7A | 0.88 (0.70) | |||||
| 7B | 0.91 (0.72) | |||||
| 7C | 0.95 (0.72) | |||||
| 12p13 | 0.97 (0.73) | |||||
| 12p12–p11 | 0.99 (0.73) | |||||
| 14q12–q13 | 0.99 (0.81) |
Fig. 3.
Variation in the measures of linkage disequilibrium, |D′| (circles) and r2 (triangles), of adjacent SNPs, between the CEPH and the MRC-OB cohorts, relative to chromosomal position along the 5-Mb region of chromosome 7q36 studied.
TagSNP analysis
The use of tagSNPs for both haplotype and whole genome tagging and the choice of selection algorithm are currently the subject of intense debate [20,21]. While an analysis of the relative merits of different strategies was outside the scope of this investigation; we used a popular and freely available algorithm (Tagger [21]) to select tagSNPs for our QTL intervals. TagSNPs were determined based on the LD patterns of the subset of shared SNPs in the CEPH cohort. These were selected for all SNPs studied and also for a set restricted to a MAF >5% (Table 4). We are merely using the concordance of tagSNPs across the two sample cohorts as a measure of LD and haplotype similarity and it is clear that other algorithms would select different subsets of SNPs as tagSNPs. Our analysis did not intend to identify the optimal set of tagSNPs across the QTL intervals.
Table 4.
Number of single-nucleotide polymorphisms (SNPs) selected as tagSNPs in the CEPH cohort and the percentage of SNPs captured in the MRC-OB cohort for the three regions of chromosome 7 studied
| Region | MAF | No. of SNPs | No. (%) of tagSNPs | % captured in MRC-OB a |
|---|---|---|---|---|
| A | 0.00 | 355 | 292 (82.3) | 98.6 |
| 0.05 | 301 | 238 (79.1) | 98.3 | |
| B | 0.00 | 162 | 122 (75.3) | 99.4 |
| 0.05 | 146 | 109 (74.7) | 96.3 | |
| C | 0.00 | 371 | 278 (74.9) | 99.7 |
| 0.05 | 338 | 253 (74.9) | 97.3 |
Percentage of SNPs captured in the MRC-OB cohort at an r2 > 0.8 when using the tagSNPs selected from the CEPH cohort.
The proportion of tagSNPs selected remained relatively high (>74%) across all regions investigated on chromosome 7 and was greater than 92% on chromosomes 12 and 14 (Supplementary Table ST4). This high rate of tagSNP selection is probably due to the initial SNP selection strategies, which used patterns of LD and an alternative tagSNP selection algorithm to determine the SNPs for investigation. The increased rate on chromosomes 12 and 14 is also likely to be influenced by the selection of SNPs only within genes and an increased emphasis on putative functional and rare SNPs.
Next, we determined the number of loci that would have been captured in the MRC-OB cohort at an r2 of 0.8 or higher had only the identified tagSNPs been genotyped. The tagSNPs identified from the CEPH cohort proved to be an efficient selection, as had just this subset of SNPs been genotyped in the MRC-OB cohort, they would have captured ~98% of the variability detected, at an r2 > 0.8 (Table 4). This proportion of sites captured was consistent across all regions investigated, demonstrating the potential benefit of selecting tagSNPs to reduce genotyping burden and maintain the levels of information generated.
Overall, the concordance between both the LD patterns and the tagSNPs selected is encouraging and would suggest that SNPs selected from the HapMap data would provide useful information in the MRC-OB cohort.
Discussion
There are few reports available comparing the LD patterns of the CEPH population used in the HapMap with other Caucasian cohorts. Rather, comparisons are more commonly made between populations from distinct geographical regions, which may reasonably be expected to have altered histories and resulting recombination patterns. However, one of the major goals of the HapMap project was to provide SNP LD data that could aid in the design of studies into the genetic causes of disease. For this, patterns detected by the HapMap must be confirmed in independent populations before they are used extensively as the “gold standard” [7]. This has been done for regions on chromosomes 1q, 4q, 6p, 10q, and 20q [8,10] in selected population cohorts, and it is likely that as more regions are investigated further data will emerge on this aspect of study.
In our study the close correspondence of LD patterns over all regions investigated between the two groups is striking, suggesting that the CEPH data indeed are useful as a reference dataset for SNP selection in our MRC-OB cohort and possibly in other populations of northern European descent. This compares well with the overall results of Evans and Cardon [10], who examined a 10 Mb stretch of chromosome 20q and demonstrated a high correlation between CEPH and UK unrelated samples. However, these authors also indicated that while measures of r2 were closely matched between the two populations, |D′| values were more variable [10]. This discrepancy in LD measures may be due to variation in the MAFs of individual SNPs [22]. The greater correspondence of |D′| values detected between the CEPH samples and our MRC-OB cohort therefore indicates not only similar patterns of LD but also a greater similarity between allele frequencies at the loci investigated. This suggests that while the MRC-OB families were selected through the initial identification of an affected obese individual, there is little or no population stratification affecting overall LD patterns. The potential variation between U.S. individuals of northern European ancestry and Europeans has been demonstrated previously [23]. These studies suggested that the European haplotype blocks may have been broken up in U.S. samples due to recent admixture with other populations. Mueller et al. [8] compared a total of 749 kb, distributed over four chromosomal regions, of the CEPH population of the HapMap to eight other European populations. They determined that LD block structure, defined on the basis of a standard algorithm [3], was similar in some cohorts, but for isolated and peripheral populations the CEPH data were less informative. Other studies of LD in European populations have consistently reported very little variation between individuals [11,24].
A further use of the HapMap is as a means of selecting tagSNPs for interrogating genomic regions in other populations, potentially reducing the amount of genotyping required.
Previous studies into the transferability of tagSNPs have yielded conflicting results, though this is likely to be related to the populations and genomic regions investigated. Carlson et al. [20], studying Europeans and African Americans, determined that tagSNPs should be selected separately for populations with different ancestries. This agrees with Nejentsev et al. [11], who indicated that tagSNPs were transferable between European populations, but not between Europeans and Gambians. However, Mueller et al. [8] demonstrated that while tagSNPs determined from the CEPH cohort performed well when used to interrogate three gene regions in Europeans, local samples were more effective at a fourth locus, suggesting some loci are conserved within subpopulations.
In the present study, over 80% of sites selected as tagSNPs from the CEPH cohort were able to capture the vast majority of SNPs in the MRC-OB cohort. This supports the use of the CEPH cohort as a reference population for studies attempting to interrogate disease-associated QTL.
To summarize, we have confirmed the patterns of LD present in the CEPH population of the HapMap in areas of chromosomes 7q, 12p, and 14q totaling 14.3 Mb, in an independent population of northern European descent. The LD patterns and identified tagSNPs are highly similar for all regions studied. While it remains to be seen whether this similarity can be confirmed for other study cohorts, our study confirms the usefulness of the CEPH population of the HapMap for selecting SNPs in disease study cohorts of similar ancestry, at least for the regions examined on chromosomes 7q, 12p, and 14q.
Materials and methods
Study populations
We have based our analyses on two populations genotyped on a corresponding set of SNPs. The first is the CEPH population genotyped by the International HapMap Consortium [6,7] and the data for 2259 markers downloaded from the HapMap Web site (http://www.hapmap.org; HapMap Public Release 16c.1); this included 30 family trios with 120 independent founder chromosomes. The second sample was a subset of the MRC-OB project cohort recruited from the TOPS membership as described previously [18,19]. Briefly, families with at least two obese siblings (body mass index (BMI) >30), the availability of one (preferably both) parent, and one or more never obese sibling (BMI <27) were recruited from the TOPS membership in 10 Midwestern U.S. states. A questionnaire gathered health information from all individuals, and phenotypic measurements included BMI, waist and hip circumferences, fasting plasma glucose levels, insulin and insulin:glucose ratio, cholesterol, LDL-cholesterol, HDL-cholesterol, and plasma triglyceride levels. From the initial population of 2209 individuals distributed over 507 Caucasian families of predominantly northern European ancestry, 22 family quads comprising both parents and two offspring (one of each gender when available) were selected without consideration of obesity or other phenotypes, giving a total of 88 independent founder chromosomes. All protocols have been approved by the Institutional Review Board of the Medical College of Wisconsin.
SNP selection
We selected four genomic regions for analysis based on previously identified QTL for traits characteristic of the metabolic syndrome. Different SNP selection strategies were used for each region.
Chromosome 7
SNPs were selected based on the LD patterns of the CEPH population genotyped as part of the HapMap project. Individual tagSNPs were chosen by applying the tagging algorithm of Carlson et al. [20] with an r2 cutoff of 0.8 to the CEPH data. In addition, SNPs were selected every 5 kb in regions not contained in an LD block, independent of any measures of LD. Also, all nonsynonymous SNPs in the region were included in the list of SNPs to be genotyped. The aim was to genotype one SNP every 5 kb.
Chromosome 12
In contrast to chromosome 7, the SNPs selected in the two regions on chromosome 12 were located only within known genes and ±5 kb of the start and stop codons. Nonsynonymous SNPs, promoter SNPs, and putative splice site-altering SNPs were selected. In addition, LD data derived from the CEPH population of the HapMap were used as the basis for selecting tagSNPs in the genic regions using the algorithm of Carlson et al. [20].
Chromosome 14
For this interval, SNPs were selected using a gene-centric approach with an emphasis on putative functional SNPs. SNPs were again selected within the coding region of all known genes and within 5 kb of the start and stop codons. Approximately five SNPs per gene were selected from the following categories (in order of priority): nonsynonymous SNPs, promoter SNPs, putative splice site-altering SNPs, and synonymous SNPs in the coding sequence.
Genotyping procedures
Genomic DNA was isolated from whole blood using commercial kits (Puregene, Gentra Systems, Minneapolis, MN, USA) and normalized to 150 ng μl−1 for genotyping on a custom-designed 3K GeneChip Universal Tag Array with the Affymetrix GeneChip Scanner 3000 7G MegAllele System (Affymetrix, Santa Clara, CA, USA) based on Molecular Inversion Probe technology [25,26] (ParAllele Bioscience, Inc.), as recommended by the manufacturer. Briefly, probes are hybridized to genomic DNA flanking the location of a SNP, an enzymatic gap-fill reaction circularizes the probes in an allele-specific manner, and the probes are then separated from unreacted and cross-reacted probes by an exonuclease reaction. The circularized (or “padlocked”) probes are inverted and amplified by PCR and an allele-specific label is added; samples are then hybridized to GeneChip Universal Tag Arrays and scanned four times (once for each allele: A, C, G, T). Samples were processed at the rate of 48/day with one positive and one negative control included in every 48 samples. Data were processed using the GeneChip Operating System version 1.3.0.031 (Affymetrix) and genotypes determined by using the Cluster Fit function of TrueCall Analyzer version 7.0.0.22 (ParAllele BioScience, Inc., Santa Clara, CA, USA).
Data analysis
Pedigree information and genotypes for the CEPH cohort were downloaded from the HapMap home page (http://www.hapmap.org), and genotypes for the MRC-OB cohort were exported from TrueCall Analyzer version 7.0.0.22 (ParAllele BioScience); data files were transformed to linkage format for further analysis. Data were initially checked for relationship and genotyping errors using PedCheck version 1.1 [27], errors were corrected by removing all genotypes for a family at a discrepant SNP as it was not possible to determine unambiguously which individual genotype was incorrect. Parental haplotypes were determined using the haplotype function of GeneHunter version 2.1_r6 (http://www.fhcrc.org/science/labs/kruglyak/Downloads/) and analyzed further using Haploview version 3.2 [28]. TagSNPs were determined using Tagger [21], implemented in Haploview version 3.2 [28]. Aggressive tagging using two- and three-marker haplotypes with a lod threshold of 3.0 was performed. These parameters were used to maintain a high tagging efficiency with minimal loss of power and ensure that selected tagSNPs were in strong LD with all predicted alleles.
To investigate the variability associated with fine-scale measures of LD between the two populations, we calculated pair-wise LD measures (|D′| and r2) between adjacent markers in each population and the Pearson correlation between the two populations.
Supplementary Material
Acknowledgments
This work was supported by Grants HL74168 (M.O.) and DK065598 (A.H.K.) from the National Institutes of Health TOPS, Inc., provided funds for establishing the family database. The authors also express their gratitude to Affymetrix for excellent technical support and assistance throughout and to Biljana Stojsavljevic (MCW) and Claire Beste (MCW) for assistance and support with genotyping.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ygeno.2006.04.004.
References
- 1.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 2.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 3.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 4.Patil N, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 5.Johnson GC, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 6.Altshuler D, et al. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 8.Mueller JC, et al. Linkage disequilibrium patterns and tagSNP transferability among European populations. Am J Hum Genet. 2005;76:387–398. doi: 10.1086/427925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La Vega FM, et al. The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res. 2005;15:454–462. doi: 10.1101/gr.3241705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans DM, Cardon LR. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am J Hum Genet. 2005;76:681–687. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, et al. Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum Mol Genet. 2004;13:1633–1639. doi: 10.1093/hmg/ddh169. [DOI] [PubMed] [Google Scholar]
- 12.Ribas G, et al. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet. 2006;118:669–679. doi: 10.1007/s00439-005-0094-9. [DOI] [PubMed] [Google Scholar]
- 13.Stankovich J, et al. On the utility of data from the International HapMap Project for Australian association studies. Hum Genet. 2006;119:220–222. doi: 10.1007/s00439-005-0120-y. [DOI] [PubMed] [Google Scholar]
- 14.Zeggini E, et al. An evaluation of HapMap sample size and tagging SNP performance in large-scale empirical and simulated data sets. Nat Genet. 2005;37:1320–1322. doi: 10.1038/ng1670. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Neira A, et al. The portability of tagSNPs across populations: a worldwide survey. Genome Res. 2006;16:323–330. doi: 10.1101/gr.4138406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer CJ, et al. Tag SNP selection for Finnish individuals based on the CEPH Utah HapMap database. Genet Epidemiol. 2006;30:180–190. doi: 10.1002/gepi.20131. [DOI] [PubMed] [Google Scholar]
- 17.Taylor JA, Xu ZL, Kaplan NL, Morris RW. How well do HapMap haplotypes identify common haplotypes of genes? A comparison with haplotypes of 334 genes resequenced in the environmental genome project. Cancer Epidemiol Biomarkers Prev. 2006;15:133–137. doi: 10.1158/1055-9965.EPI-05-0641. [DOI] [PubMed] [Google Scholar]
- 18.Kissebah AH, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci U S A. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnenberg GE, et al. Genetic determinants of obesity-related lipid traits. J Lipid Res. 2004;45:610–615. doi: 10.1194/jlr.M300474-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bakker PI, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 22.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenzel A, et al. Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet. 2004;114:377–385. doi: 10.1007/s00439-003-1075-5. [DOI] [PubMed] [Google Scholar]
- 24.Ng MC, et al. Ethnic differences in the linkage disequilibrium and distribution of single-nucleotide polymorphisms in 35 candidate genes for cardiovascular diseases. Genomics. 2004;83:559–565. doi: 10.1016/j.ygeno.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 26.Hardenbol P, et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.