Abstract
The microphthalmia-associated transcription factor (MITF) promotes melanocyte differentiation and cell cycle arrest. Paradoxically, MITF also promotes melanoma survival and proliferation, acting like a lineage survival oncogene. Thus, it is critically important to understand the mechanisms that regulate MITF activity in melanoma cells. SWI/SNF chromatin remodeling enzymes are multiprotein complexes composed of one of two related ATPases, BRG1 or BRM, and 9-12 associated factors (BAFs). We previously determined that BRG1 interacts with MITF to promote melanocyte differentiation. However, it was unclear whether SWI/SNF enzymes regulate the expression of different classes of MITF target genes in melanoma. In this study, we characterized SWI/SNF subunit expression in melanoma cells and observed down-regulation of BRG1 or BRM, but not concomitant loss of both ATPases. Re-introduction of BRG1 in BRG1 deficient SK-MEL5 cells enhanced expression of differentiation specific MITF target genes and resistance to cisplatin. Down-regulation of the single ATPase, BRM, in SK-MEL5 cells inhibited expression of both differentiation specific and pro-proliferative MITF target genes and inhibited tumorigenicity in vitro. Our data suggest that heterogeneous SWI/SNF complexes composed of either the BRG1 or BRM subunit promote expression of distinct and overlapping MITF target genes and that at least one ATPase is required for melanoma tumorigenicity.
Introduction
Microphthalmia-associated transcription factor (MITF), the master regulator of melanocyte differentiation, promotes melanocyte lineage survival and plays a key role in melanoma progression. MITF is a basic helix loop helix leucine zipper transcripton factor that binds to a conserved M box in the promoters of tyrosinase and other melanocyte specific genes that regulate melanin synthesis and melanosome structure (Hemesath et al., 1994). MITF also promotes expression of genes that regulate melanoma proliferation and survival and has been termed a “lineage survival oncogene,” being amplified in 10-20% of human melanomas (Garraway et al., 2005).
The role that MITF plays in melanoma has been controversial. MITF can inhibit proliferation by directly activating expression of the cyclin dependent kinase inhibitors, p21CIP1 and p16INK4A (Carreira et al., 2005; Loercher et al., 2005). MITF can also promote survival and proliferation by activating anti-apoptotic factors, BCL2 and MLIAP (Dynek et al., 2008; McGill et al., 2002), TBX2 (Carreira et al., 2000), and the cell cycle dependent kinase, CDK2 (Du et al., 2004). It has been proposed that the levels of MITF determine whether MITF promotes differentiation and cell cycle arrest or proliferation (Carreira et al., 2006). In addition, signaling pathways and interactions with other regulatory proteins may regulate MITF activity. We have previously determined that MITF interacts with SWI/SNF chromatin remodeling enzymes to activate differentiation specific genes (de la Serna et al., 2006a).
SWI/SNF enzymes are ATP dependent multisubunit complexes that disrupt histone-DNA contacts and promote chromatin structural changes (Sif, 2004). Mammalian SWI/SNF complexes are composed of the BRG1 or BRM catalytic ATPase subunit and 9-12 BRG1/BRM associated factors (BAFs). BRG1 and BRM have overlapping functions but are not always interchangeable and can interact with distinct transcription factors in vitro (Kadam et al., 2000). Heterogeneous SWI/SNF complexes, generated by the alternative presence of either the BRG1 or BRM ATPase as well as by a specific BAF composition, may have distinct functions. For example, BRG1 and BRM can differentially regulate gene expression (Flowers et al., 2009; Xu et al., 2007) and SWI/SNF complexes containing either the BAF250A or BAF250B subunit have opposing roles in cell cycle regulation (Nagl et al., 2007). Thus, SWI/SNF subunit composition is an important determinant of SWI/SNF specificity.
BRG1 and BRM and other components of the SWI/SNF complex have been implicated in cancer. BRG1 or BRM expression is down-regulated in a wide array of human cancers. Loss of both BRM and BRG1 expression correlates with poor prognosis of non-small cell lung cancer (Fukuoka et al., 2004; Reisman et al., 2003). Other SWI/SNF components are mutated, deleted, or not expressed in many human cancer cell lines (Decristofaro et al., 2001). Inactivating mutations in the INI1 subunit occur in the majority of malignant rhabdoid tumors (Versteege et al., 1998). Re-introduction of SWI/SNF subunits into cancer cells that lack expression induces a flat cell morphology, cell cycle arrest, apoptosis or senescence, with reversion of the transformed phenotype, indicating that loss of SWI/SNF function contributes to tumorigenicity (Dunaief et al., 1994; Wang et al., 2005). Conversely, components of the SWI/SNF complex interact with the androgen receptor to promote androgen dependent prostate cancer proliferation (Link et al., 2008; Link et al., 2005). Thus, the role that SWI/SNF plays in cancer is complex and dependent on cellular context and potentially on subunit composition.
SWI/SNF enzymes are important regulators of cellular differentiation (de la Serna et al., 2006b). In normal cells, key regulatory factors interact with SWI/SNF enzymes to promote myogenic, myeloid, erythropoietic, adipogenic, and neuronal differentiation. SWI/SNF enzymes can also promote differentiation of cancer cells. Introduction of the BRM subunit into gastric cancer cells that are deficient in BRM promotes expression of markers of intestinal differentiation and introduction of the INI1 subunit into malignant rhabdoid tumor cells differentiates these cells along the adipogenic pathway (Caramel et al., 2008; Yamamichi et al., 2007).
The status and activity of SWI/SNF components have been characterized in many cancer types, but poorly characterized in human melanoma. A recent report indicates that BRG1 is down-regulated in primary and metastatic melanomas (Becker et al., 2009). Moreover, it has recently been reported that down-regulation of the INI1 subunit of the SWI/SNF complex is important for bypassing oncogenic BRAF induced senescence in melanocytes and that the BRM subunit is associated with heterochromatic foci in senescing melanocytes (Bandyopadhyay et al., 2007; Wajapeyee et al., 2008). Because our previous work indicated that SWI/SNF enzymes are required to promote MITF mediated activation of melanocyte specific gene expression (de la Serna et al., 2006a), we hypothesized that SWI/SNF enzymes are also critical regulators of melanoma differentiation and tumorigenicity.
We previously determined that in a tissue culture model of melanocyte differentiation, MITF promotes recruitment of BRG1 to the promoter of a melanocyte specific gene, chromatin remodeling, and activation of melanocyte specific gene expression (de la Serna et al., 2006a). To determine the ramifications of the previously observed SWI/SNF/MITF interaction in melanoma, we characterized SWI/SNF subunit expression in normal human melanocytes and in established human melanoma cell lines. We found that normal melanocytes express high levels of all SWI/SNF subunits, including both the BRG1 and BRM catalytic subunits whereas several melanoma cell lines were markedly down-regulated in either BRG1 or BRM.
To characterize the role of SWI/SNF enzymes in the regulation of MITF target gene expression in melanoma and to distinguish between the roles played by BRG1 and BRM, we expressed BRG1 in BRG1 deficient SK-MEL5 cells. We found that expression of BRG1 enhanced melanoma differentiation and increased expression of MITF target genes that regulate pigmentation. Down-regulation of BRM in BRG1 deficient SK-MEL5 cells inhibited expression of many MITF target genes and reduced growth on soft agar. Our data indicate that both BRG1 and BRM containing SWI/SNF complexes interact with MITF to promote optimal expression of a subset of overlapping and distinct MITF target genes and that a functional SWI/SNF ATPase is required for aspects of melanoma tumorigenicity in vitro. Thus, we propose that SWI/SNF enzymes through their partnership with MITF are important epigenetic modulators in melanoma cells.
Results
BRG1 and BRM are not concomitantly down-regulated in melanoma cells
The loss of both BRG1 and BRM has been observed in some cancer cell lines (Decristofaro et al., 2001) while only BRG1 or BRM is down-regulated in the pancreatic cancer cell line, MiaPaCa2, and in gastric cancer (Rosson et al., 2005; Yamamichi et al., 2007). It was recently shown that BRG1 is downregulated in a subset of primary melanomas (Becker et al., 2009), however, this study did not address whether melanomas are concomitantly down-regulated for both the BRG1 or BRM ATPases nor did it evaluate the status of other SWI/SNF subunits.
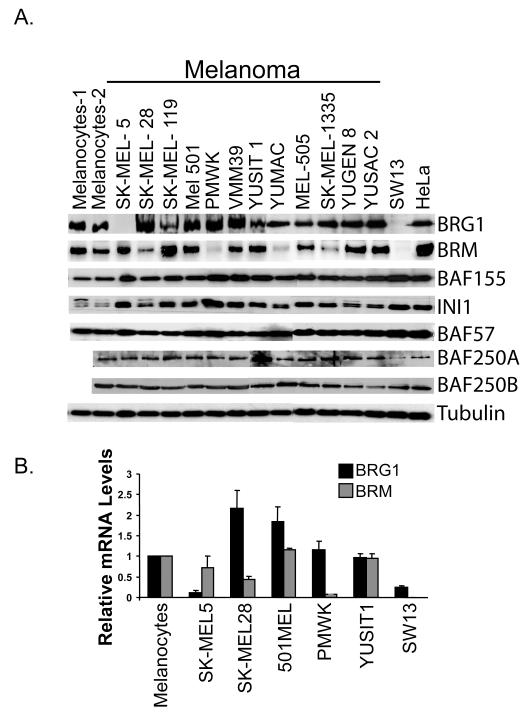
We characterized the expression of BRG1 and BRM and the expression of other SWI/SNF proteins in normal human melanocytes and established human melanoma cell lines. We found that melanocytes expressed high levels of both BRG1 and BRM when compared to HeLa cells (known to express high levels of many SWI/SNF components) and SW13 cells (known to be deficient in both BRG1 and BRM) but that BRG1 and BRM expression was variable in a number of melanoma cell lines (Fig. 1A). SK-MEL5 cells, isolated from an axillary node metastasis (Pollack et al., 1981), did not express BRG1 but had levels of BRM comparable to melanocytes. PMWK, derived from a primary melanoma (Vink et al., 1993), and YUMAC, derived from metastatic melanoma (Hoek et al., 2004), had virtually undetectable levels of BRM but expressed high levels of BRG1 (Fig. 1A). All the melanoma cells expressed high levels of several other SWI/SNF components, including the core subunits, INI1 and BAF155. Therefore, if the expressed SWI/SNF components are functional, then overall SWI/SNF activity in a subset of melanoma cells may be compromised by down-regulation of one SWI/SNF ATPase but not completely eliminated because the alternative ATPase is retained and the other core SWI/SNF components are also highly expressed.
Figure. 1.
A. Detection by Western blotting of SWI/SNF subunit expression in melanocytes from two different donors, eight human melanoma cell lines, SW13, and HeLa cells. Tubulin is a loading control. B. Detection of BRG1 and BRM mRNA levels by real time RT-PCR. mRNA levels were standardized to 18S rRNA and expressed as values relative to those in human melanocytes.
In SK-MEL5 cells, BRG1 is down-regulated at the transcriptional level
Expression of BRG1 at the protein level was consistently undetectable in SK-MEL5 cells even when cultured under many different serum conditions and at different confluencies (data not shown). To determine if BRG1 is downregulated at the transcriptional level in these cells, we compared expression of BRG1 mRNA in SK-MEL5 cells with expression in normal human melanocytes and in other melanoma cells (Fig.1B). SK-MEL5 cells expressed significantly lower levels of BRG1 mRNA than melanocytes and that there was not a compensatory increase in BRM expression. The level of BRG1 mRNA in SK-MEL5 cells was similar to that in BRG1 deficient SW13 cells (Fig. 1B).
Ectopically expressed BRG1 localizes to the nucleus and assembles into a SWI/SNF complex that includes MITF
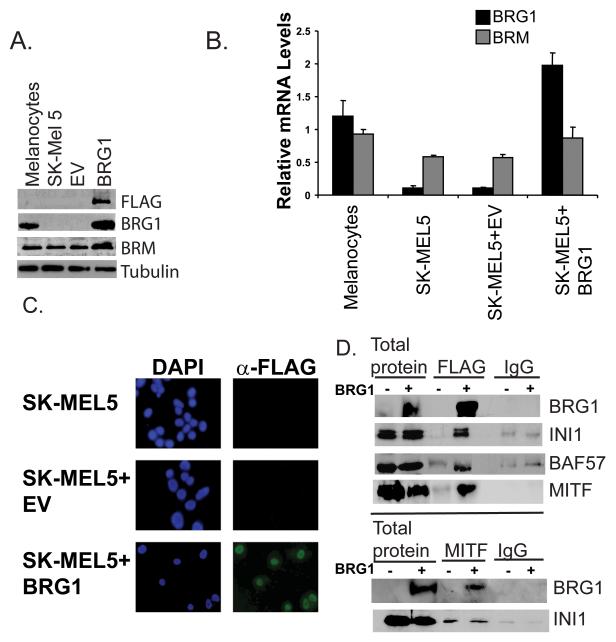
Re-introduction of SWI/SNF components into cancer cells that lack expression has been demonstrated to result in cell cycle arrest, apoptosis, or differentiation, depending on the cancer cell type and the identity of the missing SWI/SNF subunit (Caramel et al., 2008; Dunaief et al., 1994; Wang et al., 2005; Yamamichi et al., 2007). To determine how BRG1 regulates the melanoma phenotype, we expressed epitope tagged BRG1 into BRG1 deficient SK-MEL5 cells by retroviral infection and selected a pool of cells. We obtained expression levels of BRG1 that were approximately twice that present in normal melanocytes (Fig. 2A and B). SK-Mel28 melanoma cells express BRG1 at approximately twice the levels present in melanocytes (Fig. 1A and B), thus the level of BRG1 expression that we obtained in SK-MEL5 cells is within a physiologically relevant range for melanocytic cells. We found that the selected cells grew slower (50% over a 6 day period) (data not shown) but could be propagated without loss of BRG1 expression (data not shown).
Figure 2.
A. Detection by Western blotting of ectopically expressed FLAG-BRG1 protein in BRG1 deficient SK-MEL5, SK-MEL5+empty vector (EV), or SK-MEL5+BRG1. FLAG-tagged BRG1 was detected with an antibody to the FLAG epitope and an antibody to BRG1. BRM levels in BRG1 deficient and BRG1 expressing cells were also probed. Tubulin is a loading control. B. Detection of ectopically expressed FLAG-BRG1 and BRM mRNA levels by real-time RT-PCR. BRG1 and BRM mRNA levels were standardized to 18S rRNA and are shown relative to those in melanocytes. C. Detection of FLAG-BRG1 by immunocytochemistry using a FLAG antibody. D. Coimmunoprecipitation of SWI/SNF components and MITF in BRG1 deficient and BRG1 expressing SK-MEL5 cells. Top panel: Cell lysates were immunoprecipitated with M2-FLAG agarose or control mouse IgG and blotted with the indicated antibodies. Bottom panel: Cell lysates were immunoprecipitated with MITF hybridoma supernatant or control mouse IgG and blotted with the indicated rabbit antibodies.
We did not see a compensatory decrease in BRM levels as a result of BRG1 over-expression. Interestingly, we observed a small increase in BRM mRNA and protein, suggesting that the BRG1 expressing SK-MEL5 cells had increased levels of functional SWI/SNF complexes containing both of the catalytic subunits (Fig. 2A and B). Crosstalk between different subunits of the SWI/SNF complex has previously been reported but the mechanisms are not completely understood (Chen and Archer, 2005; Kang et al., 2004). Moreover, BRM levels have been shown to increase during differentiation (Machida et al., 2001). The increased levels of BRM that we observed correlated with a more differentiated phenotype (see below).
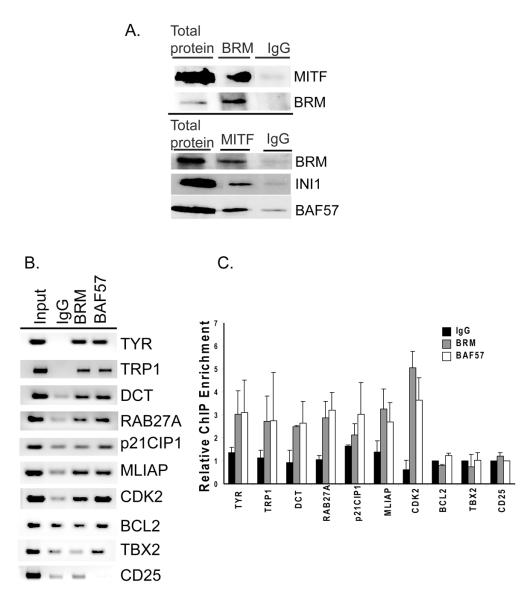
We detected epitope tagged BRG1 in approximately 100% of the selected cells and found that BRG1 localized to the nucleus (Fig. 2C). Furthermore, ectopically expressed BRG1 interacted with other SWI/SNF subunits to form potentially active SWI/SNF complexes that also contained MITF (Fig.2D). This indicated that the established SWI/SNF complexes containing the BRG1 ATPase might act as co-regulators of MITF activity in these melanoma cells.
Expression of BRG1 in SK-MEL5 cells increases pigmentation and resistance to cisplatin
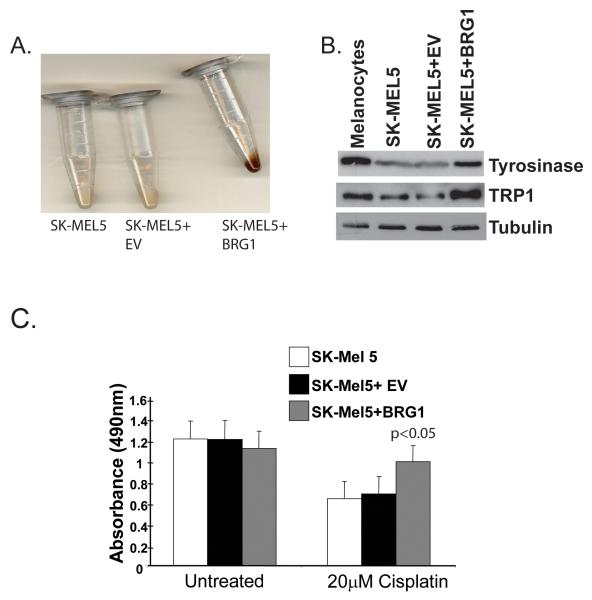
Melanoma cells are often less differentiated than their normal counterparts (Eberle et al., 1995). SK-MEL5 cells retain MITF expression but are amelanotic, suggesting that differentiation specific gene expression is suppressed downstream of MITF (note MITF expression, Fig. 2D). We found that ectopic expression of BRG1 in these cells led to a noticeable increase in pigmentation (Fig. 3A). Consistently, we detected increased expression of tyrosinase and tyrosinase related protein 1 (TRP1), two enzymes important for melanin synthesis (Fig. 3B). These results suggest that BRG1 can promote melanoma differentiation by activating MITF target gene expression. Over-expression of BRM to approximately four times the endogenous levels (supple. Fig. 1A, B, and C) did not increase visible pigmentation nor did it activate expression of most MITF target genes (supple. Fig. 1D and 1E), suggesting that when expressed at these levels, BRM cannot compensate for BRG1 in promoting differentiation of SK-MEL5 melanoma cells.
Figure 3.
A. SK-MEL5, SK-MEL5+empty vector (EV), SK-MEL5+BRG1 cell pellets. Cells were grown to 90% confluency, pelleted, and photographed. B. Detection by Western blotting of tyrosinase and TRP1 in human melanocytes, SK-MEL5, SKMEL5+empty vector (EV), and SK-MEL5+BRG1. Tubulin is a loading control. C. Sensitivity of BRG1 deficient and BRG1 expressing melanoma cells to cisplatin. SK-MEL5, SK-MEL5+empty vector (EV), SK-MEL5+BRG1 cells were treated with 20 μM cisplatin for 72 h and assayed. These results were analyzed by Student’s t test and are representative of two independent experiments performed in triplicate.
MITF is a lineage survival oncogene that is amplified in 10%-20% of human melanoma cells and can promote melanoma chemoresistance (Garraway et al., 2005). The extent of melanoma differentiation can have an impact on chemoresistance because melanin acts as a sink that sequesters cytototoxic compounds (Chen et al., 2006; Chu et al., 2000; Pak et al., 2000; Svensson et al., 2003). The MITF gene is not amplified in SK-MEL5 cells and these cells exhibit intermediate sensitivity to the alkylating agent, cisplatin (Garraway et al., 2005; Shen et al., 2007). Because ectopically expressed BRG1 interacted with MITF and led to an increase in pigmentation (Fig. 3A), we tested whether BRG1 could promote melanoma chemoresistance. We treated control and BRG1 expressing SK-MEL5 cells with cisplatin and found that expression of BRG1 significantly increased resistance as measured by a cell survival assay (Fig. 3C). Thus, consistent with previous studies (Chen et al., 2006; Chu et al., 2000; Svensson et al., 2003), we have correlated the status of melanoma differentiation with resistance to chemotherapeutic agents. Furthermore, our data is consistent with reports in other cancer types that have shown an increase in resistance to DNA damage upon re-introduction of BRG1 (Gong et al., 2008; Park et al., 2009). These data suggest that the status of SWI/SNF enzymes in melanoma cells can have a significant impact on chemoresistance, an important issue in developing therapeutics against melanoma.
BRG1 is recruited to the promoters of differentiation specific MITF target genes and activates their expression
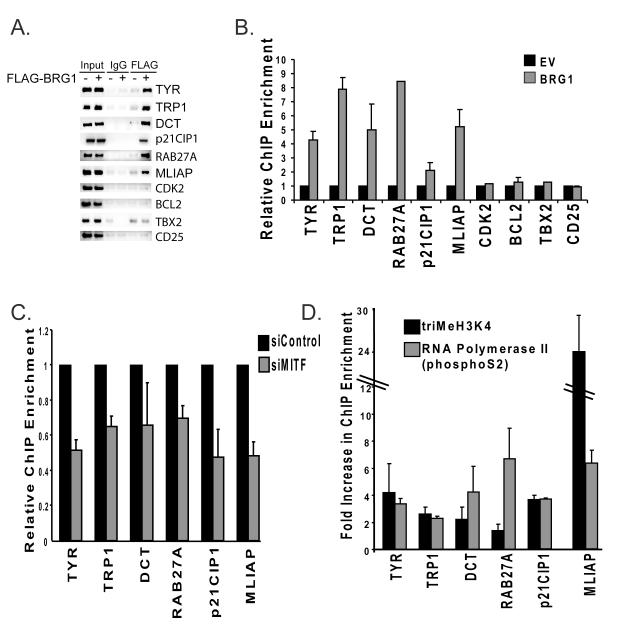
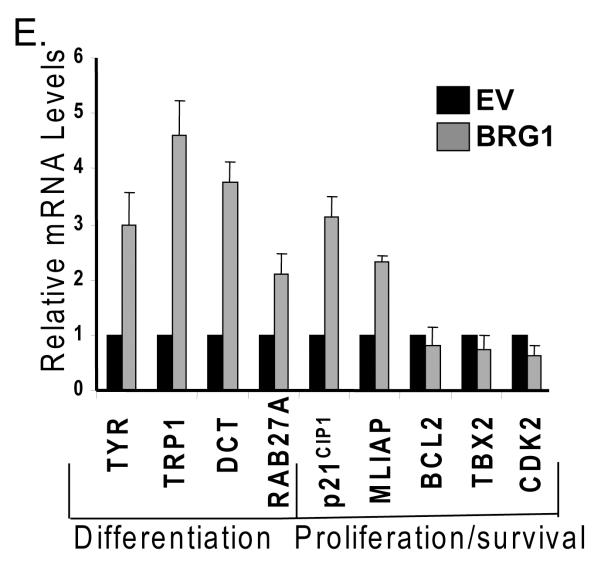
We performed chromatin immunoprecipitations (ChIPs) to determine whether ectopically expressed BRG1 in SK-MEL5 cells is recruited to different classes of MITF target genes that regulate differentiation, proliferation, and survival. We detected BRG1 on the promoters of genes that encode enzymes needed for melanin synthesis, tyrosinase, tyrosinase related protein 1 (TRP1), and dopachrome tautomerase (DCT) (Fig.4A and B). We also detected BRG1 on the RAB27A promoter. RAB27A is a recently identified MITF target gene that regulates peripheral transport of mature melanosomes (Chiaverini et al., 2008). We also detected BRG1 on the p21CIP1 promoter, an anti-proliferative MITF target gene, associated with increased melanocyte differentiation (Carreira et al., 2005). Thus, BRG1 was recruited to multiple promoters that regulate genes important for pigmentation and overall differentiation status.
Figure 4.
A. Chromatin immunoprecipitations (ChIPs) were performed in BRG1 deficient SK-MEL5+empty vector (EV) or SK-MEL5+BRG1 cells using a FLAG antibody or control IgG, and run on an agarose gel stained with ethidium bromide. 1% input is shown. MITF target promoters were amplified with primers that span the MITF binding site. CD25 is a negative control. B. ChIPs from A were quantified by real-time PCR. The results presented are an average of two or more independent experiments done in triplicate. C. Change in BRG1 recruitment to MITF target promoters in SK-MEL5+BRG1 cells expressing siRNAs that target MITF compared to SK-MEL5+BRG1 cells expressing a control siRNA. BRG1 recruitment was analyzed by ChIP analysis as in Fig. 4B. The data is the average of two independent experiments performed in triplicate. All values are statistically significant at p<.01. D. Fold increase in trimethylation at histone H3 (K4) and RNA polymerase II (phsophoS2) recruitment at MITF target promoters in SK-MEL5 expressing BRG1 compared to that in control SK-MEL5 cells. The data is the average of two independent experiments performed in triplicate. E. MITF target gene expression in SK-MEL5+empty vector (EV) and SK-MEL5+BRG1 cells. mRNA levels were quantified by real-time RT-PCR. These results are an average of greater than three independent experiments performed in triplicate.
We investigated whether BRG1 is recruited to the promoters of MITF target genes that regulate survival and proliferation. BRG1 was significantly enriched on the promoter of the melanoma inhibitor of apoptosis (MLIAP) (Fig. 4A and B), an MITF target gene and a potent anti-apoptotic factor that is highly expressed in melanoma (Dynek et al., 2008). However, we did not detect significant enrichment of BRG1 on the CDK2 promoter (Fig.4A and B), an MITF target gene that promotes proliferation (Du et al., 2004), nor on TBX2 or BCL2, other MITF target genes that promote melanoma proliferation and survival.
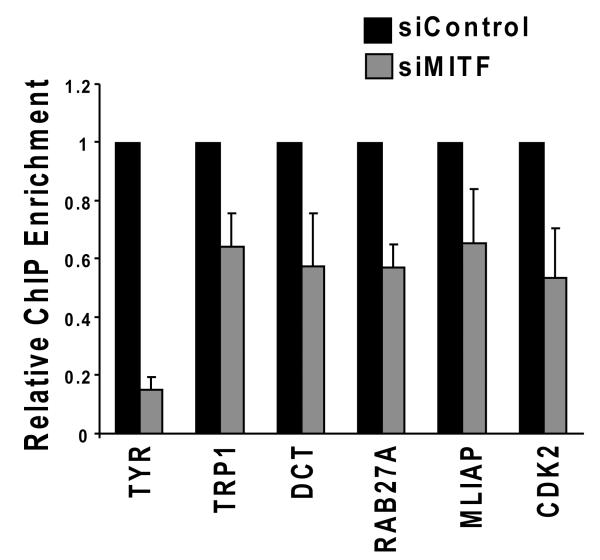
MITF plays an important role in the activation of each of the genes to which BRG1 was recruited, but is not the only transcription factor involved. To determine if MITF is required to recruit the BRG1 containing SWI/SNF complexes to these promoters, we down-regulated MITF expression with siRNA (supple. Fig. 2A and B), and performed ChIP analysis to detect BRG1. We found that down-regulation of MITF partially inhibited BRG1 recruitment to all MITF target promoters (Fig. 4C). This suggested that an important mechanism by which MITF activates gene expression is by the recruitment of BRG1 based SWI/SNF complexes to target promoters.
To determine the consequences of BRG1 recruitment to MITF target promoters, we performed ChIP analysis to detect changes in histone modifications and RNA polymerase II recruitment. BRG1 recruitment to target promoters correlated with increased levels of histone 3 tri-methylation at lysine 4 (tri-MeH3K4), an epigenetic mark associated with actively transcribed promoters and/or increased recruitment of RNA polymerase II phosphorylated at S2 (Fig. 4D). Thus, BRG1 recruitment to MITF target promoters led to changes in promoter structure and function indicative of increased transcriptional activity.
Ectopic expression of BRG1 resulted in increased tyrosinase, TRP1, DCT, RAB27A, p21CIP1, and MLIAP mRNA expression but did not have significant effects on the expression of other MITF target genes that promote proliferation (Fig.4E). Thus, for the selected MITF target genes, recruitment of BRG1 to MITF target promoters primarily led to an increase in the expression of differentiation related genes as well as to increased expression of MLIAP, but not to other MITF target genes that promote proliferation.
BRM interacts with MITF and is required for MITF target gene expression
Although ectopic expression of BRG1 promoted expression of MITF target genes associated with differentiation, these genes are not completely silenced in BRG1 deficient SK-MEL5 cells. To determine whether SWI/SNF complexes containing BRM also cooperate with MITF and contribute to the regulation of MITF target gene expression, we investigated whether BRM interacts with MITF. Fig. 5A shows that BRM and other SWI/SNF subunits can be co-immunoprecipitated with MITF in SK-MEL5 cells.
Figure 5.
A. Co-immunoprecipitation of SWI/SNF components and MITF in BRG1 deficient SK-MEL5 cells. Top panel: Cell lysates were immunoprecipitated with an antibody specific for BRM or with control rabbit IgG and blotted with the indicated antibodies. Bottom panel: Cell lysates were immunoprecipitated with MITF hybridoma supernatant or control mouse IgG and blotted with the indicated rabbit antibodies. B. Chromatin immunoprecipitations (ChIPs) were performed in BRG1 deficient SK-MEL5 cells using antibodies to BRM, BAF57, or control IgG, and run on an agarose gel stained with ethidium bromide. 1% input is shown. MITF target promoters were amplified with primers that span the MITF binding site. CD25 is a negative control. C. ChIPs from B were quantified by real-time PCR. These results are an average of two independent experiments performed in triplicate. D. Change in BRM recruitment to MITF target genes in SK-MEL5 cells expressing siRNAs that target MITF compared to SK-MEL5 cells expressing a control siRNA. These results were analyzed by Student’s t test and are representative of two independent experiments performed in triplicate. MLIAP was statistically significant at p<.05, all other values are significant at p<.01.
BRM and the associated subunit, BAF57, were enriched at several MITF target promoters. Interestingly, while ectopically expressed BRG1 was not recruited to the CDK2 promoter, we detected significant levels of BRM, and while BRM was not significantly enriched on the p21 promoter, we detected significant levels of BRG1 (compare Fig.4A and 4B with Fig. 5B and 5C). Down-regulation of MITF with siRNA (Supple. Fig. 2A and B) also partially inhibited the recruitment of BRM to its set of MITF target promoters (Fig. 5D), indicating that MITF contributes to the recruitment of BRM to the MITF target promoters examined, including CDK2. Therefore, heterogeneous SWI/SNF complexes containing either BRG1 or BRM may cooperate with MITF to regulate overlapping and distinct subsets of MITF target genes.
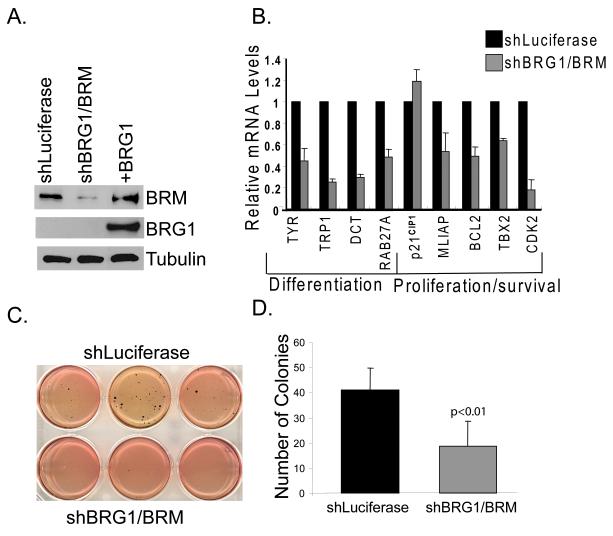
To determine whether SWI/SNF complexes containing BRM are required for expression of MITF target genes, we down-regulated BRM expression by retroviral transfer of small interfering RNAs (shRNAs) that target a sequence common in both BRG1 and BRM (Ramirez-Carrozzi et al., 2006). Introduction of these shRNAs significantly down-regulated BRM expression in SK-MEL5 cells while BRG1 levels remained undetectable (Fig. 6A). We found that in the absence of BRG1, down-regulation of BRM significantly inhibited the expression of multiple MITF target genes (Fig. 6B). Interestingly, while over-expression of BRM in the absence of BRG1 did not activate pigmentation related gene expression, down-regulation of BRM inhibited expression of these genes. Thus, although BRM cannot compensate for BRG1, BRM is also important for expression of pigmentation related genes. The expression of MITF target genes that regulate survival and proliferation was also inhibited in BRM knockdown cells, including genes that were not affected by over-expression of BRG1. Although we did not detect significant enrichment of BRM on the TBX2 and BCL2 promoters, we found that down-regulation of BRM inhibited expression of these genes. Thus, the association of BRM with the TBX2 and BCL2 promoters may be transient or BRM may be indirectly required for expression of these genes. Together, these data suggest that either or both the SWI/SNF ATPases are required directly or indirectly for expression of multiple classes of MITF target genes.
Figure. 6.
A. Detection by Western blotting of BRM and BRG1 in BRG1 deficient SKMEL5 cells that express shluciferase or shBRG1/BRM. BRG1 expressing cells were included as a positive control for BRG1. Tubulin is a loading control. B. MITF target gene expression in control SK-MEL5 (shLuciferase) cells and in cells expressing shBRG1/BRM. RNA levels were quantified by real-time RT-PCR. The results presented are an average of greater than three independent experiments done in triplicate and analyzed statistically by Student’s t test. shBRG1/BRM had a significant effect on all MITF target genes tested except for p21CIP1. C. The effect of BRG1/BRM down-regulation on the ability to grow in soft agar. These results were analyzed by Student’s t test and are representative of two independent experiments performed in triplicate. D. Quantification of control and BRG1/BRM down-regulated cells on melanoma growth in soft agar. These results were analyzed by Student’s t test and are representative of two independent experiments performed in triplicate.
We previously reported that BRG1 and other SWI/SNF subunits interact with MITF and can be detected at MITF target promoters in B16 mouse melanoma cells (de la Serna et al., 2006a). To confirm that both BRG1 and BRM are required for expression of MITF target genes in these melanoma cells, we down-regulated both BRG1 and BRM in B16 mouse melanoma cells and found that the expression of multiple MITF target genes was also inhibited (supple. Fig.3). Thus, SWI/SNF activity is required for expression of MITF target genes in multiple melanoma cell lines.
While expression of BRG1 activated p21CIP1 expression, down-regulation of BRM resulted in a slight increase in p21CIP1 mRNA. A significant stimulatory effect on p21CIP1 expression was noted when BRG1 and BRM were down-regulated in B16 mouse melanoma cells (supple. Fig.2). Regulation of p21CIP1 expression is complex and dependent on multiple transcription factors, including p53 (Gartel and Tyner, 1999). Further work will be required to determine the precise role that SWI/SNF enzymes play in regulating p21CIP1 expression in melanoma cells.
We found that all the melanoma cells we assayed retained expression of either BRG1 or BRM and expressed high levels of several other SWI/SNF subunits. Furthermore, down-regulation of SWI/SNF activity by depletion of the sole ATPase, BRM, in SK-MEL5 cells and down-regulation of both ATPases in mouse melanoma cells, significantly inhibited expression of MITF target genes that promote proliferation and survival but not p21CIP1, an inhibitor of proliferation. Based on these data and the known role of MITF in promoting melanoma proliferation and the ability to grow on soft agar (Garraway et al., 2005), we hypothesized that retention of functional SWI/SNF enzymes with at least one ATPase would also be required for melanoma tumorigenicity. Consistent with this hypothesis, we found that down-regulation of the sole ATPase, BRM, in BRG1 deficient cells significantly inhibited their ability to grow in soft agar (Fig.6C and D). Therefore, similar to MITF, a critical level of SWI/SNF activity may be required to sustain aspects of melanoma tumorigenicity.
Discussion
MITF is the master regulator of melanocyte differentiation and is also a lineage addiction oncogene in melanoma (Levy et al., 2006). Because we previously found that SWI/SNF enzymes can interact with MITF to promote melanocyte differentiation, we investigated whether SWI/SNF enzymes regulate multiple classes of MITF target genes that have been shown to be important for melanoma oncogenicity. In this study, we have characterized SWI/SNF subunit expression in established melanoma cell lines and have determined that the BRG1 or BRM ATPAse is downregulated in a subset of these cells. We found that expression of BRG1 in BRG1 deficient melanoma cells promoted melanoma differentiation and visible pigmentation. BRG1 interacted with MITF, was recruited to MITF target promoters that regulate differentiation, as well as to the promoter of the melanoma inhibitor of apoptosis (MLIAP). Furthermore, BRG1 enhanced expression of these genes. These data reveal that SWI/SNF enzymes promote expression of MITF target genes important for pigmentation and survival in human melanoma cells.
The role that SWI/SNF enzymes play in regulating the status of melanoma differentiation may have a direct bearing on melanoma therapeutics. Melanomas are notoriously resistant to cytotoxic drugs and several studies suggest that melanoma cells possess lineage specific mechanisms for drug resistance. For example, the melanocytic enzyme, DCT, confers resistance to DNA damaging agents and melanosomes contribute to resistance by sequestering cytotoxic drugs and by increasing drug export (Chen et al., 2006; Chu et al., 2000; Pak et al., 2000). Furthermore, MITF promotes expression of survival genes that also contribute to chemoresistance (Garraway et al., 2005). Consistent with our hypothesis that SWI/SNF enzymes promote pigmentation and MITF target gene expression, we found that BRG1 also increased the resistance of melanoma cells to cisplatin. In other cell types, SWI/SNF enzymes have also been shown to facilitate DNA repair and to protect cells against UV induced DNA damage (Gong et al., 2008; Gong et al., 2006; Park et al., 2006). Thus, SWI/SNF enzymes may regulate melanoma chemoresistance by multiple mechanisms.
In addition to promoting differentiation, MITF activates expression of pro-proliferative genes such as TBX2 and CDK2, and is critically important for melanoma proliferation (Carreira et al., 2000; Du et al., 2004). In melanocytic cells, terminal differentiation is associated with activation of p21CIP1 and p16INK4A expression and cell cycle arrest (Carreira et al., 2005; Loercher et al., 2005). Thus, MITF’s role in promoting both differentiation and proliferation is contradictory. Although the mechanisms have not been clearly elucidated, it has been proposed that low levels of MITF activity promote proliferation while higher levels of MITF activity are required for terminal differentiation (Carreira et al., 2006). Consistent with this model and with our hypothesis that SWI/SNF enzymes are important regulators of MITF activity, we found that higher levels of SWI/SNF activity (achieved by the expression of BRG1 in deficient melanoma cells) promoted melanoma differentiation as well as high levels of p21CIP1 expression. A lower level of SWI/SNF activity that included one functional ATPase was necessary and sufficient to promote optimal expression of proliferation related genes. Because BRM did not compensate for BRG1 in promoting melanoma differentiation and because BRG1 did not promote pro-proliferative gene expression, an intriguing possibility is that heterogeneous SWI/SNF complexes composed of a specific ATPase regulate the different classes of MITF target genes. However, additional studies investigating the role of BRG1 in BRM deficient melanoma cells will be required to determine if the two ATPases play differential roles in regulating the different classes of MITF target genes or whether the observed results are due to alterations in total SWI/SNF activity.
Like the androgen receptor (AR) in prostate cancer, MITF has been classified as a lineage addiction oncogene that can activate survival and pro-proliferative gene expression (Garraway and Sellers, 2006). AR has been shown to recruit the SWI/SNF complex to transactivate target genes that promote proliferation and tumorigenicity (Link et al., 2005). Disruption of this interaction inhibits androgen dependent prostate cancer cell proliferation (Link et al., 2008). Similar to prostate cancer, we find that in melanoma, although SWI/SNF activity may be down-regulated to some extent by down-regulation of particular subunits, at least one functional SWI/SNF ATPase must be retained to promote aspects of melanoma tumorigenicity.
Materials and Methods
Cell lines
Melanoma cells, SW13, and HeLa cells were from ATCC or from the Yale Cell Culture Core Facility (New Haven, Connecticut). Human melanocytes were from Cascade Biologics (Portland, Oregon) or Yale Cell Culture Core Facility. With the exception of melanocytes, all cells were grown in DMEM supplemented with 10% FBS. Human melanocytes were grown in Media 254 with added growth supplements (Cascade Biologics).
siRNA and shRNA
Control siRNA and siRNAs targeting MITF were purchased from Dharmacon Inc. (Chicago, Illinois). Control and shRNA constructs targeting BRG1/BRM were provided by Stephen Smale (Howard Hughes Medical Institute, UCLA). The knockdowns are described in supplemental methods.
Antibodies
Antibodies used for Westerns and chromatin immunoprecipitations are described in supplemental methods
Immunocytochemistry
Immunocytochemistry was performed as described (de la Serna et al., 2006a) using M2 FLAG antibody (Sigma) and goat anti-mouse-FITC (sc3699) (Santa Cruz). Images were taken with a Nikon Eclipse T2000-U fluorescence microscope at 40X magnification.
Cell extracts and immunoblot analysis
Cells were lysed in 20mMTris (pH 7.4),150 mM NaCl, 2mM EDTA, 1% Triton X, 10% glycerol, supplemented with a protease inhibitor cocktail (Sigma). SDS-PAGE and Western blotting were carried out as described (de La Serna et al., 2000).
Co-immunoprecipitations
Co-immunoprecipitations were performed as described (de la Serna et al., 2006a) with FLAG M2-Agarose, MITF (C5) hybridoma supernantant, or BRM (rabbit) antibody. Species matched IgG (Upstate, Billerica, MA) was used as a control.
Chromatin immunoprecipitations
Chromatin immunoprecipitations were performed as described (de la Serna et al., 2005) with the modifications described in Supplemental Methods. The purified DNA was amplified and analyzed by real-time PCR or run on an agarose gel, stained with ethidium bromide, and scanned with the Alpha Innotech FluorChem HD2 imaging system. Primers used for amplification are listed in Supplemental Methods.
RNA Isolation and Quantitative Real-time PCR
Total RNA was isolated with the Qiagen RNeasy mini kit and reverse transcribed as described (de la Serna et al., 2001). Quantitative real-time PCR was performed in SYBR Green Master Mix (Qiagen, Germantown, Maryland) with an Applied Biosystems Prism 7500 PCR system and analyzed with the SDS software. Human 18S rRNA gene was used as a control. Primer sequences are available upon request.
Cisplatin sensitivity and cell viability assay
SK-MEL-5 cells, SK-MEL-5 cells with empty vector (pBABE), and SK-MEL-5 cells with BRG1 (pBABE-BRG1) were seeded at a density of 6000 cells per well in a 96-well plate. After 24 h, cells were treated with 20 μM cisplatin for 72 h. Subsequent to drug treatment, cell viability was analyzed using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega, Madison, Wisconsin).
Soft Agar Colony Formation Assay
Each well in a 6-well plate was coated with 1 mL of 10% FBS supplemented media containing 0.6% agarose. 5000 cells/well SK-MEL-5 cells expressing shBRG1/BRM or shLuciferase were plated in triplicate in DMEM containing 0.3% agarose, overlayed onto 0.6% agarose. Cells were incubated for 21 days, stained with 10mg/ml thiazolyl blue, photographed and scored. For each cell type, ten fields were counted and averaged.
Supplementary Material
Acknowledgements
We thank Dr. David Fisher (Dana Farber) for the MITF antibody, Ruth Halaban (Yale Tissue Culture Facility) for melanocytes and melanoma cells and Stephen Smale (Howard Hughes Medical Institute, UCLA) for shRNA constructs.
Financial Support: ILD was supported by the National Institute of Environmental Health Sciences; Grant number: 5K22ES12981, Ohio Cancer Research Associates, American Cancer Society, Ohio Division
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- Bandyopadhyay D, Curry JL, Lin Q, Richards HW, Chen D, Hornsby PJ, et al. Dynamic assembly of chromatin complexes during cellular senescence: implications for the growth arrest of human melanocytic nevi. Aging Cell. 2007;6:577–91. doi: 10.1111/j.1474-9726.2007.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TM, Haferkamp S, Dijkstra MK, Scurr LL, Frausto M, Diefenbach E, et al. The chromatin remodelling factor BRG1 is a novel binding partner of the tumor suppressor p16INK4a. Mol Cancer. 2009;8:4. doi: 10.1186/1476-4598-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramel J, Medjkane S, Quignon F, Delattre O. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene. 2008;27:2035–44. doi: 10.1038/sj.onc.1210847. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–9. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–39. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Liu B, Goding CR. The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J Biol Chem. 2000;275:21920–7. doi: 10.1074/jbc.M000035200. [DOI] [PubMed] [Google Scholar]
- Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–27. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KG, Valencia JC, Lai B, Zhang G, Paterson JK, Rouzaud F, et al. Melanosomal sequestration of cytotoxic drugs contributes to the intractability of malignant melanomas. Proc Natl Acad Sci U S A. 2006;103:9903–7. doi: 10.1073/pnas.0600213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaverini C, Beuret L, Flori E, Busca R, Abbe P, Bille K, et al. Microphthalmia-associated transcription factor regulates RAB27A gene expression and controls melanosome transport. J Biol Chem. 2008;283:12635–42. doi: 10.1074/jbc.M800130200. [DOI] [PubMed] [Google Scholar]
- Chu W, Pak BJ, Bani MR, Kapoor M, Lu SJ, Tamir A, et al. Tyrosinase-related protein 2 as a mediator of melanoma specific resistance to cisdiamminedichloroplatinum(II): therapeutic implications. Oncogene. 2000;19:395–402. doi: 10.1038/sj.onc.1203315. [DOI] [PubMed] [Google Scholar]
- de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, et al. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–51. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Higashi C, Dutta C, Osias J, Kommajosyula N, et al. The microphthalmia-associated transcription factor (MITF) requires SWI/SNF enzymes to activate melanocyte specific genes. J Biol Chem. 2006a doi: 10.1074/jbc.M512052200. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006b;7:461–73. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Roy K, Carlson KA, Imbalzano AN. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J Biol Chem. 2001;276:41486–91. doi: 10.1074/jbc.M107281200. [DOI] [PubMed] [Google Scholar]
- Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186:136–45. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–76. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–30. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Chan SM, Liu J, Zha J, Fairbrother WJ, Vucic D. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 2008;68:3124–32. doi: 10.1158/0008-5472.CAN-07-6622. [DOI] [PubMed] [Google Scholar]
- Eberle J, Garbe C, Wang N, Orfanos CE. Incomplete expression of the tyrosinase gene family (tyrosinase, TRP-1, and TRP-2) in human malignant melanoma cells in vitro. Pigment Cell Res. 1995;8:307–13. doi: 10.1111/j.1600-0749.1995.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Flowers S, Nagl NG, Jr., Beck GR, Jr., Moran E. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J Biol Chem. 2009;284:10067–75. doi: 10.1074/jbc.M808782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314–24. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246:280–9. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 2008;7:1067–74. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol. 2006;13:902–7. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–80. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–51. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24:1188–99. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–14. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Link KA, Balasubramaniam S, Sharma A, Comstock CE, Godoy-Tundidor S, Powers N, et al. Targeting the BAF57 SWI/SNF subunit in prostate cancer: a novel platform to control androgen receptor activity. Cancer Res. 2008;68:4551–8. doi: 10.1158/0008-5472.CAN-07-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–15. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Murai K, Miyake K, Iijima S. Expression of chromatin remodeling factors during neural differentiation. J Biochem (Tokyo) 2001;129:43–9. doi: 10.1093/oxfordjournals.jbchem.a002834. [DOI] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–18. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Nagl NG, Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. Embo J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak BJ, Li Q, Kerbel RS, Ben-David Y. TYRP2-mediated resistance to cisdiamminedichloroplatinum (II) in human melanoma cells is independent of tyrosinase and TYRP1 expression and melanin content. Melanoma Res. 2000;10:499–505. doi: 10.1097/00008390-200010000-00013. [DOI] [PubMed] [Google Scholar]
- Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2009;8:29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. Embo J. 2006;25:3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack MS, Heagney SD, Livingston PO, Fogh J. HLA-A, B, C and DR alloantigen expression on forty-six cultured human tumor cell lines. J Natl Cancer Inst. 1981;66:1003–12. doi: 10.1093/jnci/66.6.1003. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–96. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–6. [PubMed] [Google Scholar]
- Rosson GB, Bartlett C, Reed W, Weissman BE. BRG1 loss in MiaPaCa2 cells induces an altered cellular morphology and disruption in the organization of the actin cytoskeleton. J Cell Physiol. 2005;205:286–94. doi: 10.1002/jcp.20397. [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Ahmed S, Boumber Y, Konishi K, Guo Y, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67:11335–43. doi: 10.1158/0008-5472.CAN-07-1502. [DOI] [PubMed] [Google Scholar]
- Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem. 2004;91:1087–98. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- Svensson SP, Lindgren S, Powell W, Green H. Melanin inhibits cytotoxic effects of doxorubicin and daunorubicin in MOLT 4 cells. Pigment Cell Res. 2003;16:351–4. doi: 10.1034/j.1600-0749.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Vink J, Thomas L, Etoh T, Bruijn JA, Mihm MC, Jr., Gattoni-Celli S, et al. Role of beta-1 integrins in organ specific adhesion of melanoma cells in vitro. Lab Invest. 1993;68:192–203. [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF Induces Senescence and Apoptosis through Pathways Mediated by the Secreted Protein IGFBP7. Cell. 2008;132:363–74. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Baiocchi RA, Pal S, Mosialos G, Caligiuri M, Sif S. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol Cell Biol. 2005;25:7953–65. doi: 10.1128/MCB.25.18.7953-7965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang J, Chen X. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem. 2007;282:37429–35. doi: 10.1074/jbc.M706039200. [DOI] [PubMed] [Google Scholar]
- Yamamichi N, Inada K, Ichinose M, Yamamichi-Nishina M, Mizutani T, Watanabe H, et al. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67:10727–35. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.