Abstract
Human telomeric DNA consists of tandem repeats of the DNA sequence d(GGGTTA). Oligodeoxynucletotide telomere models such as d[A(GGGTTA)3GGG] (Tel22) fold in a cation-dependent manner into quadruplex structures consisting of stacked G-quartets linked by d(TTA) loops. NMR has shown that in Na+ solutions Tel22 forms a ‘basket’ topology of four antiparallel strands; in contrast, Tel22 in K+ solutions consists of a mixture of unknown topologies. Our previous studies on the mechanism of folding of Tel22 and similar telomere analogs utilized changes in UV absorption between 270 and 325 nm that report primarily on G-quartet formation and stacking showed that quadruplex formation occurs within milliseconds upon mixing with an appropriate cation. In the current study, we assessed the dynamics and equilibria of folding of specific loops by using Tel22 derivatives in which the dA residues were serially substituted with the fluorescent reporter base, 2-aminopurine (2-AP). Tel22 folding induced by Na+ or K+ assessed by changes in 2-AP fluorescence consists of at least three kinetic steps with time constants spanning a range of ms to several hundred seconds. Na+-dependent equilibrium titrations of Tel22 folding could be approximated as a cooperative two-state process. In contrast, K+-dependent folding curves were biphasic, revealing that different conformational ensembles are present in 1 mM and 30 mM K+. This conclusion was confirmed by 1H NMR. Molecular dynamics simulations revealed a K+ binding pocket in Tel22 located near dA1 that is specific for the so-called hybrid-1 conformation in which strand 1 is in a parallel arrangement. The possible presence of this topologically specific binding site suggests that K+ may play an allosteric role in regulating telomere conformation and function by modulating quadruplex tertiary structure.
Recent analysis of the human genome reveals the presence of G-rich sequences which have a propensity to fold into quadruplex structures (1, 2). These G-rich regions include telomeres, certain transcriptional promoters, and immunoglobulin switch regions. Telomeres are nucleoprotein structures located at the ends of chromosomes (for a review see ref. (3)). In humans, telomeric DNA contains 30–50 tandem repeats of the sequence d(TTAGGG) which exist as a single-stranded overhang at the 3′ end of chromosomes (4). Telomeres maintain chromosomal integrity during cell division (3, 5, 6); the inability of most cell types to replace telomeric DNA during replication eventually results in cell death (7). Most cancer cells, by virtue of their ability to produce telomerase, are capable of maintaining telomeric DNA, thereby contributing to cellular immortality (8). Recent evidence suggests that these G-rich telomeric DNA sequences may fold into G-quadruplex structures in vivo (9).
A number of recent reviews summarize the details of quadruplex structure and function (10–13). In brief, quadruplex DNA consists of stacked planar, cyclic arrays of four dG residues linked by Hoogsteen hydrogen bonds involving the N1, N2, N7 and O6 atoms of each quartet. Quadruplex stability is markedly enhanced by coordination of a monovalent cation such as K+ or Na+ to the guanine O6 atoms which protrude into the central cavity of the G tetrad. Individual G quartets stack to form a structure with a central channel that contains a monovalent cation chelated either between quartets or within the center of the macrocyclic ring.
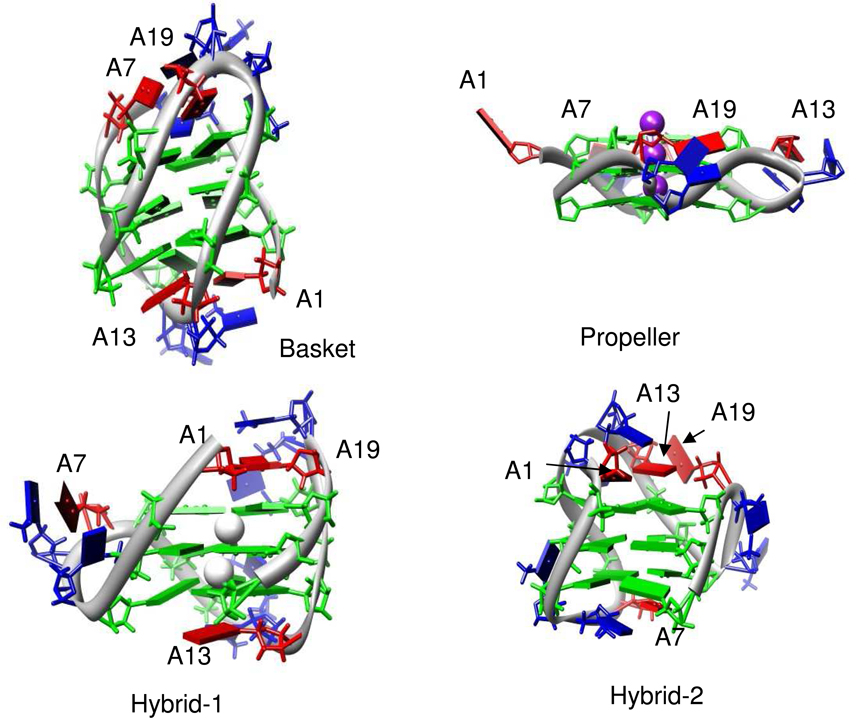
Oligonucleotides containing G quadruplexes are notable for their conformational diversity; indeed, changes in cation, loop sequence, or terminal bases may result in changes in quadruplex topology (13). A 22 nt model of the human telomeric sequence utilized in the experiments reported here, d[AG3(T2AG3)3] (Tel221), has been shown to exist in a different conformations under different conditions. From NMR data, Wang et al. (14) deduced that Tel22 in Na+-containing solution folds into a unimolecular ‘basket.’ This topology, depicted in Figure 1A, consists of an antiparallel arrangement of four strands with lateral loops connecting strand 1 to 2, strand 3 to 4, and a diagonal loop connecting strand 2 to 3. In contrast, Tel22 crystallized in K+ folds into an all-parallel structure in which the connecting loops form a ‘propeller’-like structure (Figure 1B) (15). However, NMR, as well as a variety other biophysical techniques, indicates that Tel22 in K+ solutions exists as an undefined mixture of structures (16, 17). Suggested topologies include a “chair” structure (four antiparallel strands connected by three lateral loops) and topologies in which there is a propeller-type loop connecting strand 1 to 2 or strand 3 to 4 (Figure 1, C and D) (18, 19). The latter structures in which the first two stands or the last two strands are in a parallel topology are the predominant conformers in K+ solutions of human telomeric sequences modified by addition of 5′ and 3′ nucleotides (17, 20, 21). The former has been referred to as a hybrid-1 structure and the latter as a hybrid-2 structure.
FIGURE 1.
Topological variants of G-quadruplexes formed under different conditions for the human telomeric oligodeoxynucleotide d[A(GGGTTA)3GGG] showing positions of loop adenine residues (red slabs). The basket structure is from the NMR-determined structure (PDB code 143D (14)). The propeller structure was solved by x-ray crystallography (PDB code 1KF1 (15)). The hybrid-1 and hybrid-2 structures were modeled from the NMR structures 2HY9 (43) and 2JPZ (44) as described in Methods. Green slabs represent G residues and blue slabs represent T residues. The diagrams were constructed using the molecular graphics program Chimera (48).
We recently compared the folding kinetics of oligonucleotides that form well-defined basket and hybrid-1 structures, Tel 22 in Na+ and TT-Tel22-A (d[TT(GGGTTA)3A) in K+ using rapid scanning stopped-flow spectrophotometry to assess the extent of quadruplex formation (22). For both of these sequences, a single exponential process with sequence-dependent time constants of 20–60 ms in 50 mM KCl was observed. In contrast in 100 mM NaCl, folding of both sequences consisted of three steps with relaxation times in the millisecond to second range. These kinetic data are consistent with the reaction sequences 1 and 2 below, where U represents an ensemble of unfolded conformers, I is an intermediate, and F represents the folded quadruplex structure:
| (1) |
and
| (2) |
Folding equilibria for these oligonucleotides were cooperative with respect to cation concentration and exhibited mid-point cation concentrations 10- to 20-fold less for K+ than for Na+ (22). This higher apparent affinity of K+ is consistent with the results of Hud et al. (23) who showed that the preference for K+ compared to Na+ coordination results from a higher desolvation energy required for its incorporation of Na+ into the quadruplex central channel.
In a subsequent study (24) we found that exchange of Na+ for K+ is also a multi-step process, consisting of three relaxations as determined by changes in the CD spectrum accompanying the cation-induced rearrangement shown in eq 3:
| (3) |
The changes in UV absorption in the 275–320 nm range that were used to monitor the folding transitions in the above studies most likely report predominately on G-quartet formation and may be less sensitive to loop conformation. Because the loops of Tel22 consist of dTTA triads, we reasoned that the fluorescent adenine analog, 2-aminopurine (2-AP), could be a sensitive indicator of loop conformational changes. 2-AP has been utilized extensively as a probe of oligonucleotide folding (25–29). For example, previous studies from our laboratory showed that the fluorescence properties of Tel22 analogs with 2-AP substituted serially for the four dA residues are sensitive to quadruplex folding topology (27). In addition, factors that influence 2-AP fluorescence are well characterized, thus making 2-AP-substituted Tel22 analogs excellent probes of loop structure (30–36).
EXPERIMENTAL PROCEDURES
Materials
Synthetic oligodeoxynucleotides were obtained from IDT, Inc. (Coralville, IA). Stock solutions of 500–750 µM in strand concentration were prepared by dissolving the de-salted, lyophilized oligonucleotide in 10 mM Bu4AmP, 1 mM EDTA, pH 7.0 (referred to as folding buffer). Oligonucleotide concentrations were determined from their absorbance at 260 nm using extinction coefficients supplied by IDT (228.5 mM−1 cm−1 for Tel22 and 215.5 mM−1 cm−1 for the Tel22 2-AP derivatives). NaCl, KCl, 2-aminopurine, monobasic tetrabutylammonium phosphate and tetrabutylammonium hydroxide were from Sigma, St. Louis, MO. Stock NaCl and KCl solutions for titration experiments were prepared in folding buffer.
Equilibrium measurements of cation-induced folding
The dependence of the extent of folding on cation concentration was assessed by measuring changes in UV absorption, CD, and for the 2-AP derivatives, fluorescence emission intensity. The spectrophotometric titrations were carried out as previously described (22). The absorbance at 295 nm or fluorescence intensity at 370 nm were allowed to equilibrate between additions of cation (usually 5–10 min)2. CD spectra were measured with a Jasco J-810 spectropolarimeter equipped with a magnetic mixer and a Peltier thermostat (Jasco USA, Easton, MD). Fluorometric experiments were conducted at 25 °C in a 1 cm × 1 cm quartz cuvette with a Fluromax-3 spectrofluorometer equipped with a magnetic mixer and a fluorescence polarization accessory (HORIBA Jobin Yvon Inc., Edison, NJ). 2-AP containing oligodeoxynucleotides were excited at 305 nm (2 nm bandwidth) and emission spectra were measured at 1-nm intervals from 320 to 460 nm using a 5 nm bandwidth. Fluorescence intensities were corrected for instrumental response using Fluoromax software, by subtracting a buffer spectrum. Emission spectra were corrected for day-to-day variation in excitation lamp intensity by normalizing to the intensity of the 342 nm Raman scattering peak of water. Fluorescence intensities are presented in units of counts/s/µM oligonucleotide.
Analysis of titration data
A two-state model for cation-induced folding of monomolecular G-quadruplexes is described by the equation U + nM ↔FMn where U and F designate the unfolded and folded states of the oligonucleotide and M is either Na+ or K+. The titration curves were fit to a modified Hill equation (eq. 4) corresponding to
| (4) |
where Si is the observed spectroscopic signal (e.g. absorbance, CD or fluorescence) determined at cation concentration i at the wavelength of maximum signal change. The parameters SU and SF (representing the signal of the unfolded and folded states), K0.5 (the midpoint cation concentration), and the Hill coefficient n were optimized using the non-linear least squares module in the program Origin 7.0 (OriginLab Corp., Northampton, MA). The two-state model is an oversimplification (as shown in our previous work (22) and amplified below) but it is useful for qualitative comparison of the folding isotherms.
Spectroscopic intermediates
Data matrices consisting of the wavelength-dependent cation titrations and the wavelength-dependent kinetic data were analyzed as previously described (22) using the method of singular value decomposition (SVD) to assess the presence of spectroscopic intermediates and depict their time- or [cation]-dependent profiles (37, 38). The application of SVD to multi-state DNA transitions has been previously described (39, 40); the interested reader is referred to these references for more details. The SVD data analysis was carried out with either MatLab 7.1 (The MathWorks, Natick, MA) or Specfit/32 Version 3.0.39 (Spectrum Software Associates, Marlborough, MA). The spectra and concentration profiles of the significant species in the equilibrium titrations were determined using the “model-free evolving factor analysis” module in Specfit/32 (41).
Folding kinetics
Kinetic constants for cation-induced quadruplex formation were determined by rapid scanning stopped-flow spectrophotometry using the instrument manufactured by OLIS, Inc., Bogart, GA as previously described (22). The kinetics of changes in 2-AP fluorescence was determined with the Fluoromax3 fluorometer equipped with a stopped-flow cuvette (SFA-20, High-Tech Scientific, Bradford-on-Avon, UK) or by manual addition of 3 M KCl or 3 M NaCl to the oligonucleotide solution while maintaining vigorous stirring with an in-cuvette magnetic stirrer. The nominal dead time (the time interval between the addition of cation and the initiation of data collection) was ~0.1 s and ~5 s for the stopped-flow and manual mixing methods, respectively. Fluorometric determinations of the kinetics of exchange of Na+ by K+ were assessed by rapidly adding KCl to a solution of the oligonucleotide pre-folded in NaCl as previously described (24). The progress curves for the folding and cation exchange reactions generally consisted of one or two exponentials as described by eq. 5
| (5) |
where y(t) is the fluorescence intensity at time t, y0 is the final equilibrium value of the fluorescence intensity, τ1 and τ2 are the relaxation times, and A1 and A2 are the corresponding signal amplitudes at t = 0. The values of y0, Ai and τi were optimized by fitting the experimentally determined progress curves to eq. 5 using the non-linear least squares module in Origin 7.0.
Fluorescence polarization
Steady-state fluorescence polarization values (P) of the unfolded and folded 2-AP oligonucleotides were determined at 25 °C with excitation at 305 nm and emission at 370 nm in a 1 cm × 1 cm quartz cuvette using the SpectraMax-3 fluorometer in polarization mode. Folding was initiated by rapidly adding the salt from 3 M stock solutions of either NaCl or KCl in folding buffer to a stirred solution of the unfolded oligonucleotide. P was determined at intervals of 6.7 s for 2000 s. P values for the unfolded oligonucleotide were estimated by averaging at least 100 successive measurements acquired before salt addition and for the folded oligonucleotide by averaging at least 500 measurements obtained after salt addition. The standard deviations of the mean P value were ± 0.002 to ± 0.005. Net changes in P were determined by subtracting the average P for the folded oligonucleotide from that of the unfolded oligonucleotide.
NMR experiments
Tel22 (179 µM) was dissolved in water/10% D2O + 50 µM DSS titrated to pH 6.5 with HCl. NMR spectra were recorded at 18.8 T on a 4-channel Varian Inova spectrometer (Palo Alto, CA) using an inverse triple resonance HCN probe. The intense solvent peak was suppressed using the Watergate sequence (42). The acquisition time was 1.5 s and the recycle time was 3 s. The data were zerofilled once, apodized using an unshifted Gaussian function and a 2 Hz line broadening exponential function. Chemical shifts were referenced to internal DSS. Spectra were recorded at 10 °C and 25 °C in the absence of K+ and in the presence of 0.75, 1.5, and 37 mM K+.
Molecular dynamics calculations and modeling
Starting structures of the hybrid-1 and hybrid-2 conformations were generated from the coordinates of the reported NMR structures (PDB codes 2HY9 (43) and 2JPZ (44)). Two K+ ions were placed between the adjacent G-tetrad planes in each quadruplex structure. The standard parm99.dat Amber force field was used and was modified using the frcmod.parmbsc0 parameter file (45–47). The models were solvated in a 10 Å box of TIP3P water using standard Amber 9.0 Leap rules to hydrate the systems. Potassium counter ions were added for overall charge neutrality. The systems were heated slowly and equilibrated for 250 ps with gradual removal of positional restraints on the DNA following this protocol: (i) minimize water, (ii) 50 ps MD (T = 100 °K) holding DNA fixed (100 kcal/mol Å−1), (iii) minimize water with DNA fixed (100 kcal/mol Å−1), (iv) minimize total system, (v) 50 ps MD (T = 100 °K) holding DNA fixed (100 kcal/mol Å−1), (vi) 50 ps MD (T = 300 °K) holding DNA fixed (100 kcal/mol Å−1), (vii) 50 ps MD (T = 300 °K) holding DNA fixed (50 kcal/mol Å−1), (viii) 50 ps MD (T = 300 °K) holding DNA fixed (10 kcal/mol Å−1), (ix) 50 ps MD (T = 300 °K) holding DNA fixed (1 kcal/mol Å−1). After the equilibration phase, an unconstrained production phase was then initiated and continued for 20 ns. Production runs of 20 ns after final equilibrium were used to obtain the average structures (200 snapshots in the last 2 ns), which were fully minimized. Simulations were performed in the isothermic isobaric ensemble (P = 1 atm, T = 300 °K). Periodic boundary conditions and the Particle-Mesh-Ewald algorithm were used. A 2.0 fs time step was used with bonds involving hydrogen atoms frozen using SHAKE. Molecular dynamics calculations were carried out with the AMBER program sander. The trajectories were analyzed using the PTRAJ module in AMBER and visualized using Chimera (48). Solvent accessible surface areas (SASA) were computed from structural coordinates in PDB file 143D for Tel22 in Na+ (14) or the coordinates for the Tel22 hybrid-1 or hybrid-2 computational models using NACCESS 2.1.1 (49).
RESULTS
Folding monitored by UV difference spectroscopy
We initially compared folding isotherms for Na+- and K+-induced folding of the 2-AP-oligonucletotides with those for unmodified Tel22. Quadruplex formation results in extensive changes in DNA UV absorption with a characteristic increase in absorbance at ~295 nm. The results of these titrations are summarized in Table 1; the corresponding cation-dependent difference spectra and titration curves given in “Supporting Information” (Figure S1). As previously observed (22), the folding transitions for Tel22 were cooperative with Hill coefficients of ~1.5 in KCl and ~2.8 in NaCl and K0.5(Na+) > K0.5(K+). Folding of 2-AP derivatives of Tel22 was also cooperative with respect to [M+], with n(Na+) > n(K+) and K0.5(Na+) > K0.5(K+). However, the values of the fitted parameters for some of the 2-AP derivatives were slightly different from the same parameters for folding unmodified Tel22. The most notable differences were a decrease in the n value of AP19 in NaCl, a nearly two-fold increase in K0.5(Na+) for AP7, and a two-fold decrease in K0.5(K+) for AP13 and AP19. These differences may reflect 2-AP-induced alterations in the ensemble of unfolded conformers, alterations in the conformation of the folded states, or a combination of the two (50). These differences in the binding parameters are not unexpected in view of the small differences in CD spectra of Tel22 and the four 2-AP derivatives in Na+- and K+-containing buffers along with minor differences in thermal stability of the folded structures that were previously described by Li et al. (Supplementary Information in ref. 27). It should also be noted that these fitted constants are empirical descriptions of the coupling between cation binding and oligonucleotide folding rather than pure cation binding constants or folding equilibrium constants (50).
Table 1.
Comparison Na+ and K+ concentrations for half-maximal folding (K0.5) and Hill coefficients (n) determined from titrations of cation-induced folding of Tel22 and 2-AP-Tel22 oligodeoxynucleotides. Folding was assessed from changes in absorbance at 295 nm, fluorescence at 370 nm and CD at 295 nm.
| Cation | Oligo- nucleotide |
A295 nm or a CD295 nm | F370 nm | ||
|---|---|---|---|---|---|
| K0.5 (mM) | n | K0.5 (mM) | n | ||
| Tel22 | 3.09 ± 0.05 | 2.8 ± 0.1 | na | ||
| 2.93 ± 0.04a | 2.8 ± 0.1 | ||||
| Na+ | AP1 | 5.9 ± 0.1 | 2.7 ± 0.1 | 8.3 ± 0.3 | 1.9 ± 0.1 |
| AP7 | 8.3 ± 0.08 | 2.3 ± 0.04 | 4.0 ± 0.1 | 3.1 ± 0.2 | |
| AP13 | 4.2 ± 0.1 | 1.9 ± 0.1 | 3.7 ± 0.2 | 2.1 ± 0.2 | |
| AP19 | 3.7 ± 0.1 | 1.3 ± 0.1 | 3.7 ± 0.2 | 2.8 ± 0.4 | |
| Tel22 | 0.253 ± 0.003 | 1.54 ± 0.03 | na | ||
| 0.256 ± 0.003a | 1.54 ± 0.03 | ||||
| K+ | AP1 | 0.50 ± 0.02 | 1.30 ± 0.05 | 0.50 ± 0.02 | 2.1 ± 0.2 |
| AP7 | 0.35 ± 0.004 | 1.73 ± 0.03 | 0.29 ± 0.03 | 1.5 ± 0.2 | |
| AP13 | 0.167 ± 0.004 | 1.50 ± 0.05 | 0.20 ± 0.01 | 4.5 ± 1.3 | |
| AP19 | 0.287 ± 0.005 | 1.74 ± 0.05 | 0.10 ± 0.006 | 8.9 ± 7.3 | |
Fluorescence emission spectra of 2-AP derivatives of Tel22
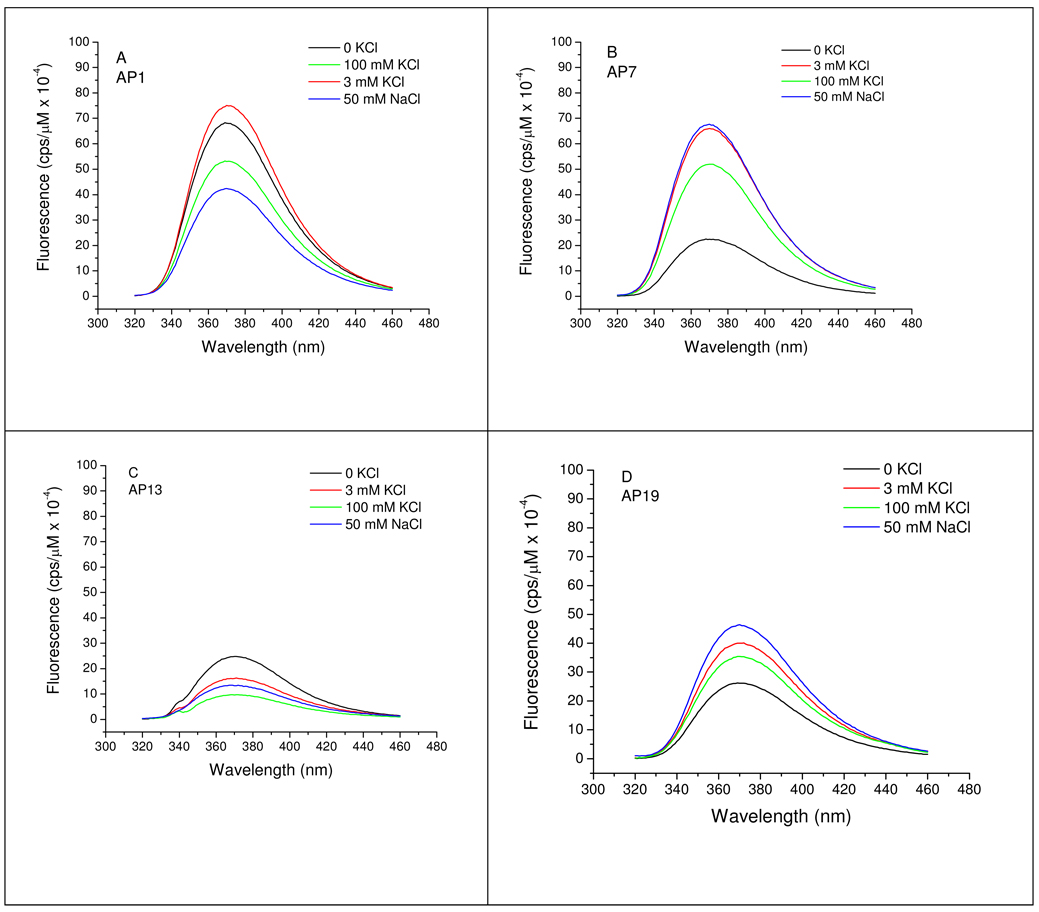
We next show cation-dependent changes in the 2-AP fluorescence emission spectra the modified Tel22 oligonucleotides. Previous studies from our laboratory (27) showed that the fluorescence of 2-AP individually substituted for the four dA residues of Tel22 depends on the substitution site and the identity of the cation. These differences were attributed to differences in positioning of the various loops connecting the G-quartets.
The emission spectra obtained in the current study for the unfolded and folded structures are shown in Figure 2. All of the spectra exhibited maxima near 370 nm, irrespective of folding state, cation or sequential location of the fluorophore. This is expected based on previous studies that show that the emission maximum of 2-AP is relatively insensitive to its local environment. However, the fluorescence quantum yield depended on the substitution position and on the cation and its concentration (Figure 2). This sensitivity of 2-AP quantum yield is expected based on studies that show that stacking 2-AP with nearest neighbor bases and solvent accessibility influence 2-AP emission intensity; in the current study, 2-AP at the 5′ end (e.g. AP1) has only a single neighboring nucleotide, whereas the other 2-AP residues positioned between T and G obviously have two neighbors and therefore exhibit higher quenching. It has also been shown that G is an especially effective quencher of 2-AP fluorescence in oligonucleotides by virtue of an electron transfer mechanism between G and 2-AP (51).
FIGURE 2.
Na+ and K+-dependent fluorescence emission spectra of 2-AP derivatives of Tel 22 as a function of cation concentration. Fluorescence intensities are normalized to 1 µM oligonucleotide and corrected for day-to-day variation in the fluorometer excitation lamp intensity by normalization to the intensity of the water Raman scatter peak at 340 nm. Conditions: [Tel22] = 0.5–0.6 µM in 10 mM Bu4AmP, 1 mM EDTA, pH 7.0, 25 °C. Excitation wavelength = 305 nm.
The relative effects of cation-induced folding on 2-AP emission intensities are summarized in Table 2. Na+-induced folding of AP1 and AP13 exhibited quenching in the folded state while folding of AP7 and AP19 resulted in enhanced fluorescence. In contrast, the fluorescence quantum yield of K+-induced folding of the 2-AP Tel22 derivatives was noticeably biphasic with respect to K+ concentration. In 3 mM KCl, AP1, AP7 and AP19 exhibited increased emission relative to the unfolded state while AP13 fluorescence was quenched in 3 mM KCl (Table 2, column 3). In 100 mM KCl, the fluorescence of AP1 and AP13 was quenched but AP7 and AP19 exhibited increased fluorescence relative to the unfolded states (Table 2, column 4). The order of quenching in the current study differs from that previously reported (27). In our previous study (in which the published spectra were determined at 5 °C) the order of emission intensity in Na+ was AP7 > AP19 > AP1 > AP13; for K+ the order was AP7 > AP1 ≈ AP19 > AP13. However, at 37 °C the order of intensities differed from that at 5 °C (Li, J. and Chaires, J. B., unpublished data) and is the same as that observed in the current experiments (conducted at 25 °C): AP7 > AP19 > AP1 > AP13 in Na+ and AP7 > AP1 > AP19 > AP13 in K+ . These results suggest temperature-dependent conformational heterogeneity at positions 1 and 19 (loop 3).
Table 2.
Relative changes in fluorescence emission at 370 nm induced by folding of 2-AP derivatives of Tel22 in NaCl and KCl.
| F/F0 | |||
|---|---|---|---|
| 50 mM NaCl | 3 mM KCl | 100 mM KCl | |
| AP1 | 0.6 | 1.2 | 0.8 |
| AP7 | 2.9 | 3 | 2.3 |
| AP13 | 0.5 | 0.7 | 0.4 |
| AP19 | 1.7 | 1.5 | 1.4 |
F is the fluorescence intensity in the indicated salt and F0 is the fluorescence intensity in the absence of added salt. The values of F and F0 were estimated from the emission spectra shown in Figure S1 in supporting information.
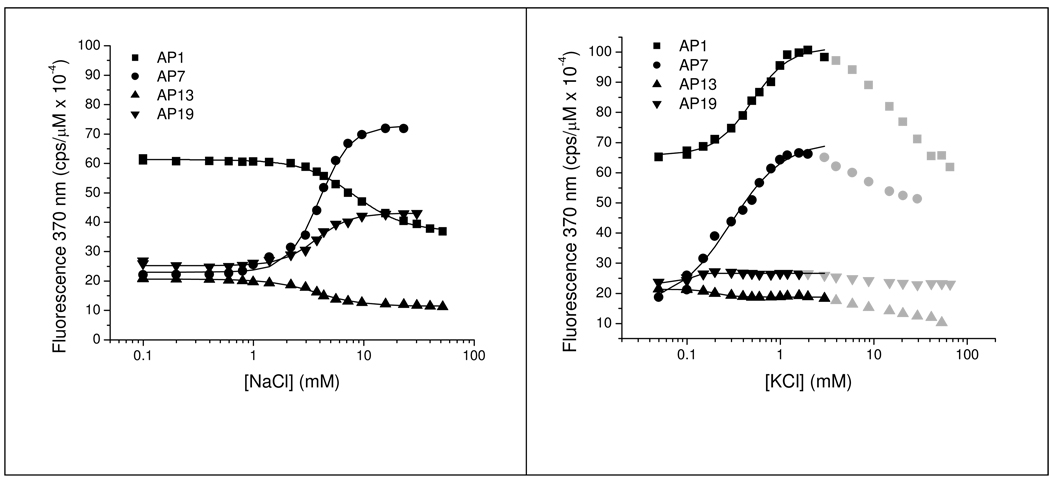
To compare loop folding with G-quartet formation, we carried out cation titrations monitored by changes in UV absorption and 2-AP fluorescence. Titration curves of AP1, AP7 and AP13 in Na+ monitored by fluorescence were approximately monophasic as shown in Figure 3A. The Na+-induced fluorescence change of AP19 was slightly biphasic, with a relatively small degree of quenching (<10%) apparent at up to ~1 mM NaCl, followed by a larger degree of fluorescence enhancement at higher NaCl concentrations. The n and K0.5 values obtained by fitting the data to eq. 4 are summarized in Table 1.
FIGURE 3.
Titration curves for cation-induced folding of 2-AP derivatives of Tel22 assessed by changes in fluorescence emission at 370 nm. Panel A shows the dependence of the fluorescence intensity on [NaCl] and panel B shows the dependence of the fluorescence intensity on [KCl] for the derivatives of Tel22 substituted with 2-AP at the indicated positions. The points represent the experimental fluorescence intensities (normalized to 1 µM oligonucleotide concentration) and the line shows the fit of the data points to eq. 3determined by non-linear least squares as described in the text. The data points were corrected for day-to-day variations in excitation intensity. The data points shown in grey in Panel B were not included in the fitting. The optimized fitting parameters are given in Table 1. Conditions: [Tel22] = 0.7 – 1.0 µM in 10 mM Bu4AmP, 1 mM EDTA, pH 7.0, 25 °C, excitation wavelength = 305 nm.
The K+ titration curves in Figure 3B reveal a clear biphasic dependence of emission intensity on [K+]. AP1 and AP19 showed increases of ~50% and ~10% in fluorescence intensity up to about 1 mM KCl followed by quenching at higher [KCl]. K+-induced folding of AP7 resulted in a 3.6-fold increase in fluorescence that was quenched at [KCl] >10 mM. AP13 fluorescence was quenched by ~20% at 1 mM KCl followed by nearly 100% near 100 mM KCl. Control experiments conducted with 2-AP showed that the fluorescence quantum yield of the free base is not affected by NaCl or KCl at concentrations up to at least to 100 mM. Thus we conclude from the biphasic titration curves that folding 2-AP Tel22 derivatives in KCl generates at least two classes of K+ binding sites. The high affinity sites are undoubtedly located within the quadruplex channel. Based on molecular dynamics simulations presented below, the lower affinity site involves residues dA1 and dT18.
Cation-induced changes in CD spectra
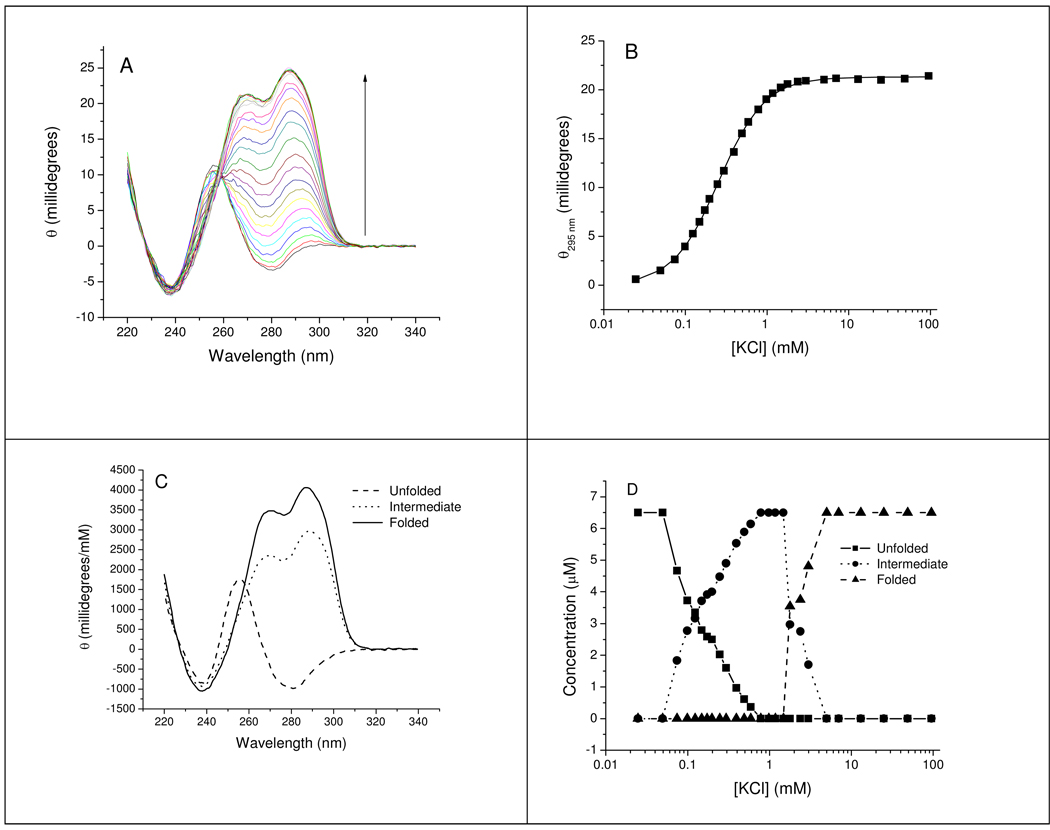
In view of the biphasic fluorescence titration data in Figure 3B, we examined the dependence of CD changes of native Tel22 on [M+]. Titrations with NaCl over the concentration range 0–100 mM monitored by CD are consistent with a folding equilibrium consisting of two significant spectroscopic species as confirmed by analysis of the wavelength-[Na+] data matrix by singular value decomposition (SVD) analysis (Figure S2). In contrast, CD titrations of K+-induced folding showed distinct spectral heterogeneity as a function of [KCl] as indicated by the lack of isodichroic points throughout the titration (Figure 4A). Figure 4B, shows changes in ellipticity at 295 nm for Tel22 over the KCl range 0–100 mM. Fitting the titration curve to eq. 4 gave a Hill coefficient of 1.5 and K0.5 of 0.26 mM KCl. Analysis of the wavelength-concentration data matrix set by SVD showed clear evidence for three significant spectroscopic species throughout the titration (Figure 4A). Evolving factor analysis allowed calculation of theoretical CD spectra for the starting species, the low K+ species and the high K+ species (Figure 4C) and their KCl-dependent concentration profiles (Figure 4D). The shape of the CD spectrum of the 1 mM KCl species resembles that of the final state formed at higher [KCl], but the intensity is lower. Both shapes are characteristic of hybrid-type structures rather than basket-type structures. Details of the SVD analysis of Na+ and K+ titrations are in Figures S3 and S4 of the supporting information.
FIGURE 4.
Titration of Tel22 with KCl determined by CD. Panel A shows the CD spectra for Tel22 at various KCl concentrations. The spectra were generated by titration with KCl between the concentrations of 0 and ~100 mM. The vertical arrows indicate the direction of the changes with increasing [K+]. The cation concentrations are summarized in Table S1 of supporting information. Panel B shows the values of ellipticity at 295 nm as a function of salt concentration. The lines show the least squares fit of the data points to eq. 3. The optimized fitting parameters are given in Table 1 for these curves. Panel C shows the theoretical CD spectra of the unfolded, intermediate and folded states species calculated by the model-free evolving factor analytical procedure as described in the text. Panel D shows the concentration profiles of the three significant spectroscopic species. The detailed results of the SVD analysis are given in the supporting information. Conditions: [Tel22] = 6.7 µM in 10 mM Bu4AmP, 1 mM EDTA, pH 7.0. Temperature = 25 °C.
KCl-dependent 1H NMR spectra of Tel22
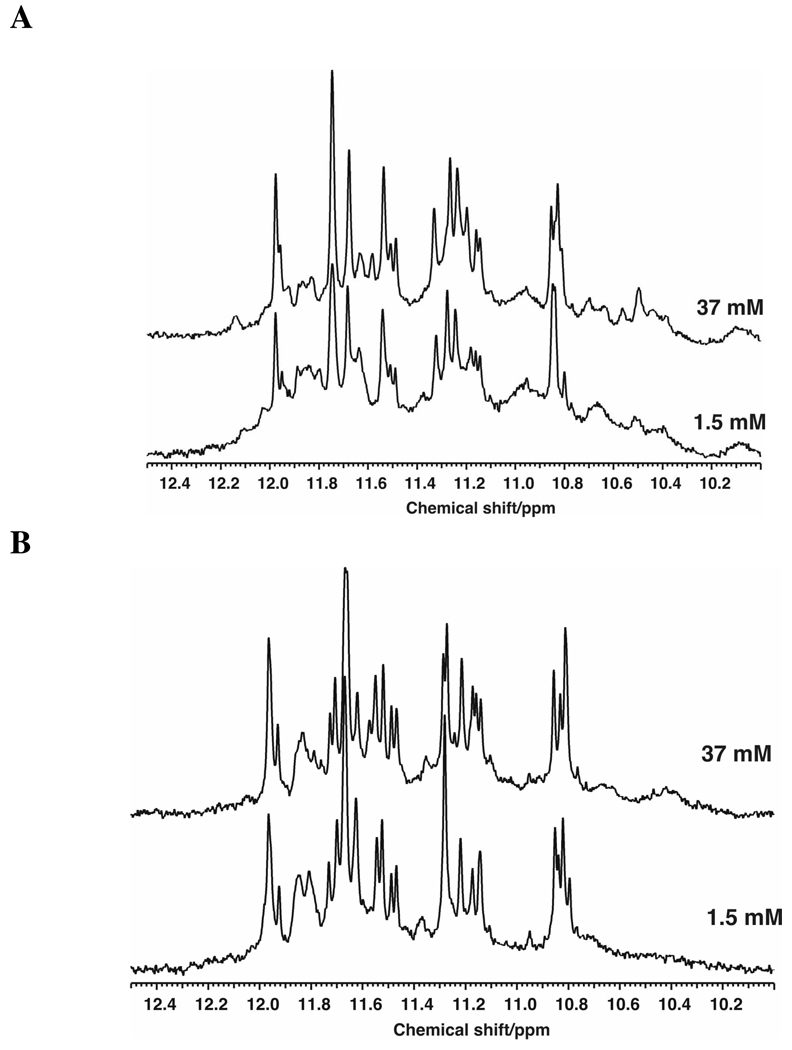
To confirm the presence of a [K+]-dependent conformational switch suggested by the fluorescence and CD titrations in Figure 3B and Figure 4C, we compared the 1-D 1H NMR spectra of Tel22 at lower and higher K+ concentrations. Figure 5 shows the imino proton region of the NMR spectra of Tel22 (179 µM) at 1.5 mM and 37 mM KCl at 10 °C and 25 °C. The spectrum at 37 mM K+ represents the “end-point” and comprises 20 different frequencies, and by intensities > 24 protons. This is roughly twice the number expected for a single 3-stack G-quadruplex such as the hybrid -1 or hybrid-2 forms (21). Indeed, the spectrum appears to be the superposition of two overlapping states in slow exchange. For example, the resonances near 10.8 ppm appear to represent species in a ratio of approximately 2:1 as assessed by the relative peak areas of the cluster. A similar spectrum was also obtained in the presence of 100 mM K+. In the absence of K+, only broad unresolved resonances were observed (data not shown). Sharpening of resonances from 10° to 25°C implies that the GN1H resonances are not exchanging significantly under these conditions, i.e. the quartets are stable even at 25°C and low salt.
FIGURE 5.
1H NMR spectra of Tel22 as a function of KCl concentration showing the imino proton region of the NMR spectra of Tel22 (178 µM) at 1.5 mM and 37 mM KCl at 10 °C (A) and 25°C (B).
Cation-induced folding kinetics monitored by UV absorbance changes
Kinetic experiments for cation-induced folding the Tel22 2-AP derivatives determined by rapid scanning stopped-flow spectrophotometry gave kinetic constants similar to those previously published for native Tel22 (22). Briefly, folding of the 2-AP-Tel22 derivatives induced by 100 mM NaCl took place in three steps with relaxation times of 20–60 ms, ~0.5 s and ~10s (Figure S5). The time constants for these steps are similar to those previously observed with Tel22. K+-induced folding of the 2-AP derivatives assessed by UV absorbance occurred in a single exponential process with relaxation times of ~40 ms in 50 mM KCl for all of the 2-AP derivatives (Figure S7). The observation of a single relaxation is the same as previously observed with native Tel22. From these two sets of experiments, we conclude that folding of the 2-AP derivatives and native Tel22 as assessed by changes in UV absorbance occurs via the same kinetic mechanisms expressed in eqs. 1 and 2.
The kinetics of both Na+- and K+-induced folding of the 2-AP-Tel22 derivatives determined by 2-AP fluorescence was multiphasic with the changes extending for milliseconds to minutes. To capture the complete range of folding times, we utilized both stopped-flow and manual mixing methods. These two procedures generated data sets for the folding kinetics of the four 2-AP derivatives in 100 mM NaCl, 3 mM KCl and 50 mM KCl. Representative progress curves are reproduced in Figure 6 for folding of AP19 in 100 mM NaCl and in Figure 7 for folding of AP7 in 3 mM and 50 mM KCl. The corresponding progress curves for Na+- and K+induced folding the remaining 2-AP derivatives of Tel22 are given in supporting information (Figure S6 and S7). The relaxation times and signal amplitudes derived from these experiments are summarized in Table 3.
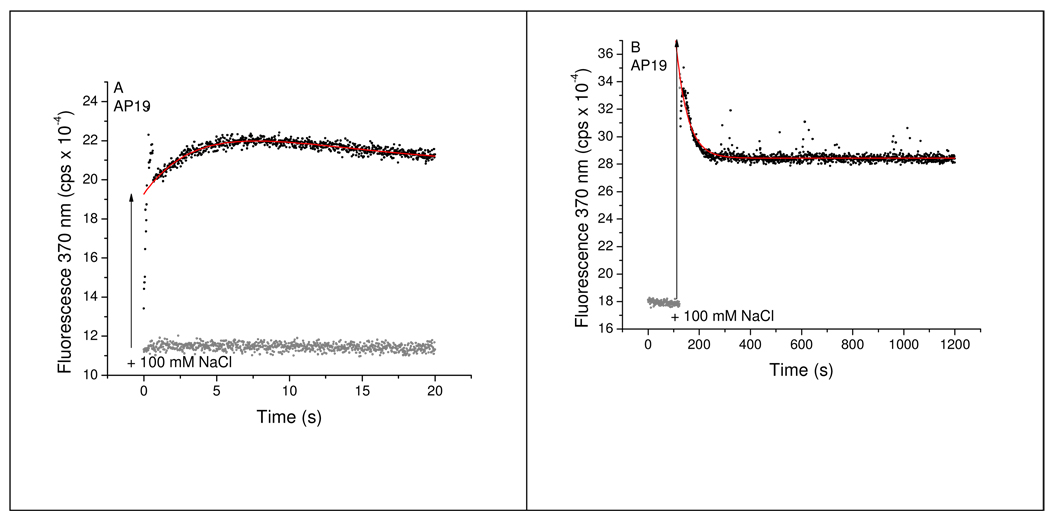
FIGURE 6.
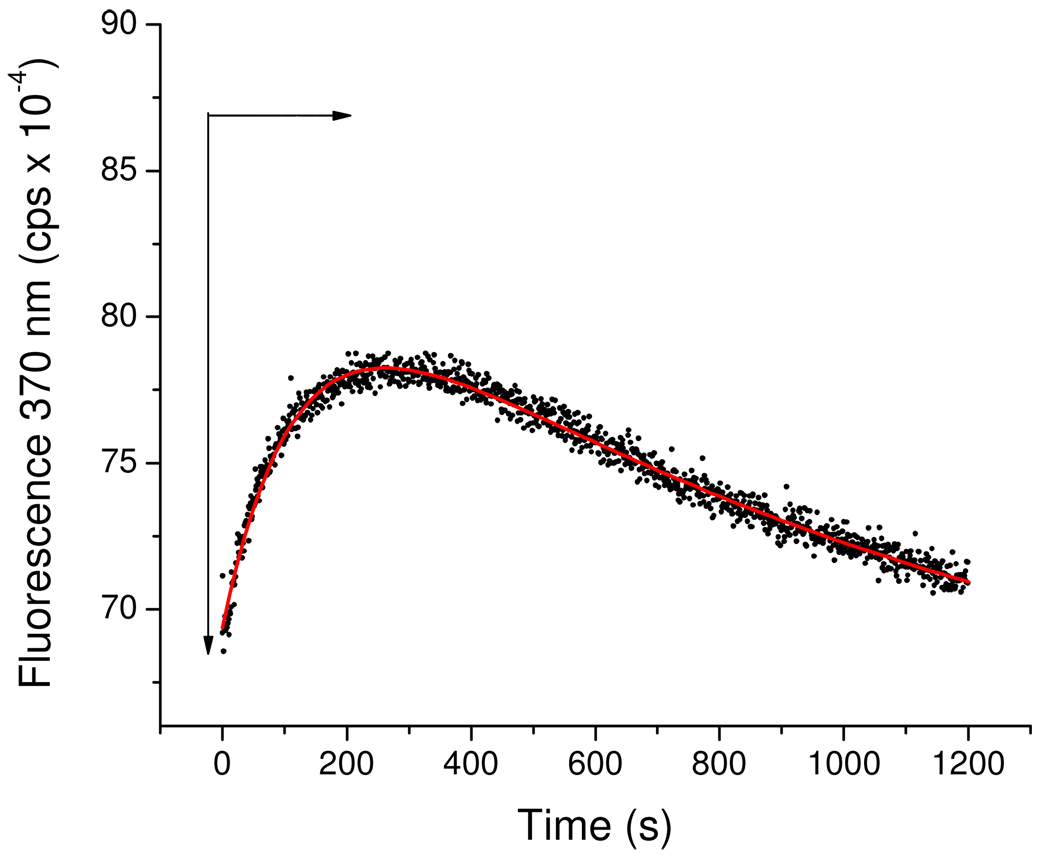
Kinetics of changes in 2-AP fluorescence accompanying Na+-induced folding of the AP19 derivative of Tel22. The data in panel A were obtained by stopped-flow mixing and the data in panel B were obtained by a manual mixing procedure. The data points in grey show the fluorescence intensity at 370 nm immediately prior to addition of NaCl (100 mM after mixing) to initiate folding and the black points show the fluorescence intensity after mixing with NaCl. Note the rapid change in emission that occurred during the mixing period (vertical arrow). The red line shows the best fit of the data points to a sum of two exponentials using the optimized parameters in Table 3.
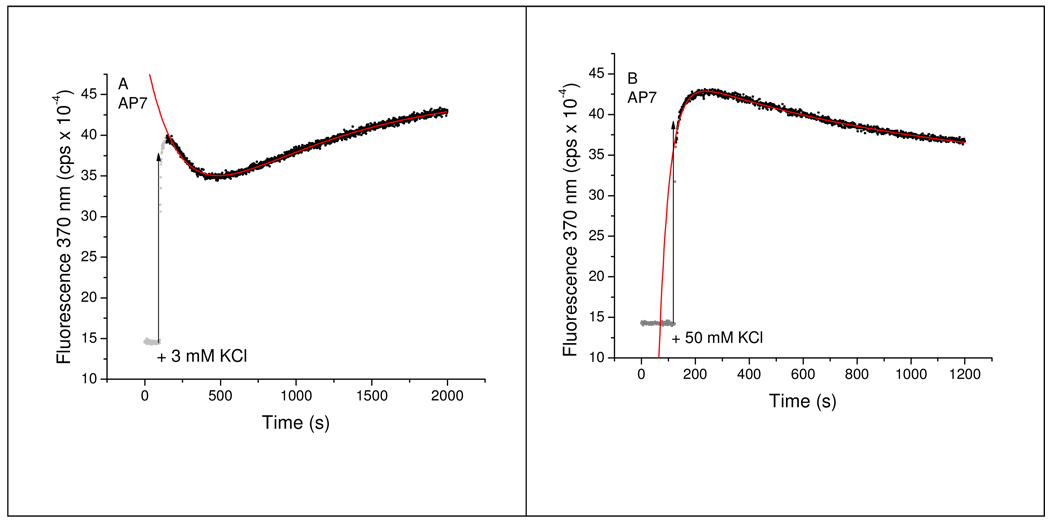
FIGURE 7.
Kinetics of changes in 2-AP fluorescence accompanying K+-induced folding of the AP7 (loop 1) Tel22 oligonucleotide. Panel A shows the folding progress curve in 3 mM KCl and panel B shows a progress curve for folding in 50 mM KCl. In both panels, the data points in grey show the fluorescence intensity at 370 nm immediately prior to addition of KCl to initiate the folding process (vertical arrow). The black points show progress curves after KCl addition and the red line shows the fit to the data points by non-linear least squares using the optimized parameters given in Table 3.
Table 3.
Fitted kinetic parameters for NaCl and KCl-induced folding of 2-AP derivatives of Tel22 determined by fluorescence.
| Position | y0 | A1 | τ1 (s) | A2 | τ2 (s) |
|---|---|---|---|---|---|
| Stopped-flow mixing: 100 mM NaCl | |||||
| AP1 | 28.84 ± 0.02 | 7.42 ± 0.06 | 4.04 ± 0.06 | 0 | |
| AP7 | 28.06 ± 0.24 | −5.2 ± 0.2 | 13 ± 1 | 0 | |
| AP13 | 6.87 ± 0.01 | 1.89 ± 0.05 | 1.47 ± 0.06 | 0 | |
| AP19 | 20.57 ± 0.41 | −7.2 ± 6.7 | 4.3 ± 1.4 | 5.9 ± 6.4 | 9.4 ± 6.2 |
| Manual mixing: 100 mM NaCl | |||||
| AP1 | 28.54 ± 0.01 | 23.9 ± 1.1 | 56 ± 1 | 0 | |
| AP7 | 35.07 ± 0.01 | −400 ± 50 | 30.3 ± 0.8 | 0 | |
| AP13 | 8.32 ± 0.00 | 1050.00 ± 1010.00 |
35 ± 4 | 0 | |
| AP19 | 28.43 ± 0.01 | 92 ± 9 | 45 ± 1 | 0 | |
| Manual mixing: 3 mM KCl | |||||
| AP1 | 71.73 ± 0.05 | −31.94 ± 0.04 | 975 ± 4 | 0 | |
| AP7 | 45.6 ± 0.2 | −24.1 ± 0.6 | 913 ± 33 | 28.9 ± 0.6 | 234 ± 6 |
| AP13 | 11.23 ± 0.01 | −2.65 ± 0.03 | 557 ± 11 | 0 | |
| AP19 | 29.36 ± 0.03 | −6.69 ± 0.03 | 788 ± 13 | 0 | |
| Stopped-flow mixing: 50 mM KCl | |||||
| AP1 | 31.5 ± 0.9 | 3.7 ± 0.1 | 1.08 ± 0.07 | 5.4 ± 0.8 | 25 ± 6 |
| AP7 | 26.55 ± 0.04 | −4.12 ± 0.06 | 5.3 ± 0.2 | 0 | |
| AP13 | 6.94 ± 0.05 | 2.18 ± 0.04 | 10.8 ± 0.5 | 0 | |
| AP19 | 13 ± 12 | 1.2 ± 0.1 | 1.8 ± 0.3 | 6 ± 11 | 92 ± 215 |
| Manual mixing: 50 mM KCl | |||||
| AP1 | 38.22 ± 0.01 | −8.7 ± 0.1 | 173 ± 2 | 0 | |
| AP7 | 32.87 ± 0.22 | −182 ± 14 | 37 ± 1 | 13.5 ± 0.1 | 923 ± 32 |
| AP13 | 6.15 ± 0.00 | 20 ± 1.5 | 41 ± 1 | 27.7 ± 0.4 | 97 ± 3 |
| AP19 | 29.79 ± 0.08 | −7.1 ± 0.3 | 480 ± 30 | 0 | |
The optimized kinetic constants associated with the progress curves in Figure 6, Figure 7, S6 and S7 were determined by fitting the progress curves to a sum of one or two exponentials by non-linear least squares analysis as described in the text. A2 values of zero indicate that the data set was adequately fit by one exponential. Positive values of the pre-exponential factor A indicate enhanced fluorescence and negative values indicate fluorescence quenching. Errors are standard deviations of the fitted parameters.
NaCl-dependent folding monitored by 2-AP fluorescence
Folding the 2-AP derivatives of Tel22 in 100 mM NaCl determined by stopped-flow fluorescence revealed that a significant fraction of the fluorescence change occurred during the ~0.1 s instrumental dead time for all of the 2-AP derivatives. For AP1 and AP13, the fluorescence initially decreased, while AP7 and AP19 exhibited an initial rapid increase in fluorescence. Independent experiments showed that changes in UV absorption at 295 nm were complete in <2 s under these conditions for all of the 2-AP-containing oligonucleotides (Figure S5). Thus we conclude that the rapid initial “burst” fluorescence change results primarily from formation of the quadruplex stack. The observation that position-specific increases or decreases in fluorescence quantum yield suggests that the 2-AP residues located within these different positions either undergo stacking (in the case of quenching) or unstacking (in the case of fluorescence enhancement) relative to the unfolded state on formation of the initial folding intermediate (I or I1 in eq. 1 or eq.2) (26).
In addition to these fast changes in fluorescence, we also observed relatively slow, position-specific changes in emission that occurred over time periods of up to several minutes. The progress curves for AP1, AP7 and AP13 are shown in Figure S6 and the kinetic constants derived from these progress curves are given in Table 3. In summary, AP1, AP7 and AP13 exhibited slow, biphasic changes in emission with relaxation times ranging from 1.5 s to 14 s (determined from the stopped-flow experiments) and slower changes with τ values of 30 s to 56 s determined by manual mixing. Depending on the loop position, both positive (fluorescence enhancement) and negative (fluorescence quenching) amplitudes were observed. Folding of AP19 (loop 3) was more complex as it occurred with three slow relaxations with τ values of ~4 s, 10 s and 45 s and signal amplitude changes of negative-positive-negative (Figure 6). The significantly different kinetic constants and directions of the fluorescence changes for the individual 2-AP derivatives suggest that the initial, rapidly formed ensemble of intermediates in NaCl slowly adjust loop conformation to form the stable basket topology.
The relaxation times described above for Na+-induced changes in fluorescence can be compared with those determined by changes in UV absorption for folding of the 2-AP-containing oligonucleotides induced by this cation. As with native Tel22, UV spectroscopic assessment of folding of the 2-AP derivatives in 100 mM NaCl revealed a rapid step (τ values of 12–43 ms, depending on the site of substitution) and two slow steps with τ values of 0.4–1 s and 5–10 s (Supporting Information, Figure S5). Comparison of these time constants with those given in Table 3 for folding assessed by fluorescence changes reveal that the folding rates assessed by the two different spectroscopic probes do not correlate exactly, suggesting that changes UV absorption and fluorescence intensity monitor different microscopic details of the structural changes. For example, it seems likely that the relatively minor, slow UV absorption changes in the 295 nm spectral region predominantly reflect subtle differences in G-quartet geometry, while the changes in 2-AP fluorescence emission may also be sensitive to local changes involving the conformation of the 2-AP residue itself. It is likely that the rate and magnitude of these changes will be different for different loops. This supposition is supported by the K+-induced folding experiments which consisted of a single major kinetic process as detected by UV spectroscopy between 275 and 320 nm, but clearly exhibited a series of relatively slow changes in fluorescence emission intensity (discussed below).
The relatively slow adjustments in tertiary structure may be interpreted to indicate a “rugged folding landscape” for quadruplex formation in which a number of kinetically accessible structures is sampled prior to slow relaxation to a thermodynamically stable structure or mixture of structures. Along this line, it is noteworthy that the triphasic adjustment of the conformation of AP19 in NaCl indicates that this loop 3 residue undergoes a more complex series of conformational rearrangements than the 2-AP residues in loops 1 and 2. This could indicate that loop3 undergoes a substantial change in topology as suggested by our proposed folding mechanism in which the basket conformation is formed from a hypothetical chair-type intermediate (22).
KCl-dependent folding monitored by 2-AP fluorescence
In view of the heterogeneity of the folding equilibria observed in equilibrium titrations, we determined progress curves for folding 2-AP-Tel22 in low (3 mM) and higher (50 mM) KCl. Surprisingly, in view of the single kinetic process observed for K+-dependent folding of Tel22 monitored by UV absorbance changes, the kinetics of fluorescence changes in KCl was heterogeneous, displaying relaxations over a period of milliseconds to minutes at all substitution positions. Representative progress curves for these fluorescence changes for AP7 are shown in Figure 7. In manual mixing experiments in 3 mM KCl, all of the 2-AP derivatives showed relatively rapid initial changes in fluorescence during the ~5 s dead time. Comparison of the progress curves with parallel stopped-flow UV experiments (shown in Figure S7) carried out under similar conditions suggest that these rapid fluorescence changes are associated with G-quartet formation and stacking. In addition, AP1, AP13 and AP7 exhibited relatively slow single exponential fluorescence quenching with τ values of ~500–900 s, reflecting site-specific adjustment of loop conformation (Table 3, Figure S8). In contrast, AP7 exhibited a triphasic response: an initial rapid, relatively large enhancement of fluorescence followed by a decrease in fluorescence which was then followed by a fluorescence increase; these slower changes were characterized by τ values of ~200 s and ~900 s, respectively (Figure 7C).
Multi-step kinetic changes in fluorescence were also noted when the 2-AP derivatives were folded in 50 mM KCl. Stopped-flow mixing experiments covering a 20-s time period revealed an initial rapid fluorescence enhancement for AP7 and AP19 and a small degree of quenching for AP1 and AP13. These rapid changes were followed by relatively slow fluorescence enhancement for AP7 and slow quenching for AP13 and AP19. There was little initial fluorescence change for AP1 on folding in 50 mM KCl, suggesting that initial formation of G-quartets in this case does not result in a significant change in AP1 environment relative to the unfolded state. The observed slow biphasic increase-decrease sequence of changes in fluorescence signal suggests a complex repositioning of AP1 after quartet stacking. The time constants associated with these intermediate steps were 1–10 s and 25–90 s. Manual mixing experiments with 50 mM KCl revealed further slow adjustments in fluorescence occurring with τ values of ~40 s to ~900 s. These results suggest that the initial, rapidly formed ensemble of conformers slowly rearranges by adjusting loop folding to give a stable mixture of conformers. A major conclusion from the fluorescence studies of K+-driven folding of Tel22-2-AP derivatives is that monitoring the folding reaction by site-specific fluorescence changes reveals intermediate steps in folding that did not give appreciable changes in UV absorbance in the 270–320 nm wavelength range.
Fluorescence depolarization
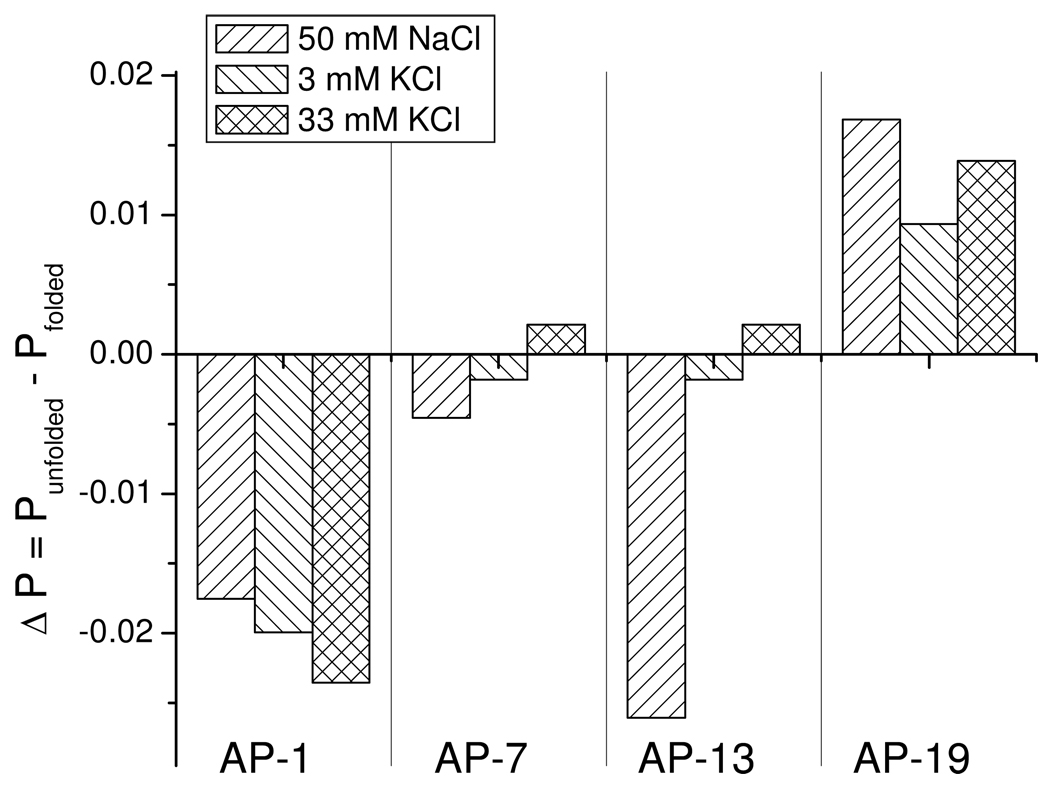
Determination of depolarization of fluorescence when a fluorophore is excited with polarized light can reveal the rotational mobility of the fluorophore provided that the lifetime of the excited state is comparable to the rotational relaxation time of the fluorophore. P, the steady-state degree of emission polarization, is defined as (I‖ - I⊥)/(I‖ + I⊥) where I‖ and I⊥ represent the intensities of the emitted light in directions parallel and perpendicular to the direction of polarization of the exciting radiation. P consists of two components: (a) an intrinsic polarization (P0) that depends on the geometric relationship between the excitation and emission dipoles and (b), a component that depends on the rotational motion of the fluorophore. For macromolecules, a change in P may result from a change in the local mobility of the fluorophore and/or a change in rotational diffusion resulting from a change in hydrodynamic volume, e.g. one that accompanying a transition from a random coil to a compact, folded structure.
The average steady-state values of P were within a range of 0.07 to 0.14 depending on the 2-AP derivative and the nature of the cation. These relatively small P values are expected given the short lifetime of the 2-AP excited states in quadruplexes (~0.5 ns (52)) and the rotational relaxation time of the folded quadruplex (calculated to be 4 ns at 25 °C using the program Hydropro (53)). Except for AP7 in NaCl and KCl, and AP-13 in KCl, significant changes in P were induced by folding for each oligonucleotide (summarized in Figure 8). Folding of AP1 in NaCl and KCl and folding of AP13 in NaCl were characterized by an increase in P while folding of AP19 was accompanied by a decrease in P. Thus, motion of AP1 and AP13 in NaCl appeared to be more restricted in the folded ensemble compared to the unfolded ensemble, while AP19 gained mobility in the folding transition.
FIGURE 8.
Changes in fluorescence polarization measured at 370 nm induced by cation-induced folding of 2-AP derivatives of Tel22. Folding was initiated by rapidly adding a small volume of a 3 M salt solution to a rapidly stirring solution of oligonucleotide to give the final salt concentrations indicated in each panel.
Na+/K+ exchange kinetics
We expected that a comparison of fluorescence changes accompanying the Na+ → K+ cation exchange reaction for the four 2-AP-Tel22 derivatives might indicate which loops undergo a change in orientation in the transition from the Na+-antiparallel basket structure to the K+-dependent mixture of topologies. A representative progress curve for cation exchange in AP7 is shown in Figure 9 and the fitted time constants associated with the exchange of Na+ for K+ for each of the four 2-AP derivatives of Tel22 are collected in Table 4. The progress curves for cation exchange with AP1, AP13 and AP19 are reproduced in Figure S9. For all of the 2-AP-Tel22 derivatives there was an initial rapid fluorescence change that occurred during the ~5 s mixing time when KCl was rapidly added to the NaCl-folded structures. For AP1, the fluorescence intensity rapidly increased followed by a slower decrease that fit first-order kinetics. For AP7, AP13 and AP19, an initial rapid quenching in fluorescence was observed. For AP13 the rapid step was followed by slow quenching, while for AP19, the rapid quenching step was followed by slow fluorescence enhancement. AP7 uniquely exhibited biphasic slow fluorescence changes in which enhanced fluorescence was followed by quenching (Figure 9). This complex series of fluorescence changes for AP7 suggests that when switching from the Na+-bound basket conformation to the K+-bound conformations, loop 1 undergoes an additional isomerization that does not occur with loops 2 and 3. We speculate that this additional step may result from a switch in loop 1 in the Na+-basket from an antiparallel topology to the parallel arrangement characteristic of the hybrid-1 structure. As shown below, our molecular dynamics simulation suggests a mechanism of stabilization of the hybrid-1 structure by specific binding an additional potassium cation.
FIGURE 9.
Na+/K+ exchange kinetics for the AP7 derivative of Tel22. The exchange reaction was initiated by adding KCl from a 3M stock solution to a rapidly stirred solution of the oligonucleotide pre-equilibrated with 75 mM NaCl. The data points show the change in fluorescence upon adding 150 mM KCl. The horizontal arrow indicates the starting value of the fluorescence in NaCl immediately prior to the addition of KCl. The vertical arrows indicate the rapid fluorescence change that occurred during the ~5 s mixing time. The red line shows the fit of the experimental data to a double exponential relaxation using the optimized parameters in Table 4.
Table 4.
Fitted kinetic constants for Na+/K+ exchange for 2-AP derivatives of Tel22.
| y0 | A1 | τ1(s) | A2 | τ2 (s) | |
|---|---|---|---|---|---|
| AP1 | 27.74 ± 0.03 | 7.63 ± 0.04 | 364 ± 5 | 0 | |
| AP7 | 64.3 ± 0.7 | −14.60 ± 0.03 | 122 ± 3 | 19.9 ± 0.4 | 1086 ± 86 |
| AP13 | 8.47 ± 0.03 | 1.92 ± 0.04 | 189 ± 10 | 0 | |
| AP19 | 33.88 ± 0.03 | −9.74 ± 0.04 | 296 ± 3 | 0 |
The optimal kinetic constants describing the progress curves in Figures 9 and S8 were determined by fitting the data sets to a sum of one or two exponentials by non-linear least squares analysis as described in the text. A2 values of zero indicate that the data set was adequately described by a single exponential. Positive values of A indicate enhanced fluorescence and negative values indicate fluorescence quenching. Errors are standard deviations of the fitted parameters.
It is of interest to compare the data obtained in monitoring fluorescence changes within loop 1 with the results of our study in which CD was used to monitor the Na/K exchange reaction for native Tel22 (24). The CD change induced by switching cations occurs in at least three steps: an initial rapid step within the 5-s mixing time followed by slower steps of with τ values of ~50 s and ~800 s. The rapid CD change was interpreted to reflect an exchange of K+ for Na+ within the central channel of the quadruplex, a process known to take place on a µs time scale (54, 55). The intermediates associated with the two slower steps were suggested to consist of transiently formed triplex structures.
Molecular modeling
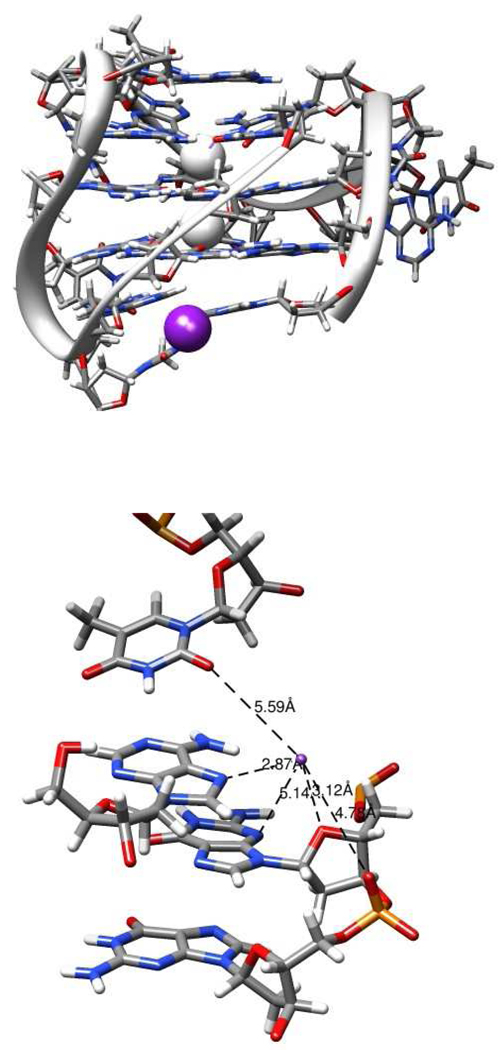
Numerous studies (reviewed in (54)) have shown that K+ ions are bound within the quadruplex central channel by coordination to the O6 atoms of the G residues; because of their larger size, these K+ are located between the individual tetrads rather than within the tetrad plane as suggested for Na+. A number of studies also show that in certain quadruplex structures, cations are bound within the connecting loops. As described in more detail in the discussion below, these coordination sites often involve thymine O2 atoms. The biphasic changes in loop fluorescence observed in our K+ titrations (Figure 2 and Figure 3) imply the existence of K+ coordination sites in addition to the channel sites. To further investigate this possibility, we examined the K+ distribution in unrestrained molecular dynamics trajectories calculated for models with various Tel22 topologies. In addition to the expected K+-binding sites in the quadruplex core, we noted an increase in K+ ion density exclusively within a region of the hybrid-1 model adjacent to A1. This region was manifested by extended K+ residence times of up to approximately 2.5 ns compared to other sites; a similar increase in K+ density was not found in the hybrid-2 structure. These results suggest the existence of an “external” K+-binding site specific for the hybrid-1 structure. Figure 10 shows characteristics of this site.
FIGURE 10.
Hypothetical external K+ binding site in Tel22 hybrid-1 topology. The figure was generated from a snapshot taken at the conclusion of the molecular dynamics simulation of the hybrid-1 form of Tel22 described in the text. The grey spheres represent the channel-bound K+ and the purple sphere shows the position of the externally bound K+. The bottom panel shows a detailed representation of the K+-binding site with potential coordinating groups (and their distance in Å from K+): dT18-O2 (5.59), dA1-N7 (2.87), dG20-O4′ (3.12), dG20-N3 (5.14), dG21-O1P (4.78). The figures were constructed using Chimera (41).
DISCUSSION
This study compares the kinetics of quadruplex folding previously deduced from UV-dependent stopped-flow studies with the folding kinetics monitored by loop-specific changes in 2-AP fluorescence. We expected that these folding-induced fluorescence changes would report the dynamics of formation of stable loops and might reflect changes in strand topology. The telomeric model oligodeoxynucleotide chosen for the study, Tel22, has a well-defined solution structure in Na+ that consists of an antiparallel basket with two lateral loops and one diagonal loop (Figure 1A). In contrast, in solution in the presence of the physiologically interesting cation K+, Tel22 consists of a mixture of unknown topologies with different loop arrangements and strand orientations (17, 19, 21, 56, 57).
Our experimental strategy involved comparing the folding equilibria and kinetics of derivatives of Tel22 in NaCl and KCl in which the 5′ dA residue and the three loop dA residues were serially replaced with the fluorescent adenine analog, 2-aminopurine. Factors that influence 2-AP fluorescence quantum yield have been extensively studied. Among the most important of these factors is base stacking which quenches 2-AP fluorescence; it has also been reported that the 2-AP quantum yield is sensitive to the presence of nearby cations (28). Previous studies from our laboratory (27) showed that the fluorescence of Tel22 2-AP derivatives are sensitive to differences in quadruplex folding topology induced by different cations, and, moreover, the studies provide a rational approach to distinguishing among various quadruplex folding topologies in solution.
Loop dynamics
Our earlier work on the cation-induced folding of Tel22 as assessed by changes in UV absorption indicated a multi-step folding mechanism (22). In both Na+ and K+, a folding intermediate, possibly hairpin structure(s), was formed in a relatively fast step that became rate-limiting at high cation concentrations. In K+, there were no significant changes in UV absorption in the 275–320 nm range subsequent to this rapid step. In Na+, however, two slower UV relaxations indicated the presence of additional kinetic intermediates on the pathway to formation of the basket topology prevalent with this cation. A similar mechanism was apparent for folding TT-Tel22-A in Na+, a sequence known to form the hybrid-1 structure in K+ solution (20). These results show that the folding mechanism determined by monitoring changes in UV absorption depends on the cation and is independent of the flanking sequences for Tel22 and these 5′ and 3′ variants. Furthermore, since TT-Tel22-A forms the hybrid-1 topology in K+ whereas Tel22 in K+ forms of a mixture of structures, the folding mechanism determined by changes in UV absorption in the 275–320 nm range is apparently independent of the nature of the final equilibrium ensemble of folded structures.
The 2-AP fluorescence changes observed here support a multi-step mechanism for Tel22 folding induced by both Na+ and K+ and the fluorescence data indicate that the slower steps involve loop rearrangements. K+-driven folding exhibits a series of slow steps not evident in the UV kinetic studies but were obvious from fluorescence changes in the loops. In addition, the observation of sequential positive and negative changes in fluorescence emission intensity induced in KCl-dependent folding for AP19 (Figure 5) and for folding of AP1, AP7 and AP19 in NaCl (Figure 6) support a sequential folding pathway (eq. 2) as opposed to a branched pathway (eq. 5):
| (5) |
This conclusion is illustrated by a simulation of the reaction sequences given in eqs 2 and 5 with the assumption that each species is characterized by a different fluorescence quantum yield show that only eq. 2 gives a reaction profile characterized by increasing and decreasing exponentials (Figure S10).
Folding equilibria
Our previous equilibrium titrations monitored by cation-induced changes in UV absorption suggested the presence of small amounts of intermediate species, possibly partially folded structures and/or structures containing an incomplete complement of bound cations. In the current study, the fluorescence experiments conducted in KCl with the 2-AP-Tel22 derivatives and the CD studies conducted with Tel22 reveal the presence of species with different fluorescence and CD properties as a function of [K+]. It is likely that these species are fully folded, stable quadruplexes with differing topographies rather than partially folded intermediates. It is clear from the fluorescence KCl titration curves in Figure 3B taken in conjunction with the KCl-dependent NMR spectra in Figure 5 that a different structural ensemble is present in ~1 mM [K+] compared to that at [K+] >10 mM for a period of several hours after mixing the cation solution with the unfolded oligonucleotide at 25 °C. The solution structure of these conformers and their relative concentrations is unknown, but a possible interpretation is that the predominant conformers are the hybrid-1 and hybrid-2 structures. Based on the presence of an additional K+ binding site unique to the hybrid-1 structure suggested by our molecular dynamics simulations, the prevailing conformer at low K+ concentrations may be the hybrid-2 structure (which lacks this site).
External potassium binding sites in quadruplexes
Several studies have been published that provide precedent for the existence of cation binding sites within the loops of G-quadruplex structures (see Hud et al. (54) for a comprehensive review). For example, Jing et al. (58, 59) reported that K+-induced formation of a unimolecular quadruplex by the anti-HIV oligonucleotide d[GTGGT(GGGT)3] is a two-step process in which one K+ is incorporated into the two-stack quadruplex core to form a high-affinity complex while two K+ ions bind in a slower reaction to loop sites with lower affinity. These investigators observed biphasic folding isotherms in CD and UV titrations of K+ similar to those presented here (Figure 2 and Figure 3). For titrations of Na+-induced folding, the isotherms were monophasic, suggesting that Na+ binds only within the quadruplex channel but not to loop sites for the anti-HIV quadruplex. In another example of cation binding within a loop, Marathias and Bolton (60) characterized K+-induced folding of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG). These authors reported two K+ binding sites: one within the quadruplex channel and one in a loop region involving the thymine O2 atoms and the O6 atoms of one of the G-quartets. More recently, Phan et al. (21) determined the solution structure of the two human telomeric sequences d[TAGGG(TTAGGG)3] and d[TAGGG(TTAGGG)3TT] by NMR. In K+ solution, the former consisted of 70% hybrid-1 and the latter consisted of 70% hybrid-2. K+-induced changes in NOEs for the longer sequence were explained by coordination of K+ (but not Na+) within the T12-T13-A14 loop. A molecular dynamics simulation indicated that the T1, T12, A14 residues and the O6 of the adjacent quartet chelate one K+ ion.
Cation binding sites located within quadruplex loops have also been demonstrated in bimolecular and tetramolecular quadruplexes. Bouaziz et al. (61) determined the solution structure of the bimolecular quadruplex formed by d[G3CT4G3C] in Na+ and K+. The two different cations stabilize structures with different loop configurations. Modeling identified potential cation binding sites specific for K+ involving the O2 atoms of T6 and T8, the sugar ring O of T7, T8 and G9, and the backbone O atoms of T7–T8 and G8–G9 located within the two T4 loops. In pointing out the potential functional significance of loop cation binding, the authors suggested that these K+ binding to these sites resulted in “a defined loop architecture whose outwardly pointing groups provide a unique folded topology that can target potential receptor sites” (61).
The crystal structure of d(G4T4G4), which forms a bimolecular structure consisting of four G-stacks, reveals three K+ coordinated between quartets and two K+ bound within the T-loops (62). Recently, Ida and Wu (55) provided NMR evidence of Na+ binding within the T4 loops of d(G4T4G4). The coordination site was proposed to involve the four O6 of guanine residues of the last quartet, the O2 of a loop thymine, and a water molecule.
In an example of K+-dependent conformational heterogeneity in monomeric G-quadruplexes, Lee et al. (63) analyzed the distribution of conformational states in a solid-state, single molecule FRET system. They presented evidence for three inter-convertible states: an unfolded conformation and two folded conformations, each characterized by a different FRET efficiency. The proportion of each conformation depended on [K+] and temperature. One folded state was favored at low K+ (0.1– 2 mM) and different one was favored at 100 mM KCl. The folding kinetics observed when 2 mM KCl was added to a bulk solution of the unfolded structure was biphasic with τ values of 8.8 and 253 s, relaxation times which are similar to the relaxation times for the K+-dependent loop conformational changes noted in our study. It was suggested that the slow step involved a change in strand topology. The two studies are therefore complementary in that both independently point to a K+-dependent conformational isomerization of quadruplex structures.
Loop structures
We next turn to a structural rationalization for the changes in fluorescence properties of 2-AP induced by folding in NaCl, 3 mM KCl and 30 mM KCl. For reference, Table 5 provides a qualitative summary of the changes in the fluorescence properties associated with each substitution position. It is important to note at the outset that signals such as fluorescence are measured with respect to the unfolded oligonucleotide, which probably consists under our starting conditions of an ensemble of partially collapsed states with varying degrees of interaction between adjacent bases rather than an extended structure dictated by the electrostatic repulsion between phosphate groups. An additional complicating factor is the known heterogeneity of the folded state of Tel22 in KCl. The spectroscopic signals observed for the starting and final states will represent an average of the signals for each conformer weighted according to its relative concentration. This structural heterogeneity almost certainly falls into two categories: micro-heterogeneity at the site of the 2-AP residue and topological heterogeneity due to different folded structures in the presence of K+. The observed changes in the fluorescence properties of a fluorophore in solution will thus be an average weighted in proportion to the conformers present in the starting and final states and the extent of the change associated with each conformational change. Since the mechanisms driving quenching and changes in polarization are different, there may not always a direct correlation between the two parameters. The net change in either fluorescence quantum yield or polarization is therefore not predictable without prior knowledge of the degree of change for a given conformer and its concentration within the sample. With these caveats, we next discuss the relationship between the folded structures and the fluorescence quantum yield and polarization at each position in the basket, hybrid-1 and hybrid-2 topologies.
Table 5.
Summary of the direction of fluorescence changes for cation-induced folding of Tel22.
| ODN | Location | Stackeda | Mobility | Change in fluorescence intensity (U → F) |
Change in polarization (U → F) |
Change in fluorescence intensity for kinetic steps |
|---|---|---|---|---|---|---|
| Na+ (50 mM) | ||||||
| AP1 | core | yes | fixed | b↓ | ↑ | c↓↓↓ |
| AP7 | loop 1 | no | mobile | ↑ | ↑ | ↑↑ |
| AP13 | loop 2 | yes | fixed | ↓ | ↑ | ↓↓ |
| AP19 | loop 3 | yes | mobile | ↑ | ↓ | ↓↑ |
| K+ (3 mM) | ||||||
| AP1 | core | ↓ | ↑ | ↑↓→ | ||
| AP7 | loop 1 | ↑ | → | ↑↓↓ | ||
| AP13 | loop 2 | ↓ | → | ↑↓↓ | ||
| AP19 | loop 3 | ↑ | ↓ | ↑↓↑ | ||
| K+ (33 mM) | ||||||
| AP1 | core | → | ↑ | ↑↓ | ||
| AP7 | loop 1 | ↑ | → | ↑↓ | ||
| AP13 | loop 2 | ↓ | → | ↓ | ||
| AP19 | loop 3 | ↑ | ↓ | ↑↓ | ||
Determined from the NMR structure of Tel22 (14)
↑ = increase, ↓ = decrease, → = no significant change.
In order of steps 1, 2 and 3.
5′-Cap (A1)
In the basket conformation, the topology in Na+ solution, A1 stacks over G2 (14). It is the least solvent-exposed of the four dA residues (SASA = 104 Å2). The NMR-derived structures in PDB file 143D (14) suggest that A1 is relatively fixed with respect to the molecular framework. In the titrations with Na+, AP1 fluorescence was quenched, which is consistent with an increase in base stacking interactions relative to the unfolded state. Fluorescence polarization increased on folding, indicating that AP1 becomes less mobile on folding which is consistent with the restricted movement suggested by the ensemble of NMR structures. The triphasic folding transition assessed by fluorescence is similar that observed previously by UV absorption, suggesting that the changes in fluorescence reflect changes in the local environment at the 5′ position resulting from relatively slow optimization of the tertiary structure in the vicinity of the first G-quartet. In addition, these changes were reported by the (rather small) changes in UV absorption (22).
The interpretation of the fluorescence changes in KCl are not straight-forward because, as noted, Tel22 in K+ exists as a mixture of unknown topologies in unknown proportions. However, it is of interest to compare the hybrid-1 and hybrid-2 computational models of Tel22. In the hybrid-2 model, A1 is sandwiched between T12 and G10. It is the least exposed of all dA residues (SASA of 85 Å2). Experimentally, we found an increase in AP1 fluorescence on folding up to ~3 mM KCl, and an increase in steady-state polarization. The increase in polarization is consistent with sequestration of AP1 relative to the unfolded state but the observed increase in quantum yield is at odds with an increase in stacking. An increase in polarization and a corresponding increase in quantum yield may seem inconsistent; however, we suggest that folding could lead to a decrease in rotational motion of AP1 due to its sequestration while simultaneously providing less stacking interactions in the folded state. This suggests the hybrid-2 model does not account for the behavior of AP1 in low KCl. The folding kinetics in 3 mM KCl was biphasic, exhibiting a small decrease followed by a large increase in fluorescence. The absence of a large rapid change in emission (probably associated with quartet formation) suggests that the environment of AP1 in the initially formed structure is not much different than in the unfolded ensemble; consequently, most of the fluorescence change takes place during the final positioning of AP1.
A1 also forms a 5′ cap in the hybrid-1 model. It is stacked with G20 and has intermediate solvent accessibility (SASA = 183 Å2). In high [K+], there was net quenching fluorescence. Polarization increased, suggesting that 2-AP1 experiences a net decrease in flexibility on folding to its equilibrium conformational ensemble in high K+. Thus, the fluorescence data are consistent with the predictions suggested by biasing the conformational equilibrium toward the hybrid-1 state.
Loop 1 (T5-T6-A7)
In the Na+-basket form, A7 is located in a lateral loop and is not stacked with other bases. A7 is the most exposed to solvent (250 Å2) of the four dA residues. The NMR-derived structures in 143D (14) indicate that a variety positions is consistent with the data, suggesting flexibility at this position. Indeed, the fluorescence changes on folding are consistent with higher flexibility within loop 1 since folding was accompanied by an increase in quantum yield and little or no change in polarization relative to the unfolded state. These results are consistent with decreased stacking interactions. Loop formation was biphasic, with both steps resulting in increased fluorescence, suggesting formation of folding intermediates with decreased base stacking.
In the hybrid-2 model, A7 is located in a lateral loop where it is imperfectly stacked on the G4-G8-G16-G22 quartet. It has a relatively high exposure (SASA = 215 Å2). In 3 mM K+, folding was characterized by an increase in quantum yield and no net change in polarization. The kinetics of the changes in emission was triphasic, suggesting successive intermediates characterized by decreases in fluorescence with the final equilibrium ensemble showing an increase in fluorescence.
In the hybrid-1 model, AP7 stacks on top of the G4-G10-G14-G20 quartet. Its SASA is 236 Å2. In 30 mM K+, AP7 showed an identical set of changes as in 3 mM KCl except that the rapid phase in fluorescence emission, corresponding to quartet formation, was not apparent, probably having occurred during the instrumental dead-time.
Loop 2 (T11-T12-A13)
In Na+, loop 2 is in a diagonal arrangement with A13 stacked over G22. A13 is relatively exposed (SASA = 170 Å2). The ensemble of NMR structures indicates that this residue has restricted mobility. The Na+-induced changes in AP13 emission are consistent with this picture. There was a net decrease in quantum yield on folding and an increase in polarization. The kinetics of the change in emission was biphasic, with two intermediates of lower fluorescence. This is consistent with formation of the diagonal loop occurring in a step-wise fashion with two kinetic intermediates.
In hybrid-2, A13 in loop 2 stacks on top of the G2-G10-G14-G20 tetrad. It has intermediate exposure to solvent (SASA = 139 Å2). Experimentally, in 3 mM K+, loop 2 was formed with a net decrease in fluorescence and a negligible change` in polarization. The kinetics of the fluorescence change was triphasic with the intermediates showing successively decreased, decreased, and increased fluorescence.
A13 in the hybrid-1 structure is stacked over G22. The accessible surface area is ~223 Å2. In 30 mM K+, formation of loop 2 occurred with a net decrease in fluorescence, suggesting increased stacking with respect to the unfolded state, little or no change in polarization, and a single step decrease in fluorescence.
Loop 3 (T17-T18-A19)
In the basket structure, A19 is stacked over G16 and exhibits flexibility as suggested by the variety of orientations evident in the NMR-generated conformational ensemble. SASA is 140 Å2. The folding-induced increase in fluorescence and decrease in polarization are consistent with this structure. As with loops 1 and 2, the kinetics of formation of lateral loop 3 was biphasic, showing an increase followed by a decrease in fluorescence.
In hybrid-2, A19 packs into a groove. It is relatively more exposed to solvent (SASA = 217 Å2) and is not stacked with other bases. In 3 mM K+, formation of loop 3 was accompanied by an increase in fluorescence and decrease in polarization, consistent with unstacking and an increase in rotational mobility. The kinetics of the fluorescence emission change was triphasic, suggesting changes in loop conformation that are associated with variations in base stacking.
In hybrid-1, A19 lies under the G2-G8-G16-G20 tetrad and over T18. It is relatively inaccessible to solvent (SASA = 135 Å2). Folding in 100 mM KCl occurred with an increase in fluorescence and a decrease in polarization relative to the unfolded ensemble. These observations are consistent with an unstacking of A19 relative to the unfolded state.
In conclusion, the observed position-specific changes in 2-AP fluorescence correlate well with those expected from the antiparallel basket structure formed in Na+. Detailed correlation of the changes in fluorescence with specific structures in KCl is not possible due to the topological heterogeneity of the folded state. Nevertheless, it is clear from the differential effects of KCl concentration that changes in the concentration of this cation leads to different populations of conformers. Similarly, the K+ → Na+ conformational switch occurs on a slow time scale, suggesting that both reactions require adjustment of strand topologies.
Cation-driven allosterism in quadruplex DNA
The studies reported here show that changes in cation identity and concentration induce changes in quadruplex tertiary structure. These observations suggest the possibility of K+-driven allosteric control mechanisms. Our studies define the energetics and kinetics of these conformational switches as driven by either change in cation from Na+ to K+ or a change in K+ concentration.
The ability of biological macromolecules to undergo conformational changes in response to ligand binding is fundamental to the theory of allosteric regulation. The original allosteric theory of Monod, Wyman and Changeaux (64) postulated that regulatory proteins exist in equilibrium between two conformations with different intrinsic activities and different affinities for regulatory molecules. According to thermodynamic linkage theory, binding a molecule to a site specific for one conformation will bias the population of conformers toward a state with a characteristic activity profile. Recently, the mechanism of allostery has been reformulated to take into account current theories of protein structure (65, 66) which postulate that proteins exist as an ensemble conformational states in which relatively high energy states may be transiently visited due to thermal motion. An allosteric regulatory molecule may bind to one of these rare states, thereby lowering its free energy and thus increasing its representation in the population. In this model, understanding the molecular basis of allosteric regulation requires knowledge of the relative populations of individual states and their rates of their inter-conversion.
It is less widely appreciated, but none-the-less equally important, to recognize that polynucleotides may also utilize allosteric mechanisms for functional regulation (67, 68). As with proteins, ligand binding to a particular DNA conformation may drive structural changes that may result in changes in affinity of the DNA sequence for effector molecules such as small ligands, polymerases, transcription factors, etc. Several examples of nucleic acid allosterism have recently been summarized (68). Among the non-classical structural motifs amenable to study of allosterically-mediated DNA regulatory mechanisms are conformational transitions in G-quadruplex structures. These structures display a high degree of conformational heterogeneity in which the individual topographies are separated by relatively low energy barriers. These features make G-quadruplex structures attractive candidates for the evolution of sensitive and facile regulatory mechanisms.
Conclusions
We demonstrated that 2-AP fluorescence is a sensitive monitor of the equilibria and kinetics of changes in the conformation of specific loop regions in model oligonucleotides that mimic the human telomeric sequence. Changes in 2-AP fluorescence resulting from K+ binding clearly indicates the presence of high-affinity binding sites within the quadruplex channel and low-affinity binding sites specific for K+. Molecular dynamics simulations revealed a cation binding site specific for the hybrid-1 form of the human telomeric sequence that involves the A1 residue. This model suggests that elevated [K+] will increase loop rigidity and stabilize a particular topography. We propose that this low-affinity site falls within the classical definition of an allosteric site. Thus a K+-dependent switching mechanism could be utilized to control quadruplex tertiary structure, thereby favoring the binding of conformation-specific effector molecules.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Prof. A. N. Lane for assistance with NMR experiments and the reviewers for thoughtful comments and suggestions.
Footnotes
Supported by grant GM 077422 from the National Institutes of Health and the James Graham Brown Foundation. NMR spectra were recorded at the James Graham Brown Cancer Center NMR facility, supported in part by NIH grant P20RR018733 from the National Center for Research Resources and National Science Foundation EPSCoR grant EPS-0447479.
Abbreviations: 2-AP, 2-aminopurine; Bu4AmP, tetrabutylammonium phosphate; cps, fluorescence intensity in counts/s; DSS, 4,4-dimethyl-4-silapentane-1-sulfonic acid; MD, molecular dynamics; FRET, fluorescence resonance energy transfer; NOE, nuclear Overhauser effect; SASA, solvent accessible surface area; SVD, singular value decomposition; Tel22, d[A(GGGTTA)3GGG]; TT-Tel22-A, d[TT(GGGTTA)3GGGA]; TT-Tel22, d[TT(GGGTTA)3GGG].
After completion of this manuscript, we observed additional very slow changes in UV absorption and ellipticity in the spectral region between 220 nm and 265 nm that occurred over a period of up to eight hours when KCl was mixed at room temperature with unfolded Tel22. The implications of these slow changes are currently under investigation.
SUPPORTING INFORMATION AVAILABLE.
Figures S1–S10 and Table S1 illustrating spectrophotometric titrations, kinetic data, SVD analyses and simulated fluorescence progress curves. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Huppert JL. Hunting G-quadruplexes. Biochimie. 2008;90:1140–1148. doi: 10.1016/j.biochi.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 3.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu. Rev. Genetics. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 4.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 6.Palm W, de Lange T. How shelterin protects mammalian telomeres. Ann. Rev. Genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 7.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nature Rev. Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;8:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 15.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 16.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hud NV, Smith FW, Anet FAL, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: A thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 24.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall KB, Williams DJ. Dynamics of the IRE RNA hairpin loop probed by 2-aminopurine fluorescence and stochastic dynamics simulations. RNA. 2004;10:34–47. doi: 10.1261/rna.5133404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jean JM, Hall KB. Stacking-unstacking dynamics of oligodeoxynucleotide trimers. Biochemistry. 2004;43:10277–10284. doi: 10.1021/bi049701p. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballin JD, Bharill S, Fialcowitz-White EJ, Gryczynski I, Gryczynski Z, Wilson GM. Site-specific variations in RNA folding thermodynamics visualized by 2-aminopurine fluorescence. Biochemistry. 2007:13948–13960. doi: 10.1021/bi7011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballin JD, Prevas JP, Bharill S, Gryczynski I, Gryczynski Z, Wilson GM. Local RNA conformational dynamics revealed by 2-aminopurine solvent accessibility. Biochemistry. 2008;47:7043–7052. doi: 10.1021/bi800487c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stivers JT. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menger M, Eckstein F, Porschke D. Multiple conformational states of the hammerhead ribozyme, broad time range of relaxation and topology of dynamics. Nucleic Acids Res. 2000;28:4428–4434. doi: 10.1093/nar/28.22.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl. Acad. Sci. U.S.A. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neill MA, Barton JK. 2-Aminopurine: a probe of structural dynamics and charge transfer in DNA and DNA:RNA hybrids. J. Am. Chem. Soc. 2002;124:13053–13066. doi: 10.1021/ja0208198. [DOI] [PubMed] [Google Scholar]
- 34.Jean JM, Hall KB. 2-Aminopurine electronic structure and fluorescence properties in DNA. Biochemistry. 2002;41:13152–13161. doi: 10.1021/bi020308y. [DOI] [PubMed] [Google Scholar]
- 35.Jean JM, Krueger BP. Structural fluctuations and excitation transfer between adenine and 2-aminopurine in single-stranded deoxytrinucleotides. J. Phys. Chem. B. 2006;110:2899–2909. doi: 10.1021/jp054755+. [DOI] [PubMed] [Google Scholar]
- 36.Wilson JN, Cho Y, Tan S, Cuppoletti A, Kool ET. Quenching of fluorescent nucleobases by neighboring DNA: the "insulator" concept. Chembiochem. 2008;9:279–285. doi: 10.1002/cbic.200700381. [DOI] [PubMed] [Google Scholar]
- 37.Hendler RW, Shrager RI. Deconvolutions based on singular value decomposition and the pseudoinverse: a guide for beginners. J. Biochem. Biophys. Methods. 1994;28:1–33. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 38.DeSa RJ, Matheson IB. A practical approach to interpretation of singular value decomposition results. Meth. Enzymology. 2004;384:1–8. doi: 10.1016/S0076-6879(04)84001-1. [DOI] [PubMed] [Google Scholar]
- 39.Sheardy RD, Suh D, Kurzinsky R, Doktycz MJ, Benight AS, Chaires JB. Sequence dependence of the free energy of B–Z junction formation in deoxyoligonucleotides. J. Mol. Biol. 1993;231:475–488. doi: 10.1006/jmbi.1993.1295. [DOI] [PubMed] [Google Scholar]
- 40.Antonacci C, Chaires JB, Sheardy RD. Biophysical characterization of the human telomeric (TTAGGG)4 repeat in a potassium solution. Biochemistry. 2007;46:4654–4660. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- 41.Gampp H, Maeder M, Meyer CJ, Zuberbuhler AD. Calculation of equilibrium constants from multiwavelength spectroscopic data. IV. Model-free least-squares refinement by use of evolving factor analysis. Talanta. 1986;33:943–951. doi: 10.1016/0039-9140(86)80233-8. [DOI] [PubMed] [Google Scholar]
- 42.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 43.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 46.Kollman PA, et al. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 47.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Hubbard SJ, Thornton JM. NACCESS. London: University College; 1993. [Google Scholar]
- 50.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 51.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 52.Kimura T, Kawai K, Fujitsuka M, Majima T. Monitoring G-quadruplex structures and G-quadruplex-ligand complex using 2-aminopurine modified oligonucleotides. Tetrahedron. 2007;63:3585–3590. [Google Scholar]
- 53.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hud NV, Plavec J. The role of cations in determining quadruplex structure and stability. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 100–130. [Google Scholar]
- 55.Ida R, Wu G. Direct NMR detection of alkali metal ions bound to G-quadruplex DNA. J. Am. Chem. Soc. 2008;130:3590–3602. doi: 10.1021/ja709975z. [DOI] [PubMed] [Google Scholar]
- 56.Gaynutdinov TI, Neumann RD, Panyutin IG. Iodine-125 radioprobing of intramolecular quadruplex conformation of human telomeric DNA in the presence of cationic porphyrin TMPyP4. Int. J. Radiation Biol. 2008;84:984–990. doi: 10.1080/09553000802415747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008;36:4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jing N, Gao X, Rando RF, Hogan ME. Potassium-induced loop conformational transition of a potent anti-HIV oligonucleotide. J. Biomol. Struct. Dyn. 1997;15:573–585. doi: 10.1080/07391102.1997.10508967. [DOI] [PubMed] [Google Scholar]
- 59.Jing N, Rando RF, Pommier Y, Hogan ME. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- 60.Marathias VM, Bolton PH. Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA. Nucleic Acids Res. 2000;28:1969–1977. doi: 10.1093/nar/28.9.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouaziz S, Kettani A, Patel DJ. A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 1998;282:637–652. doi: 10.1006/jmbi.1998.2031. [DOI] [PubMed] [Google Scholar]
- 62.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 63.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 65.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 66.Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol. Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaires JB. Long-range allosteric effects on the B to Z equilibrium by daunomycin. Biochemistry. 1985;24:7479–7486. doi: 10.1021/bi00346a067. [DOI] [PubMed] [Google Scholar]
- 68.Chaires JB. Allostery: DNA does it, too. ACS Chem. Biol. 2008;3:207–209. doi: 10.1021/cb800070s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.