Abstract
Micro-RNAs (miRNAs) are estimated to regulate 30% of the human genome primarily through translational repression. In 2005–2008, the first series of observations establishing the key significance of miRNAs in the regulation of vascular biology came from experimental studies involved in arresting miRNA biogenesis to deplete the miRNA pools of vascular tissues and cells. Dicer-dependent biogenesis of miRNA is required for blood vessel development during embryogenesis and wound healing. miRNAs regulate redox signaling in endothelial cells, a key regulator of vascular cell biology. miRNAs that regulate angiogenesis include miRNA 17–5p, cluster 17–92, 21, 27a&b, 126, 130a, 210, 221, 222, 378 and the let7 family. miRNAs also represent a new therapeutic target for the treatment of proliferative vascular diseases as well as hypertension. Evidence supporting the regulation of inducible adhesion molecules by miRNA supports a role of miRNAs in regulating vascular inflammation. Productive strategies to safely up-regulate as well as down-regulate miRNAs in vivo are in place and being tested for their value in disease intervention. Prudent targeting of non-coding genes such as miRNAs, which in turn regulates large sets of coding genes, holds promise in gene therapy. Recent developments in miRNA biology offer lucrative opportunities to manage vascular health.
Keywords: Angiogenesis, MicroRNA, Non-coding genes, Redox control, RNA, Wound healing
Introduction
The central dogma in molecular biology has been that DNA replicates its genetic information contained within nucleotide (nt) sequences and transcribes it to RNA where it codes for the production of mRNA. mRNA is processed essentially by splicing and translocates from the nucleus to the cytoplasm. mRNA carries nt-coded information to the ribosomes. Ribosomes translate the code for protein synthesis. Do the vast majority of nts in RNA carry the code to synthesize protein? No. However, almost all means of gene identification assume that genes encode proteins. The central dogma failed to consider a significant part of the genetic code, which remained behind a veil for decades. Non-coding RNA (ncRNA) genes produce functional RNA molecules rather than encoding proteins. Several different systematic screens have identified a surprisingly large number of ncRNA genes. ncRNAs seem to be particularly abundant in roles that require highly specific nucleic acid recognition without complex catalysis, such as in directing posttranscriptional regulation of gene expression or in guiding RNA modifications. Although it has been generally assumed that most genetic information is transacted by proteins, recent evidence suggests that the majority of the genomes of mammals and other complex organisms is in fact transcribed into ncRNAs, many of which are alternatively spliced and/or processed into smaller products [1]. These RNAs (including those derived from introns) appear to comprise a heretofore hidden layer of internal signals that control various levels of gene expression in physiology and development, including chromatin architecture/epigenetic memory, transcription, RNA splicing, editing, translation and turnover. This hidden layer of internal signals is now emerging to be of such critical significance that lack of consideration of that layer poses the serious risk of clouding our ability to understand the molecular basis of health and disease [1–4]. In all forms of life, ncRNA includes ribosomal, transfer, small nuclear and small nucleolar RNA, interference RNA (RNAi), short interfering RNA (siRNA) and micro-RNA (miRNA).
miRNAs are powerful regulators of gene expression and fine-tune the synthesis of proteins encoded by a large number of genes [Baek, 2008 #257; Selbach, 2008 #258]. It is estimated that about 3% of human genes encode for miRNAs [5]. miRNAs are approximately 22-ribonucleo-tide-long ncRNAs, with a potential to recognize multiple mRNA targets guided by sequence complementarity and RNA-binding proteins. Recent evidence suggests that the number of unique miRNA genes in humans exceeds 1,000 [6]. The numbers of miRNAs and their targets turn out to be much greater than what we previously thought [7]. miRNAs are functionally versatile, with the capacity to specifically inhibit translation initiation or elongation as well as induce mRNA destabilization, predominantly by targeting the 3′-untranslated regions (UTR) of mRNA. Briefly, miRNA are transcribed in the nucleus by conventional mechanisms and are exported to the cytoplasm [8], where, after biological processing, they form the mature miRNA that can interact with matching mRNAs by RNA-RNA binding. This binding, with the assistance of the RNA-induced silencing complex, leads to modes of action, resulting in mRNA degradation or translational inhibition (fig. 1) [9]. This mechanism of action is termed posttranscriptional gene regulation. Contrary to plants, in animals there is not 100% nt match between miRNA and its target mRNA, leading to a mode of action causing mRNA translational inhibition [10] as well as mRNA destabilization [Baek, 2008 #257, Selbach, 2008 #258]. The interaction between the miRNA and its matching mRNA occurs between the 5′-UTR of the miRNA to the 3′-UTR region of the mRNA by a matching seed element in the miRNA. Utilizing these data, computational prediction approaches estimate that miRNAs can target 30% of the human genome [5, 11–13]. Other estimates claim that over 50% of human protein-coding genes might be regulated by miRNAs [14]. Furthermore, one miRNA can regulate hundreds of genes [14], and that one gene can be regulated by a number of miRNAs. Tissue specificity for miRNA expression adds to the role of miRNA in developmental biology and to cell and tissue phenotyping [15–17]. The complexities underlying tissue-dependent miRNA biogenesis and target selection processes are currently under investigation [7].
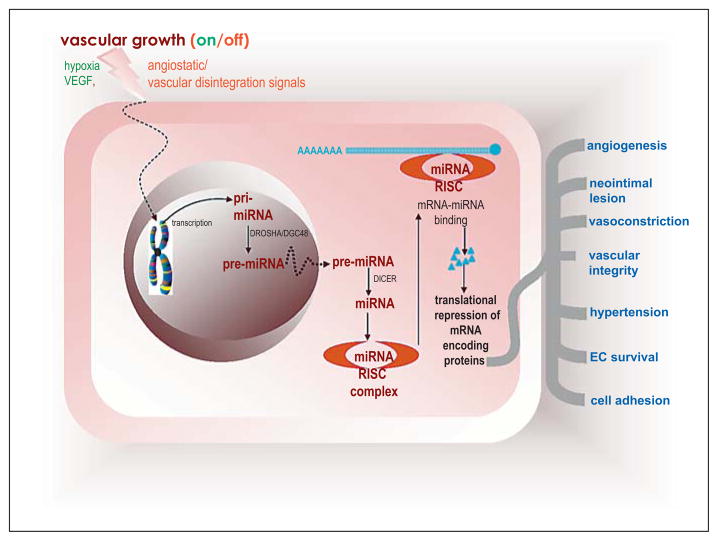
Fig. 1.
miRNAs in vascular biology. pri-miRNA is synthesized in the nucleus by RNA pol II. Then by the RNA endonuclease Drosha and its cofactor DGC48, pri-miRNA is cleaved to produce pre-miRNA that is about 70 nts long. This product is exported to the cytosol by Exportin 5, where it is cleaved again by the second RNA endonuclease Dicer, to form the approximately 20–22-nt-long mature miRNA. Then one of the ds miRNA is incorporated into the RNA-induced silencing complex (RISC), where by base matching with the 3′UTR end of mRNA it will capture the target mRNA in the complex, and as a result inhibits its translation by the ribosome. Depending on the specific mRNA targeted by any given miR (table 1), several aspects of vascular biology may be influenced. EC = Endothelial cell.
Vascular Consequences in Response to Arrest of miRNA Biogenesis
In 2005–2008, the first series of observations establishing the key significance of miRNAs in the regulation of mammalian vascular biology came from experimental studies involved in arresting miRNA biogenesis to deplete the miRNA pools of vascular tissues and cells [26–29]. Dicer, a member of the ribonuclease III family of enzymes, processes double-stranded RNA substrates into approximately 21- to 27-nt products that trigger sequence-directed gene silencing by RNA interference. The mechanism of RNA recognition and length-specific cleavage by Dicer has been established. Dicer represents a key enzyme involved in miRNA biogenesis [30]. In the study of embryonic angiogenesis, it has been reported that the Dicer gene is significantly expressed in 11-day embryos and remains constant to embryonic day 17, being evenly expressed throughout the embryonic tissues [26]. The dicerex1/2 mutant mouse lacking the first two exons of dicer essential for the function of the protein represents a tool that has shed light on embryonic angiogenesis. Of note, these mutant mice are hypomorphic for Dicer function and do not represent the Dicer null state. Homozygous mutant mice were not viable, therefore the embryos were examined. Starting from embryonic day 11.5, virtually all dicerex1/2 embryos were growth and developmentally retarded compared with their wild-type or heterozygous litter mates. The embryos that were still viable at this stage had thin and suboptimally developed blood vessels providing first evidence for the involvement of miRNAs in angiogenesis [26]. Microscopic examination of the yolk sacs from the mutant embryos revealed that there were fewer blood vessels in the dicerex1/2 yolk sacs and that these vessels were thin, small and less organized than those of control yolk sacs. These observations indicate that Dicer-dependent biogenesis of miRNA is required for blood vessel development during embryogenesis.
Support for the involvement of miRNAs in angiogenesis also comes from in vitro studies [31]. Large-scale analysis of miRNA expression in human umbilical vein endothelial cells (HUVEC) led to the observation that among those miRNAs that are highly expressed, there are 15 that may target the receptors of angiogenic factors. In particular, miR-221 and miR-222 were identified as regulating c-Kit expression as well as the angiogenic properties of the c-kit ligand: stem cell factor. The miR-221/2 and c-Kit interaction represents an integral component of a complex circuit that controls the ability of endothelial cells to form new capillaries [31]. Inhibition of c-kit results in reduced vascular endothelial growth factor (VEGF) expression [32]. Furthermore, we know that c-kit is involved in neovascularization [33, 34]. In addition to Dicer, Drosha is an miRNA processing enzyme required for the maturation of miRNAs. The maintenance and regulation of endogenous miRNA levels via Dicer-mediated processing is critical for endothelial cell gene expression and functions in vitro [28]. The significance of both Dicer and Drosha in driving angiogenesis in vitro has been reported [27, 28]. Silencing of Dicer and Drosha significantly attenuated capillary sprouting of endothelial cells and tube-forming activity. Migration of endothelial cells was significantly decreased in Dicer siRNA-transfected cells, whereas Drosha siRNA had no effect. Silencing of Dicer, but not of Drosha, attenuates the angiogenic response of endothelial cells in vitro. The let7 family and miR-27b are recognized as key regulators of angiogenic responses of endothelial cells.
Several studies have demonstrated a central role of NADPH oxidase-derived reactive oxygen species (ROS) as signaling messengers in driving angiogenesis [35–38]. Whether such redox control of angiogenesis is subject to regulation by miRNA remains to be determined. A recent study in our laboratory presents first evidence that a cellular redox state, a key driver of cell signaling, is regulated by miRNAs. In this work, a Dicer knockdown approach was used to test the significance of miRNA in regulating the redox state and angiogenic response of human microvascular endothelial cells (HMECs). Lowering of the miRNA content by Dicer knockdown induced VEGF expression but diminished the angiogenic response of HMECs determined by cell migration and Matrigel tube formation. Such impairment of the angiogenic response in Matrigel was rescued by exogenous low-micromolar H2O2. Dicer-knockdown HMECs demonstrated lower inducible production of ROS when activated with either phorbol ester, tumor necrosis factor (TNF)-α or VEGF. Limiting ROS production by antioxidant treatment or NADPH oxidase-knockdown approaches impaired angiogenic responses. Experiments aiming to identify how ROS production is limited by Dicer knockdown revealed a lower expression of p47phox protein in these cells. This decreased cellular miRNA content induced expression of the transcription factor HBP1, a suppressor transcription factor that negatively regulates p47phox expression. Knockdown of HBP1 restored the angiogenic response of miRNA-deficient HMECs [29, 39]. Results of this study lead to the hypothesis that miRNA regulates wound angiogenesis and may therefore influence wound healing outcomes. A possible role for miRNA in wound healing has been stated by another independent line of investigation examining the function of argonaute proteins. The argonautes (Agos) represent a cadre of evolutionarily conserved proteins that directly associate with and are required for miRNA function. The core miRNA-ribonucleic-protein complex, composed of the miRNA and an Ago protein, functions as a highly modifiable scaffold that associates with specific mRNAs via the bound miRNA and facilitates localized activity of a variety of accessory proteins [40]. The expression of Ago-2, critically important in mammalian RNAi, is regulated by the epidermal growth factor receptor (EGFR)/mitogen-activated protein kinase (MAPK) signaling pathway. The pharmacological inhibitor of MAPK kinase, U0126, or an siRNA directed against EGFR limits Ago-2 expression [41]. Ago-2 overexpression enhances cell proliferation, attenuates cell-cell adhesion and increases cell migration in an in vitro wound healing setting [41].
Early embryonic lethality of Dicer1 null alleles in mice limits our ability to address the role of Dicer1 in normal mouse growth and development. Studies on mice mutants with a hypomorphic Dicer1 allele (Dicerd/d) demonstrate that Dicer1 deficiency causes female infertility. This defect in female Dicerd/d mice was caused by corpus luteum insufficiency and resulted, at least in part, from the impaired growth of new capillary vessels in the ovary. miR17–5p and let7b represent two specific miRNAs that participate in angiogenesis by regulating the expression of the anti-angiogenic factor tissue inhibitor of metalloproteinase (TIMP)-1. Injection of miR17–5p and let7b into the ovaries of Dicerd/d mice partially normalized TIMP-1 expression and corpus luteum angiogenesis [42]. More recent evidence supports that endothelial miRNAs regulate postnatal angiogenesis and that VEGF induces the expression of miRNAs implicated in the regulation of an integrated angiogenic response [43]. Reduction of endothelial miRNAs by cell-specific inactivation of Dicer attenuates postnatal angiogenic responses to a variety of stimuli, including exogenous VEGF, tumors, limb ischemia and wound healing [43].
Emergence of a Complex Gene Regulatory Network
In a setting where one miRNA can regulate hundreds of genes [14] and one gene can be regulated by a number of miRNAs, a major issue is to improve the knowledge on the regulatory loops that govern miRNA-miRNA as well as miRNA-mRNA interactions. In addition, factors contributing to the control of inducible or repressible miRNA expression need to be understood. At present, we are far from understanding these processes in all their complexity. In particular, miRNA-coordinated expression with other regulatory molecules, e.g. transcription factors, is an area under development. When miRNA 221 and 222 were depleted in HUVEC, the entire miRNA profile was affected in these cells. Nine miRNAs were up-regulated and 23 miRNAs were down-regulated. Such observations lead to the concept of a complex network involving co-expressed miRNAs and transcription factors being affected by single miRNA variations [44]. miRNAs are important components of the p53 transcriptional network. Global miRNA expression analyses identify a cohort of miRNAs that exhibit p53-dependent up-regulation following DNA damage. One such miRNA, miR-34a, is commonly deleted in human cancers and is frequently absent in pancreatic cancer cells [45]. Characterization of the miR-34a primary transcript and promoter demonstrated that this miRNA is directly transactivated by p53. Expression of miR-34a caused reprogramming of gene expression and promoted apoptosis. Much like the known set of p53-regulated genes, miR-34a-responsive genes are highly enriched for those that regulate cell-cycle progression, apoptosis, DNA repair and angiogenesis. Therefore, it is likely that the modulation and fine-tuning of the gene expression program initiated by p53 is an important function of miR-34a [45]. An miRNA polycistron induced by the oncogenic transcription factor c-myc is involved in an unusually structured network of interactions. This network includes the seemingly paradoxical transcriptional induction and translational inhibition of the same molecule, the E2F1 transcription factor. This miRNA cluster has been implicated in inhibiting proliferation, as well as inhibiting apoptosis, and promoting angiogenesis [46].
Recent evidence shed new light on how cytokines may regulate miRNA biogenesis. Regulation of miRNA biogenesis by ligand-specific SMAD proteins is critical for the control of the vascular smooth muscle cell phenotype and potentially for SMAD4-independent responses mediated by transforming growth factor (TGF)-β and bone morphogenetic protein (BMP) signaling pathways. The TGF-β and BMP family of growth factors orchestrates fundamental biological processes in the development and homeostasis of adult tissues, including the vasculature. Induction of a contractile phenotype in human vascular smooth muscle cells by TGF-β and BMPs is mediated by miR-21. miR-21 down-regulates PDCD4 (programmed cell death 4), which in turn acts as a negative regulator of smooth muscle contractile genes. TGF-β/BMP signaling rapidly increases the expression of mature miR-21 through a posttranscriptional step, promoting the processing of primary transcripts of miR-21 (pri-miR-21) into precursor miR-21 (pre-miR-21) by the Drosha (also known as RNASEN) complex. TGF-β- and BMP-specific SMAD signal transducers are recruited to pri-miR-21 in a complex with the RNA helicase p68 (also known as DDX5), a component of the Drosha microprocessor complex [47].
Tumor Vascular Biology
The vast majority of the current literature on miRNA addresses cancer and developmental biology. miRNAs may act as oncogenes and/or tumor suppressor genes within the molecular architecture of gene regulatory networks, thereby contributing to the development of cancer. Bioinformatic analyses of putative miRNA binding sites have identified several novel potential gene targets of cancer-associated miRNAs that function in aspects of cell adhesion, neovascularization and tissue invasion. It has been proposed that miRNAs regulate E-cadherin, integrin αvβ3, hypoxia-inducible factor-1α, syndecan-1, lysyl oxidase, adamalysin metalloproteinase-17, TIMP-3, c-Met and CXCR-4 that underpin the tissue architectural changes associated with malignancy [48]. Aberrant expression of miRNA occurs in human chronic lymphocytic leukemias, where miRNA signatures are associated with specific clinical manifestations. miRNAs are also aberrantly expressed in human breast cancer. The overall miRNA expression profile clearly separates normal from cancer tissues, with the most significantly deregulated miRNAs being miR-125b, miR-145, miR-21 and miR-155. In breast cancer tissue, miRNAs regulate tumor vascular invasion [49]. Intravasation of tumor cells into the vasculature and/or lymphatics represents a key stage in cancer metastases. miR-378 enhances cell survival, reduces caspase-3 activity, and promotes tumor growth and angiogenesis by attenuating the expression of suppressor of fused (Sufu), a potential target of miR-378 and Fus-1, a gene responsible for β-lactamase production by Gram-negative anaerobic bacteria, but which may have other functions as well [50].
Human adenocarcinomas commonly harbor mutations in the K-Ras and Myc proto-oncogenes and the TP53 tumor suppressor gene. All three genetic lesions are potentially pro-angiogenic, as they sustain VEGF production. Interestingly, Kras-transformed mouse colonocytes lacking p53 form indolent, poorly vascularized tumors, whereas additional transduction with a Myc-encoding retrovirus promote vigorous vascularization and growth [51]. In addition, VEGF levels were unaffected by Myc, but enhanced neovascularization correlated with down-regulation of anti-angiogenic thrombospondin-1 (Tsp1) and related proteins, such as connective tissue growth factor (CTGF). Both Tsp1 and CTGF are predicted targets for repression by the miR-17–92 miRNA cluster, which is up-regulated in colonocytes co-expressing K-Ras and c-Myc. miR-17–92 knockdown partly restores Tsp1 and CTGF expression. Furthermore, transduction of Ras-only cells with a miR-17–92-encoding retrovirus attenuated Tsp1 and CTGF levels. Cells transduced with miR-17–92, also known as Oncomir-1, formed larger, better-perfused tumors. These findings establish a role for the miR-17–92 cluster in non-cell-autonomous Myc-induced tumor vascular biology [51–53]. More recent work demonstrates that VEGF regulates the expression of several miRNAs, including cluster miR-17–92. Transfection of endothelial cells with components of the miR-17–92 cluster, induced by VEGF treatment, rescued the induced expression of Tsp1 and the defect in endothelial cell proliferation and morphogenesis initiated by the loss of Dicer [43]. The miR-17–92 cluster composed of seven mature miRNAs (miR-17–5p, miR-17–3p, miR-18a, miR-19a, miR-20a, miR-19b and miR-92–1), residing in intron 3 of the C13orf25 gene at 13q31.3, is markedly and frequently overexpressed in lung cancers, with occasional gene amplification, especially in those with small-cell lung cancer histology [54].
Genomic estrogen receptor (ER) associates with other transcription factors such as the activating protein-1 complex, nuclear factor-κB and specificity proteins (Sp) to modulate ligand-dependent gene expression. Members of the Sp family of transcription factors exert differential effects on gene transcription. For example, Sp1 and Sp4 act as transcriptional activators, whereas Sp3 antagonizes Sp1 activation by competing for promoter occupancy. Sp1, Sp3 and Sp4 are overexpressed in tumors and contribute to the proliferative and angiogenic phenotype associated with cancer cells [55]. Sp1, Sp3 and Sp4 are expressed in a panel of ER-positive and ER-negative breast cancer cell lines and are subject to the regulation by miR-27a, which is also expressed in these cell lines and has been reported to regulate the zinc finger ZBTB10 gene, a putative Sp repressor. Experimental down-regulation of miR-27a using an anti-sense approach increased the expression of ZBTB10 mRNA and decreased the expression of Sp1, Sp3 and Sp4 at the mRNA and protein level, and also decreased activity in cells transfected with constructs containing Sp1 and Sp3 promoter inserts. Such responses were accompanied by decreased expression of Sp-dependent survival and angiogenic genes, including survivin, VEGF, and VEGF receptor 1 [55]. Thus, miR-27a supports angiogenesis in the context of breast cancer.
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the eighth human herpesvirus. Its formal name according to the International Committee on Taxonomy of Viruses is HHV-8. This virus causes Kaposi’s sarcoma, a cancer commonly occurring in AIDS patients, as well as primary effusion lymphoma and some types of multicentric Castleman’s disease. KSHV-encoded miRNAs directly contribute to the pathogenesis by down-regulation of Tsp1, a major regulator of cell adhesion, migration and angiogenesis [56]. The study of gene expression profiling in cells stably expressing KSHV-encoded miRNAs has identified a set of 81 genes whose expression is subject to control by miRNAs. For three of these genes, characterization of miRNA-dependent regulation led to the observation that protein levels of Tsp1 were decreased >10-fold. Tsp1 is known to be down-regulated in Kaposi sarcoma lesions and functions as a potent tumor suppressor and anti-angiogenic factor, exerting its anti-angiogenic effect in part by activating the latent form of TGF-β. Attenuated Tsp1 expression in the presence of viral miRNAs translated into decreased TGF-1β activity demonstrating that TGF-β function is subject to miRNA control [56].
A homeobox is a DNA sequence found within genes that are involved in the regulation of patterns of development or morphogenesis in animals, fungi and plants. Genes that have a homeobox are called homeobox genes and form the homeobox gene family. The most studied and the most conserved groups of homeodomain proteins are the Hox genes, which control segmental patterning during development; however, not all homeodomain proteins are Hox proteins. The homeobox gene GAX inhibits angiogenesis and induces p21(WAF1/CIP1) expression in vascular endothelial cells. GAX activates p21(WAF1/CIP1) through multiple upstream AT-rich sequences [57]. miR-130a regulates the angiogenic phenotype of vascular endothelial cells largely through its ability to modulate the expression of GAX and HOXA5 [58]. HOXA5 is anti-angiogenic and regulates vascular patterning [59].
Vascular Diseases
miRNAs represent a new therapeutic target for the treatment of proliferative vascular diseases such as atherosclerosis, restenosis after angioplasty, transplantation arteriopathy and stroke. The first evidence describing a role of miRNAs in vascular disease originated from microarray analyses demonstrating that miRNAs are aberrantly expressed in the vascular walls after balloon injury [52, 60]. Modulating an aberrantly overexpressed miRNA, miR-21, via antisense-mediated knockdown, attenuated neointimal lesion formation. In vitro, miR-21 expression in dedifferentiated vascular smooth muscle cells was higher than that in freshly isolated differentiated cells. Depletion of miR-21 resulted in decreased cell proliferation and increased cell apoptosis in a dose-dependent manner. miR-21-mediated cellular effects were further confirmed in vivo in balloon-injured rat carotid arteries. PTEN and Bcl-2 are involved in miR-21-mediated cellular effects.
miRNAs are also directly implicated in mechanisms by which the +1166 A/C polymorphism can lead to increased angiotensin II (AngII) type 1 receptor (AT1R) densities and possibly cardiovascular disease [61]. The silent polymorphism (+1166 A/C) in the human AT1R gene has been associated with cardiovascular disease, possibly as a result of enhanced AT(1)R activity. Because this polymorphism occurs in the 3′-UTR of the human AT1R gene, the biological importance of this mutation has always been questionable. Recently, it has been observed that a +1166 A/C polymorphism occurs in a cis-regulatory site which is recognized by miR-155. When the +1166 C allele is present, base-pairing complementarity is interrupted, and the ability of miR-155 to interact with the cis-regulatory site is decreased. As a result, miR-155 no longer attenuates translation as efficiently. Mature miR-155 is abundantly expressed in the same cell types as the AT1R (e.g. endothelial and vascular smooth muscle). Down-regulation of miR-155 expression in human primary vascular smooth muscle cells induced endogenous human AT1R expression and AngII-induced ERK1/2 activation [61]. Compared with that of Wistar-Kyoto rats, miR-155 expression was decreased in the aorta of adult spontaneously hypertensive rats and is negatively correlated with blood pressure, suggesting that miR-155 is possibly involved in the development and pathologic progress of hypertension. miR-155 expression is significantly lower in the aorta of 16-week-old spontaneously hypertensive rats than in age-matched Wistar-Kyoto rats. Furthermore, miR-155 expression is negatively correlated to blood pressure. miR-208 expression in the aorta is also negatively correlated with blood pressure as well as age [62]. Whether age-dependent changes in miR-208 expression in the rat aorta influence blood vessel development in the aged remains an open question at present.
miRNA Regulation of Stem Cell Biology
Stem cells in the blood and the vessel wall have the ability to repair endothelial cells after extensive loss. Stem cell self-renewal and differentiation are defined by the dynamic interplay between transcription, epigenetic control and posttranscriptional regulators, including miRNAs, whose key role in stem cell biology is just emerging [63, 64]. One of the key characteristics of stem cells is their capacity for self-renewal for long periods of time. In this respect, stem cells are similar to cancer cells, which also have the ability to escape cell cycle stop signals. Some miRNAs are specifically expressed in stem cells, and control stem cell self-renewal and differentiation via negative regulation of the expression of certain key genes in stem cells [65]. miRNAs are implicated in the proper control of germline stem cell (GSC) division in Drosophila melanogaster. Analysis of GSCs mutant for dicer-1 (dcr-1), the double-stranded RNaseIII essential for miRNA biogenesis, revealed a marked reduction in the rate of germline cyst production. These dcr-1 mutant GSCs exhibit a normal phenotype but are defective in cell cycle control. On the basis of cell cycle markers and genetic interactions, it may be understood that dcr-1 mutant GSCs are delayed in the G1 to S transition, which is dependent on the cyclin-dependent kinase inhibitor Dacapo, suggesting that miRNAs are required for stem cells to bypass the normal G1/S checkpoint. Hence, the miRNA pathway might be part of a mechanism that makes stem cells insensitive to environmental signals that normally stop the cell cycle at the G1/S transition [66]. In addition, miRNAs signal cell fate [67] and stem cell phenotype [68].
miRNAs are components of the molecular circuitry that controls mouse hematopoiesis. In one of the earlier studies, three miRNAs that are specifically expressed in hematopoietic cells were identified. The expression of these miRNAs was dynamically regulated during early hematopoiesis and lineage commitment. One of these miRNAs, miR-181, was preferentially expressed in the B-lymphoid cells of mouse bone marrow, and its ectopic expression in hematopoietic stem/progenitor cells led to an increased fraction of B-lineage cells in both tissue culture differentiation assays and adult mice [69]. Some human miRNAs are linked to leukemias: the miR-15a/miR-16 locus is frequently deleted or down-regulated in patients with B-cell chronic lymphocytic leukemia, and miR-142 is at a translocation site found in a case of aggressive B-cell leukemia [70]. miRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation [71]. miRNA-155 transduction greatly reduces both myeloid and erythroid colony formation of normal human CD34+ hematopoietic stem-progenitor cells [72]. In sum, miRNA-mediated posttranscriptional regulation influences the development and function of blood cells.
miRNAs target megakaryocytic transcription factors and regulate megakaryocytopoiesis. Megakaryocytic differentiation is associated with down-regulation of miR-10a, miR-126, miR-106, miR-10b, miR-17 and miR-20. miR-130a targets the transcription factor MAFB, which is involved in the activation of the GPIIB promoter, a key protein for platelet physiology. In differentiated megakaryocytes, miR-10a expression is inverse to that of HOXA1, a direct target of miR-10a [73]. Innoculating mice with lentiviral vectors encoding target sequences of endogenous miRNAs has shown that miRNAs are highly efficient at segregating gene expression among different tissues. Transgene expression from vectors incorporating target sequences for miR-142–3p effectively suppressed intravascular and extravascular hematopoietic lineages, whereas expression was maintained in non-hematopoietic cells [74]. Bone marrow mesenchymal stem cells participate in myocardial repair following myocardial infarction. However, their in vivo reparative capability is limited due to lack of survival in the infarcted myocardium. Adenoviral miRNA targeting CXCR4 plus SDF-1α blocked the engraftment of mesenchymal stem cells to the site of injury and compromised neomyoangiogenesis [75]. These observations underscore the significance of miRNA-dependent regulation of CXCR4 as well as SDF-1α in influencing neomyoangiogenesis at the site of myocardial infarction.
miRNA-Based Therapeutic Opportunities for the Treatment of Vascular Disorders
Efforts to biologically validate computationally predicted target genes for specific miRNAs are in progress. To examine the regulatory role of small RNAs at the transcriptional level, 21-nt dsRNAs targeting selected promoter regions of human genes, E-cadherin, p21(WAF1/CIP1) and VEGF, have been designed and synthesized. Transfection of these dsRNAs into human cell lines caused long-lasting and sequence-specific induction of targeted genes [76]. Hypoxia, when modest and sub-extreme, serves as a cue for angiogenesis [77]. miRNAs seem to have emerged as an important component of that cue. A significant proportion of the hypoxia-sensitive miRNAs are also overexpressed in human cancers, suggesting a role in tumorigenesis. Current evidence leads to the hypothesis that hypoxia-sensitive miRNAs could affect important processes such as apoptosis, proliferation and angiogenesis. The expression of several hypoxia-sensitive miRNAs are induced in response to hypoxia-inducible factor activation, thus extending its repertoire of targets beyond translated genes [78]. Specifically, miR-210 is hypoxia inducible and supports the angiogenic properties of endothelial cells by down-regulating its target gene ephrin receptor A3 [79]. Ephrin receptor A3, also known as EPHA3, is a human gene that belongs to the ephrin receptor subfamily of the protein tyrosine kinase family. Soluble EPHA receptors are known to inhibit tumor angiogenesis and progression in vivo [80]. EPHA3 encodes a protein that binds EPHA ligands. miR-210 overexpression in endothelial cells stimulates the formation of capillary-like structures on Matrigel and VEGF-driven cell migration. Conversely, miR-210 blockade inhibits the formation of capillary-like structures stimulated by hypoxia and decreased cell migration in response to VEGF [79].
Nitrate-containing compounds have been used for medicinal purposes for over 150 years. Today, nitric oxide (NO) is broadly recognized as a major vascular signaling molecule with functions such as smooth muscle relaxation, inhibition of platelet and leukocyte aggregation, promotion of angiogenesis and attenuation of vascular smooth muscle cell proliferation. NO influences two key aspects of O2 supply and demand. It regulates vascular tone and blood flow by activating soluble guanylate cyclase in the vascular smooth muscle, and it controls mitochondrial O2 consumption by inhibiting cytochrome c oxidase [81]. In the late 1980s, an enzyme that produces NO from L-arginine was described. In 1990, the first of such enzymes was purified. Today, NO synthase (NOS; EC 1.14.13.39) represents one of the best-studied enzymes primarily because of its impact on health and disease. NOS catalyzes the production of NO and L-citrulline from L-arginine, O2 and NADPH-derived electrons in the presence of several specific cofactors and prosthetic groups such as thiolate-bound heme, FAD and FMN, calmodulin and Ca2+ (for constitutive NOS only), and (6R)-5,6,7,8-tetrahydro-l-biopterin (tetrahydrobiopterin; BH4). In mammalian species, three types of NOSs orchestrate the production of NO from L-arginine, i.e. nNOS (neuronal NOS or NOS1), iNOS (inducible NOS or NOS2) and eNOS (endothelial NOS or NOS3) [82, 83]. The endothelium plays a key role in the maintenance of vascular homeostasis, and increased oxidative stress in vascular disease leads to reduced NO bioavailability and impaired endothelium-dependent relaxation of resistance vessels. NOS3 expression is vital for endothelial function. Numerous physiological and pathophysiological stimuli modulate NOS3 expression via mechanisms that alter steady-state NOS3 mRNA levels. These mechanisms involve changes in the rate of NOS3 gene transcription and alterations in NOS3 mRNA processing and stability. Repeats (27 nts) in intron 4 play a cis-acting role in NOS3 promoter activity. Intron-based miRNAs in NOS3 induce significant gene-specific transcriptional suppression, which could be an effective negative feedback regulator for NOS3 expression [84]. Soluble CD40 ligand (sCD40L) is contained in platelet granules and thus its presence in the blood is a marker of platelet activation. By interacting with CD40, which is found on endothelial and smooth muscle cells, sCD40L trigger the release of inflammatory mediators leading to increased activity of matrix metalloproteinases and activation of the coagulation cascade. In studies examining the effects and mechanisms of sCD40L on endothelial dysfunction in both human coronary artery endothelial cells and porcine coronary artery rings, it has been noted that sCD40L reduces eNOS levels via mechanisms involving miRNAs [85].
Although down-regulation of the miRNA-processing enzymes Dicer and Drosha impairs angiogenesis, only a few specific miRNAs targeting endothelial cell function and angiogenesis have been identified [86]. miR-221 and miR-222 block endothelial cell migration, proliferation and angiogenesis in vitro by targeting the stem cell factor receptor c-Kit and indirectly regulating eNOS expression. A pro-angiogenic function has been established for the miR-17–92 cluster, which promotes tumor angiogenesis in vivo. Expression of let7f and miR-27b contributes to in vitro angiogenesis. miR-126 regulates the response of endothelial cells to VEGF. This was observed in endothelial cells derived from mouse embryonic stem cells and in developing mouse embryos. Loss of vascular integrity and hemorrhage during embryonic development were noted in response to knockdown of miR-126 in zebra fish. Such angiogenic effects of miR-126 are mediated by directly repressing negative regulators of the VEGF signaling pathway such as Sprouty-related protein 1 (SPRED1) and phosphoinositol-3 kinase regulatory subunit 2 [87]. SPRED1 negatively regulates VEGF-induced ERK activation. SPREDs also play an important role in vascular development by negatively regulating VEGF-C/VEGFR-3 signaling [88]. Increased expression of SPRED1 or inhibition of VEGF signaling in zebra fish causes defects similar to miR-126 knockdown [87]. In endothelial cells, miR-126 mediates developmental angiogenesis in vivo. It enhances the angiogenic functions of VEGF and fibroblast growth factor as well as promotes blood vessel formation by repressing the expression of SPRED1. Targeted deletion of miR-126 in mice causes leaky vessels, hemorrhaging and partial embryonic lethality due to loss of vascular integrity and defects in endothelial cell proliferation, migration and angiogenesis. The surviving subset of mutant animals displays defective cardiac neovascularization following myocardial infarction. The vascular abnormalities of miR-126 mutant mice resemble the consequences of diminished signaling by angiogenic growth factors, e.g. VEGF and fibroblast growth factor [89].
Negative regulation of inducible adhesion molecule expression represents a mechanism by which miR-126 exhibits anti-inflammatory functions. In endothelial cells, miR-126 negatively regulates inducible VCAM-1 expression. Lowering of miR-126 expression has been reported to enhance TNF-α-stimulated VCAM-1 expression. Conversely, overexpression of the precursor to miR-126 increases miR-126 levels and subsequently decreases inducible VCAM-1 expression. Functionally, decreasing endogenous miR-126 levels increases leukocyte adherence to endothelial cells [90]. E-selectin, also known as CD62E, is a cell adhesion molecule expressed only on cytokine-activated endothelial cells. Like other selectins, CD62E plays an important role in inflammation. miRNA constructs to suppress E-selectin expression productively diminish leukocyte recruitment into activated endothelial cells [91]. The constructs tested were composed of miRNAs (miR-E1 and miR-E2) complementary to the human E-selectin cDNA. Delivery of these constructs to human aorta endothelial cells (HAECs) revealed that the E-selectin promoter was sufficiently activated in response to TNF-α, and that miR-E1 and miR-E2 effectively suppress inducible E-selectin expression resulting in the significant inhibition of leukocyte adhesion [91].
While most miRNAs are transcribed from their own dedicated genes, some map to introns of ‘host’ transcripts, the biological significance of which remains unknown. Intronic miRNAs from tissue-specific transcripts, or their natural absence, make cardinal contributions to cellular gene expression and phenotype. These findings also open the door to tissue-specific miRNA therapy [92]. Prostate cells, naturally devoid of EGF-like domain 7 (Egfl7) transcripts, are also deficient in miR-126* generated from splicing and processing of its ninth intron. Use of recombinant and synthetic miRNAs or a specific antagomir established a role of miR-126* in silencing prostein in non-endothelial cells. Two miR-126*-binding sites in the 3′-UTR of the prostein mRNA required for translational repression have been mapped. Prostein is a recently described molecule expressed at the mRNA level in a prostate-specific manner. Transfection of synthetic miR-126* into prostate cancer LNCaP cells strongly reduced the translation of prostein. Loss of prostein expression was associated with a reduction in LNCaP cell migration and invasion [92]. Identification of more specific miRNAs regulating specific aspects of angiogenesis will help formulate effective strategies to manipulate angiogenic outcomes through miRNA-based tools.
miRNA-Based Therapeutic Tools
miRNA-based therapeutics now represents one of the major commercial hot spots in today’s biotechnology market space [93]. The discovery of miRNAs as regulators of gene expression has created new options for the design of therapeutic agents that could modify gene expression in disease. The ability to modulate miRNA activity in vivo may have tremendous impact on disease therapy and on in vivo research opportunities. The direction towards this has been actually set. There are, essentially, two possible therapeutic approaches: either overexpression or silencing of the prospective miRNA. For the former, delivery of corrective synthetic miRNAs in the form of (siRNA-like) dsRNA will be beneficial. If a disease phenotype derived from abnormal inhibition of mRNAs, e.g. due to excessive expression of miRNAs or shutting down an miRNA, will be positive, oligonucleotides complementary to either the mature miRNA or its precursors can be designed, so that the miRNAs will be arrested and will not be able to compliment the target mRNA. The fundamental properties of these therapeutic oligonucleotides (for either up- or down-regulation of miRNA activity) will be delivery in vivo, degradation avoidance, specificity and high binding affinity to RNA. This can be achieved by modification of the nts, especially the addition of chemical groups to the 2′-hydroxyl group, which has been shown to be effective at knocking down the function of specific miRs. Three forms of chemically modified oligonucleotides may be used to silence miRNAs [94]. These include 2′-O-methyl-group (OMe)-modified oligonucleotides, 2′-O-methoxyethyl (MOE)-modified oligonucleotides, which have higher affinity and specificity to RNA than their OMe-analogs, and locked nucleic acid (LNA)-modified oligonucleotides (LNA-antimiR), in which the 2′-O-oxygen is bridged to the 4′-position via a methylene linker to form a rigid bicycle, locked into a C3′-endo (RNA) sugar conformation. Recently, striking data demonstrating the therapeutic power of LNA-antimiR technology have been reported in non-human primates. Simple systemic delivery of unconjugated PBS-formulated LNA-antimiR potently antagonized the liver-expressed miR-122 in non-human primates. Acute administration by intravenous injections of 3 or 10 mg/kg LNA-antimiR to African green monkeys resulted in uptake of the LNA-anti-miR in the cytoplasm of primate hepatocytes and formation of stable heteroduplexes between the LNA-antimiR and miR-122. This was accompanied by depletion of mature miR-122 and dose-dependent lowering of plasma cholesterol. Efficient silencing of miR-122 was achieved in primates by three doses of 10 mg/kg LNA-antimiR, leading to a long-lasting and reversible decrease in total plasma cholesterol without any evidence for LNA-associated toxicities or histopathological changes in the study animals [95].
Specific miRNA silencing is achieved by antisense targeting. The genetic approaches include knockout of miRNA genes in mice, mutation of miRNA target sites in protein-encoding genes and conditional alleles of the miRNA-processing gene Dicer1, leading to a deficiency in all mature miRNAs. The non-genetic approaches utilize the OMe-modified oligonucleotides that go through further modifications to be compatible for in vivo delivery. This method was successfully demonstrated in vivo using modified oligonucleotides named ‘antagomirs’ [96, 97]. Chemically modified, cholesterol-conjugated single-stranded RNA analogues complementary to miRNAs, antagomirs, have been designed. They are synthesized starting from a hydroxyprolinol-linked cholesterol solid support and 2′-OMe phosphoramidites. Antagomirs harbor optimized phosphorothioate modifications, require >19-nt length for highest efficiency and can discriminate between single nt mismatches of the targeted miRNA. Degradation of different chemically protected miRNA/antagomir duplexes in mouse livers and localization of antagomirs in a cytosolic compartment that is distinct from processing bodies indicates a degradation mechanism independent of the RNAi pathway. Antagomirs, although incapable of silencing miRNAs in the central nervous system when injected systemically, efficiently target miRNAs when injected locally into the mouse cortex [98]. Antagomir treatment is effective in abolishing tumor growth in vivo, specifically in therapy-resistant neuroblastoma [99]. Intravenous injection of antagomir-122 decreased miR-122 levels specifically. Antagomir-122 resulted in a complete loss of miR-122 signal, and miR-122 levels were undetectable for as long as 23 days after injection. Antagomir-16 silences miR-16 in all body tissues besides the brain. Therefore, when using antagomirs, one can silence efficiently miRNAs in vivo in almost all body tissues. In the case of neuroblastoma, silencing miR-122 resulted in changes in cholesterol biosynthesis genes with down-regulation of HMGCR (3-hydroxy-3-methylgluta-ryl-CoA-reductase), a rate-limiting enzyme of endogenous cholesterol biosynthesis. Plasma cholesterol levels were decreased >40% in antagomir-122-treated animals. Moreover, antagomir injection did not seem to have any toxic effect. These lines of evidence validate the efficacy of antagomirs in vivo and should facilitate future studies to silence miRNAs for functional analysis and in clinically relevant settings.
Another form of modified oligonucleotides, 2′-MOE phosphorothioate-modified antisense oligonucleotide (ASO), has also been successful in the silencing of miRNAs in vivo [100]. Using this approach, miR-122 was successfully silenced by mere intraperitoneal injection of ASO. Verification of miR-122 silencing was proven by the increase in mRNA levels of four target genes of miR-122. No target mRNA changes were observed in mice treated with control ASO demonstrating specific inhibition of miR-122 activity in the liver. This study has taken another step forward by applying this technique in a disease model of obesity in mice. C57BL/6 mice fed a high-fat diet for 19 weeks were treated with miR-122 ASO. Blocking miR-122 resulted in a 35% decrease in plasma cholesterol levels compared to control mice, although the authors explained that there was a maximal relief threshold from the effect of silencing miR-122. Over-expression of miRNAs in vivo can be achieved [101]. pre-miR-1 plus flanking sequence was subcloned into –MHCclone26 or –MHCclone32 vectors and introduced into mice. Northern blots of the transgenic mice confirmed that they expressed miR-1. Next, Western blots of transgenic heart were performed. A significant decrease in Hand2 protein levels compared with non-transgenic littermates was observed, with no change in mRNA levels of Hand2. These observations confirmed that Hand2 is an miR-1 target in vivo and that up-regulation of a single miRNA using its precursor can elevate specific proteins levels.
Targeting non-coding genes such as miRNAs, which have the capacity to regulate large sets of coding genes, represents the future of gene therapy. Targeting a single coding gene to obtain desirable clinical outcomes has generated very modest results primarily limited to disorders caused by a single genetic mutation. Functional outcomes in vivo are often regulated by a large number of gene products which act in concert. For example, excessive delivery of VEGF may not be effective in stimulating functional angiogenesis and may end up exhibiting adverse outcomes such as enhanced vascular leakiness. Because single miRNAs regulate large sets of gene clusters, focus on miRNA-directed gene therapy seems prudent.
Table 1.
miRNAs and vascular function
| Vascular function and miR | Transcriptional regulator | 1st target | 2nd target | Ref. |
|---|---|---|---|---|
| Angiogenesis | ||||
| miR17-92 | Tsp1, CTGF | 51–53 | ||
| VEGF | Tsp1 | 43 | ||
| miR-27a | ZBTB10 | Sp1, Sp3, and Sp4 | 55 | |
| ? | KSHV | Tsp1 | TGF-β | 56 |
| miR-130a | GAX, HOXA5 | 58 | ||
| miR-378 | Fus-1 | 50 | ||
| miR-210 | hypoxia | EPHA3 | 79 | |
| sCD40L | NOS3 | 85 | ||
| miR-221, miR-222 | c-Kit | NOS3 stem cell factor | 31, 86 | |
| let7f miR-27b | 86 | |||
| miR-126 | SPRED1 | VEGF repressors and phosphoinositol 3 kinase regulatory subunit 2 | 87 | |
| Vasoconstriction, angiogenesis | ||||
| Intron-based miRNAs | NOS3 | 84 | ||
| Angiogenesis, vascular integrity | ||||
| miR-126 | SPRED1 | 89 | ||
| Cell adhesion, inflammation | ||||
| miR-126 | VCAM1 | TNF-α effects | 90 | |
| miR-E1, miR-E2 | CD62E | 91 | ||
| Cell migration, tumor invasion | ||||
| miR-126 | prostein | 92 | ||
| Pro-injury, cell proliferation, cell survival | ||||
| miR-21 | Bcl-2 | |||
| PTEN | 52 | |||
| Hypertension | ||||
| miR-155 | AT1R | AngII-induced ERK activation inverse association with blood pressure inverse association with blood pressure and age |
61 62 |
|
| miR-208 | 62 | |||
| Plasma cholesterol | ||||
| miR-122 | HMGCR | 95 | ||
Acknowledgments
This study was supported by NIH awards RO1 GM 077185 and GM 069589 to C.K.S., and by RO1 DK076566 to S.R.
References
- 1.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 2.Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 3.Tomaru Y, Hayashizaki Y. Cancer research with non-coding RNA. Cancer Sci. 2006;97:1285–1290. doi: 10.1111/j.1349-7006.2006.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Racz Z, Hamar P. Can siRNA technology provide the tools for gene therapy of the future? Curr Med Chem. 2006;13:2299–2307. doi: 10.2174/092986706777935186. [DOI] [PubMed] [Google Scholar]
- 5.Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perera RJ, Ray A. MicroRNAs in the search for understanding human diseases. Bio-Drugs. 2007;21:97–104. doi: 10.2165/00063030-200721020-00004. [DOI] [PubMed] [Google Scholar]
- 7.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miR-NAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smalheiser NR, Torvik VI. Complications in mammalian microRNA target prediction. Methods Mol Biol. 2006;342:115–127. doi: 10.1385/1-59745-123-1:115. [DOI] [PubMed] [Google Scholar]
- 14.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 15.Song L, Tuan RS. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res C Embryo Today. 2006;78:140–149. doi: 10.1002/bdrc.20070. [DOI] [PubMed] [Google Scholar]
- 16.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 19.McHale NA, Koning RE. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell. 2004;16:1730–1740. doi: 10.1105/tpc.021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong R, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 2004;45:369–385. doi: 10.1093/pcp/pch051. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Narry Kim V, Chua NH, Park CM. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005;132:3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- 23.Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007;48:391–404. doi: 10.1093/pcp/pcm008. [DOI] [PubMed] [Google Scholar]
- 24.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 25.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 26.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 27.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 28.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 29.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:471–477. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 30.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 31.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 32.Litz J, Krystal GW. Imatinib inhibits c-Kit-induced hypoxia-inducible factor-1α activity and vascular endothelial growth factor expression in small cell lung cancer cells. Mol Cancer Ther. 2006;5:1415–1422. doi: 10.1158/1535-7163.MCT-05-0503. [DOI] [PubMed] [Google Scholar]
- 33.Strumberg D. Preclinical and clinical development of the oral multikinase inhibitor sorafenib in cancer treatment. Drugs Today (Barc) 2005;41:773–784. doi: 10.1358/dot.2005.41.12.937959. [DOI] [PubMed] [Google Scholar]
- 34.Roboz GJ, Giles FJ, List AF, Cortes JE, Carlin R, Kowalski M, Bilic S, Masson E, Rosamilia M, Schuster MW, Laurent D, Feldman EJ. Phase 1 study of PTK787/ZK 222584, a small molecule tyrosine kinase receptor inhibitor, for the treatment of acute myeloid leukemia and myelodysplastic syndrome. Leukemia. 2006;20:952–957. doi: 10.1038/sj.leu.2404213. [DOI] [PubMed] [Google Scholar]
- 35.Cave AC, Brewer AC, Narayanapanicker A, Ray R, Grieve DJ, Walker S, Shah AM. NADPH oxidases in cardiovascular health and disease. Antioxid Redox Signal. 2006;8:691–728. doi: 10.1089/ars.2006.8.691. [DOI] [PubMed] [Google Scholar]
- 36.Roy S, Khanna S, Nallu K, Hunt TK, Sen CK. Dermal wound healing is subject to redox control. Mol Ther. 2006;13:211–220. doi: 10.1016/j.ymthe.2005.07.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen CK, Khanna S, Babior BM, Hunt TK, Ellison EC, Roy S. Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J Biol Chem. 2002;277:33284–33290. doi: 10.1074/jbc.M203391200. [DOI] [PubMed] [Google Scholar]
- 38.Ushio-Fukai M. VEGF signaling through NADPH oxidase-derived ROS. Antioxid Redox Signal. 2007;9:731–739. doi: 10.1089/ars.2007.1556. [DOI] [PubMed] [Google Scholar]
- 39.Shilo S, Roy S, Khanna S, Sen CK. Micro-RNA in cutaneous wound healing: a new paradigm. DNA Cell Biol. 2007;26:227–237. doi: 10.1089/dna.2006.0568. [DOI] [PubMed] [Google Scholar]
- 40.Hammell CM. The microRNA-argonaute complex: a platform for mRNA modulation. RNA Biol. 2008;5:123–127. doi: 10.4161/rna.5.3.6570. [DOI] [PubMed] [Google Scholar]
- 41.Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2008;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, Han J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118:1944–1954. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuccoli A, Poliseno L, Rainaldi G. miRNAs regulate miRNAs: coordinated transcriptional and post-transcriptional regulation. Cell Cycle. 2006;5:2473–2476. doi: 10.4161/cc.5.21.3422. [DOI] [PubMed] [Google Scholar]
- 45.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feld mann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coller HA, Forman JJ, Legesse-Miller A. ‘Myc’ed messages’: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 49.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 50.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 53.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 55.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 56.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Leal AD, Patel S, Gorski DH. The homeobox gene GAX activates p21WAF1/CIP1 expression in vascular endothelial cells through direct interaction with upstream AT-rich sequences. J Biol Chem. 2007;282:507–517. doi: 10.1074/jbc.M606604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3:240–252. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
- 60.Silvestri P, Rigattieri S, Loschiavo P. Does the effect of microRNAs in vascular neointimal formation depend on cell cycle phase? Circ Res. 2008;102:e101. doi: 10.1161/CIRCRESAHA.108.175661. author reply e102. [DOI] [PubMed] [Google Scholar]
- 61.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microRNA-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Xu CC, Han WQ, Xiao B, Li NN, Zhu DL, Gao PJ. Differential expression of microRNAs in the aorta of spontaneously hypertensive rats (in Chinese) Sheng Li Xue Bao. 2008;60:553–560. [PubMed] [Google Scholar]
- 63.Yang Z, Wu J. MicroRNAs and regenerative medicine. DNA Cell Biol. 2007;26:257–264. doi: 10.1089/dna.2006.0548. [DOI] [PubMed] [Google Scholar]
- 64.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 65.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 67.Raftopoulou M. microRNA signals cell fate. Nat Cell Biol. 2006;8:112. doi: 10.1038/ncb0206-112. [DOI] [PubMed] [Google Scholar]
- 68.Micro-RNA’s function to maintain stem cell phenotype. Cancer Biol Ther. 2007;6:305. [PubMed] [Google Scholar]
- 69.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 70.Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 71.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Georgantas RW, 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, Calin GA, Liu CG, Bloomfield CD, Andreeff M, Croce CM. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 75.Zhang D, Fan GC, Zhou X, Zhao T, Pasha Z, Xu M, Zhu Y, Ashraf M, Wang Y. Over-expression of CXCR4 on mesenchymal stem cells augments myoangiogenesis in the infarcted myocardium. J Mol Cell Cardiol. 2008;44:281–292. doi: 10.1016/j.yjmcc.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen CK. Wound healing essentials: let there be oxygen. Wound Repair Regen. 2009;17:1–18. doi: 10.1111/j.1524-475X.2008.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–671. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 79.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brantley DM, Cheng N, Thompson EJ, Lin Q, Brekken RA, Thorpe PE, Muraoka RS, Cerretti DP, Pozzi A, Jackson D, Lin C, Chen J. Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene. 2002;21:7011–7026. doi: 10.1038/sj.onc.1205679. [DOI] [PubMed] [Google Scholar]
- 81.Chen K, Pittman RN, Popel AS. Nitric oxide in the vasculature: where does it come from and where does it go? A quantitative perspective. Antioxid Redox Signal. 2008;10:1185–1198. doi: 10.1089/ars.2007.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Knott AB, Bossy-Wetzel E. Nitric oxide in health and disease of the nervous system. Antioxid Redox Signal. 2008 doi: 10.1089/ars.2008.2234. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia Y. Superoxide generation from nitric oxide synthases. Antioxid Redox Signal. 2007;9:1773–1778. doi: 10.1089/ars.2007.1733. [DOI] [PubMed] [Google Scholar]
- 84.Zhang MX, Ou H, Shen YH, Wang J, Wang J, Coselli J, Wang XL. Regulation of endothelial nitric oxide synthase by small RNA. Proc Natl Acad Sci USA. 2005;102:16967–16972. doi: 10.1073/pnas.0503853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen C, Chai H, Wang X, Jiang J, Jamaluddin MS, Liao D, Zhang Y, Wang H, Bharadwaj U, Zhang S, Li M, Lin P, Yao Q. Soluble CD40 ligand induces endothelial dysfunction in human and porcine coronary artery endothelial cells. Blood. 2008;112:3205–3216. doi: 10.1182/blood-2008-03-143479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci. 2008;29:12–15. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 87.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taniguchi K, Kohno R, Ayada T, Kato R, Ichiyama K, Morisada T, Oike Y, Yonemitsu Y, Maehara Y, Yoshimura A. Spreds are essential for embryonic lymphangiogenesis by regulating vascular endothelial growth factor receptor 3 signaling. Mol Cell Biol. 2007;27:4541–4550. doi: 10.1128/MCB.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshizaki K, Wakita H, Takeda K, Takahashi K. Conditional expression of microRNA against E-selectin inhibits leukocyte-endothelial adhesive interaction under inflammatory condition. Biochem Biophys Res Commun. 2008;371:747–751. doi: 10.1016/j.bbrc.2008.04.160. [DOI] [PubMed] [Google Scholar]
- 92.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mack GS. MicroRNA gets down to business. Nat Biotechnol. 2007;25:631–638. doi: 10.1038/nbt0607-631. [DOI] [PubMed] [Google Scholar]
- 94.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 95.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 96.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 97.Yang M, Mattes J. Discovery, biology and therapeutic potential of RNA interference, microRNA and antagomirs. Pharmacol Ther. 2008;117:94–104. doi: 10.1016/j.pharmthera.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 98.Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and sub-cellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D. Antagomir-17–5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 101.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]