Abstract
• Background and Aims The Brassicaceae family encompasses numerous species of great agronomic importance, belonging to such genera, as Brassica, Raphanus, Sinapis and Armoracia. Many of them are characterized by extensive intraspecific diversity of phenotypes. The present study focuses on the polymorphism of number, appearance and chromosomal localization of ribosomal DNA (rDNA) sites and, when possible, in relation to polyploidy, in 42 accessions of Brassica species and ten accessions of Diplotaxis, Eruca, Raphanus and Sinapis species.
• Methods Chromosomal localization of ribosomal DNA was carried out using dual colour fluorescence in situ hybridization (FISH) with 5S rDNA and 25S rDNA sequences as probes on enzymatically digested root-tip meristematic cells.
• Key Results Loci for 5S and 18S–5.8S–25S rDNA were determined for the first time in six taxa, and previously unreported rDNA constellations were described in an additional 12 accessions. FISH revealed frequent polymorphism in number, appearance and chromosomal localization of both 5S and 25S rDNA sites. This phenomenon was most commonly observed in the A genome of Brassica, where it involves exclusively pericentromeric sites of 5S and 25S rRNA genes. The intraspecific polymorphism was between subspecies/varieties or within a variety or cultivar (i.e. interindividual).
• Conclusions The number of rDNA sites can differ up to 5-fold in species with the same chromosome number. In addition to the eight previously reported chromosomal types with ribosomal genes, three new variant types are described. The extent of polymorphism is genome dependent. Comparing the A, B and C genomes revealed the highest rDNA polymorphism in the A genome. The loci carrying presumably inactive ribosomal RNA genes are particularly prone to polymorphism. It can also be concluded that there is no obvious polyploidization-related tendency to reduce the number of ribosomal DNA loci in the allotetraploid species, when compared with their putative diploid progenitors. The observed differences are rather caused by the prevailing polymorphism within the diploids and allotetraploids. This would make it difficult to predict expected numbers of rDNA loci in natural polyploids.
Keywords: 5S rDNA, 25S rDNA, 18S–5.8S–25S ribosomal RNA genes, Brassicaceae, chromosomes, FISH
INTRODUCTION
The Brassicaceae family comprises about 350 genera and more than 3000 species and is one of the most important crop plant families. To date, detailed chromosome analyses in Brassicaceae have been almost exclusively limited to the leading crop species of Brassica, Sinapis and Raphanus, while knowledge of the genome organization in the wild species is still fragmentary (Ali et al., 2005) or non-existent. Investigated species of Brassica and other related genera possess small and morphologically uniform chromosomes. The number of chromosomes is often relatively high due to widespread allopolyploidy. These features make karyotype analysis difficult when simple chromatin staining and morphometric studies of chromosome organization are carried out (Olin-Fatih and Heneen, 1992).
The two types of ribosomal RNA genes, 5S rDNA and 45S rDNA encoding for 18S–5.8S–25S ribosomal RNAs, are widely used as probes for fluorescence in situ hybridization (FISH), providing chromosome markers in experiments performed on various plant species (Maluszynska, 2002). In species of Brassica, Raphanus and Sinapis, the simultaneous use of these probes revealed a high number of ribosomal RNA loci at different chromosomal locations, which in some genomes offers simple but reliable chromosome landmarks (Kamisugi et al., 1998; Schrader et al., 2000; Snowdon et al., 2000, 2002; Hasterok et al., 2001; Kulak et al., 2002).
Analysis of the chromosomal localization of 45S ribosomal DNA sites (Maluszynska and Heslop-Harrison, 1993; Cheng et al., 1995; Snowdon et al., 1997; Fukui et al., 1998; Hasterok and Maluszynska, 2000) followed later by 5S rDNA sites (Snowdon et al., 2000, 2002; Hasterok et al., 2001; Kulak et al., 2002) was performed independently in several different institutions, both for the key crop species of Brassica and also, but to a lesser extent, for other Brassicaceae species (Fransz et al., 1998; Schrader et al., 2000; Ali et al., 2005). Since the first FISH-based studies, discrepancies regarding the number and distribution of rDNA loci have been reported both for diploid and allotetraploid species, confirming similar observations made in earlier molecular analyses (Delseny et al., 1990; McGrath et al., 1990; Kianian and Quiros, 1992). Importantly, in the studies above, determination of rDNA chromosomal distribution was usually based on a single variety or cultivar. Such variation was also reported in some other plant species, e.g. rice (Fukui et al., 1994) and common bean (Pedrosa et al., 2003), and in the case of Brassica species was usually briefly attributed to different origins of the material studied, and differences in technical approaches and sensitivity of FISH carried out by different groups (Maluszynska and Heslop-Harrison, 1993; Snowdon et al., 1997, 2000; Fukui et al., 1998; Hasterok et al., 2001).
The aim of the present study was to analyse the chromosomal distribution of 5S and 45S ribosomal DNA loci in the key crop species of Brassica, their wild relatives and in species closely related to the Brassica genus. Special emphasis is placed on the analysis of polymorphism of rDNA loci, a variation that seems to be particularly widespread in this group of plants.
MATERIALS AND METHODS
Plant material
Fifteen species of the Brassicaceae family belonging to five genera and comprising 52 accessions were analysed in this study. Seeds were obtained from botanical gardens, plant breeding stations, research centres and commercial sources. Information on the origins of this plant material is provided in Table 1.
Table 1.
Origins, chromosome numbers, total numbers of 5S and 25S rDNA sites, and the fraction of these sites at which the two types of rDNA are co-localized, of the 52 accessions of the 15 Brassicaceae species studied*
| Number of rDNA sites |
|||||
|---|---|---|---|---|---|
| Taxon (common name; genome) | 2n | 5S | 25S | (5S + 25S) | Origin† |
| Brassica | |||||
| B. fruticulosa (Mediterranean cabbage; F) subsp. fruticulosa | 16 | 4 | 4 | (2) | a |
| B. nigra (L.) Koch (black mustard group; B) | |||||
| var. occidentalis ‘128’ | 16 | 2 | 6 | – | b |
| var. occidentalis ‘1858’ | 16 | 2 | 6 | – | – |
| var. occidentalis | 16 | 2 | 6 | – | c |
| B. cretica Lam. (Cr) | 18 | 2 | 4–5 | – | d |
| B. oleracea group (cabbage group; C) | |||||
| subsp. oleracea convar. botrytis var. alboglabra (Bail.) | 18 | 2 | 4 | – | – |
| subsp. oleracea convar. botrytis var. alboglabra (Bail.) ‘No. 4003’ | 18 | 2 | 4‡ | – | e |
| subsp. oleracea convar. botrytis var. italica Plenck (broccoli) | 18 | 2 | 4 | – | f |
| subsp. oleracea convar. botrytis var. botrytis L. (cauliflower) | 18 | 2 | 4 | – | g |
| subsp. oleracea convar. acephala var. gongylodes L. ‘Delikates Biala’ (kohlrabi) | 18 | 2 | 4–5 | – | h |
| subsp. oleracea convar. acephala var. gongylodes L. ‘Niebieska Maslowa’ (kohlrabi) | 18 | 2 | 4 | – | g |
| subsp. oleracea convar. acephala var. viridis L. ‘Jarmuz Sredniowysoki’ (kale) | 18 | 2 | 4 | – | i |
| subsp. oleracea convar. capitata var. capitata L.‘Kissendrup Swe’ (common cabbage) | 18 | 2 | 4 | – | g |
| subsp. oleracea convar. capitata var. capitata L.‘Koda’ (common cabbage) | 18 | 2 | 4 | – | g |
| subsp. oleracea convar. gemmifera var. gemmifera DC.‘Maczuga’ (Brussels sprouts) | 18 | 2 | 4 | – | g |
| subsp. oleracea convar. capitata var. sabauda L.‘Langediijker Dauer’ (Savoy cabbage) | 18 | 2 | 4 | – | g |
| subsp. oleracea convar. capitata var. capitata L.‘Kamienna Glowa’ (common cabbage) | 18 | 2 | 4 | – | j, k |
| subsp. oleracea convar. capitata var. capitata L.‘Amager’ (common cabbage) | 18 | 2 | 4 | – | g |
| subsp. oleracea (ornamental form) | 18 | 2 | 4 | – | g |
| ‘Bpl 3-1’ (rapid-cycling form) | 18 | 2 | 4 | – | l |
| B. oxyrrhina (O) | 18 | 4 | 4 | (2) | d |
| B. rapa group (turnip/bird rape group; A) | |||||
| subsp. oleifera (DC.) ‘Schneeball’ (oilseed turnip) | 20 | 6 | 6 | (4) | g |
| subsp. pekinensis (Lour.) (Chinese cabbage) | 20 | 6 | 10 | (4) | g |
| subsp. rapa ‘Goldball’ (turnip) | 20 | 6 | 10 | (4) | g |
| subsp. rapa ‘Goldball’ (turnip) | 20 | 8 | 10 | (6) | m |
| ‘Bpl 1-1’ (rapid-cycling form) | 20 | 9 | 10 | (7) | l |
| subsp. trilocularis (Roxb.) ‘K-151’ (Indian colza) | 20 | 10 | 10 | (8) | e |
| B. tournefortii Gouan ‘1412’ (Asian mustard; T) | 20 | 4 | 2 | (2) | a |
| B. carinata group (mustard collard group; BC) | |||||
| ‘s-67’ | 34 | 4 | 8–10 | – | n |
| ‘Yellow Dodolla’ | 34 | 4 | 8–10 | – | n |
| B. juncea group (India mustard group; AB) | |||||
| var. crispa ‘685’ | 36 | 8–10 | 12 | (4–6) | o |
| ‘684’ | 36 | 10 | 14 | (6) | o |
| ‘Muasiao’ | 36 | 10 | 16 | (6) | p |
| ‘9565’ | 36 | 10 | 16 | (6) | o |
| ‘Malopolska’ | 36 | 10 | 16 | (6) | q |
| B. napus group (rape; AC) | |||||
| ‘429’ | 38 | 9–11 | 10–11 | (5–7) | o |
| subsp. napus ‘Kana’ (rapeseed) | 38 | 10 | 12 | (6) | r |
| subsp. napobrassica ‘Brukiew Wilhelmburska’ (swede) | 38 | 10 | 14 | (6) | g |
| ‘Bpl 5-1’ (rapid-cycling form) | 38 | 10 | 14 | (8) | l |
| ‘K-151’ × ‘No. 4003’ (resynthesized) | 38 | 12 | 14 | (8) | e |
| subsp. napus ‘Licosmos 00’ (rapeseed) | 38 | 12 | 14 | (8) | s |
| subsp. napus ‘Marita’ (rapeseed) | 38 | 12 | 14 | (8) | r |
| Diplotaxis | |||||
| D. muralis ‘690’ (wall rocket; Dm) (putative allotetraploid) | 42 | 6 | 6 | (2) | t |
| Eruca | |||||
| E. vesicaria (garden rocket; E) subsp. sativa | 22 | 2 | 7 | (2) | o |
| Raphanus | |||||
| R. sativus (radish; R) | |||||
| convar. sativus ‘186’ | 18 | 4 | 6 | (2) | b |
| convar. sativus ‘Opolanka’ | 18 | 5 | 5 | (3) | g |
| convar. sativus ‘Agata’ (autotetraploid) | 36 | 8 | 8 | (4) | g |
| Sinapis (mustard; S) | |||||
| S. arvensis L. ‘1524’ (charlock; Sar) | 18 | 4 | 6 | (2) | a |
| S. alba (white mustard; Sal) | |||||
| subsp. alba ‘755’ | 24 | 4 | 8 | (2) | t |
| subsp. alba ‘Kamis’ | 24 | 4 | 8 | (2) | u |
| subsp. dissecta (Lag.) | 24 | 4 | 8 | (2) | a |
| subsp. alba var. oleifera ‘774’ (autotetraploid) | 48 | 8 | 16 | (4) | o |
Botanical nomenclature used in this study follows Mansfeld's Taxonomic Database of Crop Plants (http://mansfeld.ipk-gatersleben.de/mansfeld/Query.htm).
Five individuals from each accession, except for the two accessions of B. carinata from which 20 individuals, were studied.
a, Botanical Garden (BG) Berlin, Dahlem, Germany; b, Conservatorie et jardins Botaniques de Nancy, France; c, BG Powsin, Poland; d, Coimbra, Portugal; e, Swedish University of Agricultural Sciences, Alnarp, Sweden; f, CNOS Wroclaw, Poland; g, PNOS Ozarow Mazowiecki, Poland; h, Coleus Mosina, Poland; i, CNOS Poznan, Poland; j, PNOS Torun, Poland; k, Flora-Samen sp. z o.o., Jazgarzew k/Piaseczna, Poland; l, CrGC Madison, USA; m, PNOS Krakow, Poland; n, Holetta R. Center, Ethiopia; o, UMCS BG Lublin, Poland; p, Glasgow, UK; q, PHN Lublin, Poland; r, ZDHiAR Malyszyn, Poland; s, PHPU Kruszewnica, Poland; t, BG Gottingen, Germany; u, Kamis, Przyprawy SA, Wolka Kosowska, Poland.
Polymorphism regarding the localization of two sites.
Somatic chromosome preparations
All seeds were germinated on filter paper moistened with tap water at 20–22 °C in the dark for 3–5 d. Further treatment was carried out as described earlier (Hasterok et al., 2001). Briefly, to ensure optimal chromatin condensation at metaphase, whole seedlings with roots 1.0–2.0 cm long were immersed for 2 h at room temperature in 2 mm 8-hydroxyquinoline, then fixed in 3 : 1 (v/v) methanol : glacial acetic acid and stored at –20 °C. Excised roots were washed in 0.01 m citric acid–sodium citrate buffer (pH 4.8) for 20 min prior to digestion in an enzyme mixture of 20 % (v/v) pectinase (Sigma), 1 % (w/v) cellulase (Calbiochem) and 1 % (w/v) cellulase ‘Onozuka R-10’ (Serva) for 1.5–2 h at 37 °C. Meristems were dissected out from root tips, squashed in drops of 45 % acetic acid and frozen. After removal of coverslips, the preparations were post-fixed in 3 : 1 ethanol : glacial acetic acid, followed by dehydration in absolute ethanol and air-dried.
Successful chromosome preparations made from at least five individual plants from each accession were subjected to FISH analysis. In the case of the two accessions of B. carinata, 20 individuals from each were studied for a detailed comparison of interindividual variation. One root tip per seedling (individual) was used to make one preparation.
DNA probes and FISH
The following ribosomal DNA sequences were used as probes: 5S rDNA (pTa794) (Gerlach and Dyer, 1980) labelled using PCR with tetramethyl-rhodamine-5-dUTP (Roche), and a 2.3-kb ClaI subclone of the 25S rDNA coding region of Arabidopsis thaliana (Unfried and Gruendler, 1990) labelled by nick translation using digoxigenin-11-dUTP (Roche). The latter probe was used to determine the chromosomal localization of 18S–5.8S–25S rRNA genes (45S rDNA). The labelling procedure followed the methods described in detail by Hasterok et al. (2002).
The hybridization mixture consisted of 50 % deionized formamide, 20 % dextran sulphate, 2× SSC and salmon sperm blocking DNA in 50–100× excess of labelled probes. The ribosomal DNA probes were mixed to a final concentration of about 2.5 ng μl−1 and pre-denatured (80 °C for 10 min). The slides with chromosome material were then denatured along with the hybridization mixture (70 °C for 4.5 min) and allowed to hybridize for 12–18 h in a humid chamber at 37 °C. The post-hybridization washes were carried out in two changes of 20 % deionized formamide in 0.1× SSC, 5 min each, at 42 °C, which is equivalent to 85 % stringency. Immunodetection of digoxigenated probes was done according to standard protocol using FITC-conjugated anti-digoxigenin antibodies (Roche). The preparations were mounted and counterstained in Vectashield (Vector Laboratories) containing 2.5 μg ml−1 of DAPI (4′,6-diamidino-2-phenylindole; Serva).
Image capturing and processing
All images were acquired using either an Olympus Camedia C-4040Z digital camera attached to a Leica DMRB epifluorescence microscope or a Hamamatsu C5810 CCD camera attached to an Olympus Provis AX epifluorescence microscope and processed uniformly using Micrografx (Corel) Picture Publisher software.
RESULTS AND DISCUSSION
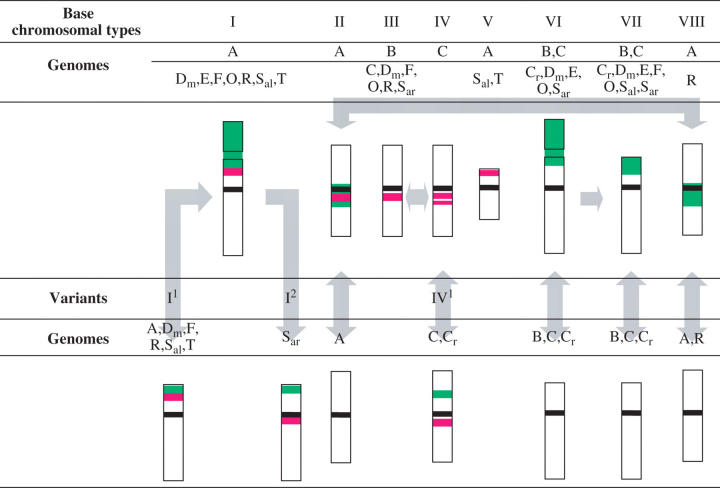
Total numbers of 5S rDNA and 25S rDNA sites in all 52 accessions of the 15 species studied are summarized in Table 1. The fractions of sites at which the two classes of ribosomal genes are co-localized are also indicated. Figures 1–5illustrate selected chromosomal distributions of rDNA sites in 18 accessions representing 13 species of Brassicaceae. Table 2 provides detailed comparative information about types and distributions of rDNA-bearing chromosomes found in these selected examples, without listing other distributions recorded in these accessions (Table 1). Emphasis in Table 2 has been placed on taxa or rDNA distributions not reported before. The idiograms of the base types of chromosomes bearing 5S rDNA and 25S rDNA loci and their variants are presented in Table 3. The base chromosomal types, numbered I–VIII, have been introduced and described in detail by Hasterok et al. (2001), with the exception that the 5S rDNA site in chromosome type V is now assigned to the short arm. These types have originally been used to describe the chromosomal distribution of rDNA sites in the three genomes (A, B and C) that constitute the six most studied crop species of Brassica. Most of these types have also been found in related species and genera. To be emphasized here is the fact that a certain type may refer to a specific chromosome in a defined complement (Hasterok et al., 2005). It may also refer to undefined chromosomes in the same complement and/or homoeologues or non-homoeologues in different complements.
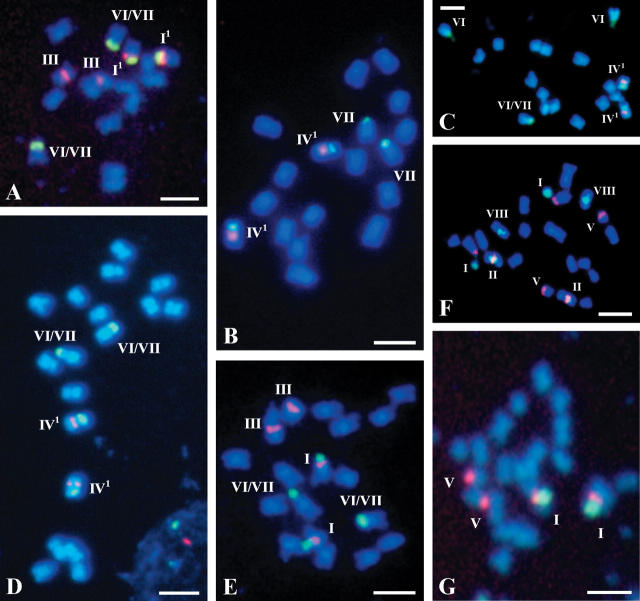
Fig. 1.
Fluorescence in situ hybridization of 5S (red) and 25S (green) ribosomal DNA probes to somatic metaphase chromosomes of (A) Brassica fruticulosa, (B and C) B. cretica, (D) B. oleracea ‘No. 4003’, (E) B. oxyrrhina, (F) B. rapa `Schneeball’, (G) B. tournefortii ‘1412’. The nomenclature of rDNA-bearing chromosomal types (Roman numerals) in Fig. 1–5 is according to Hasterok et al. (2001), supplemented by the new and variant chromosomal types identified in the present study. The chromosomes are counter-stained with DAPI (blue). White arrows visible on some photographs help to link the Roman numerals with their rDNA-bearing chromosomes. All scale bars = 5 μm.
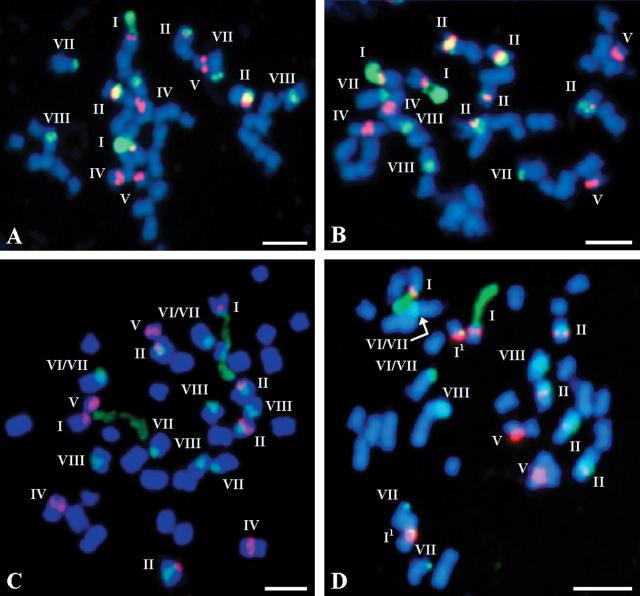
Fig. 2.
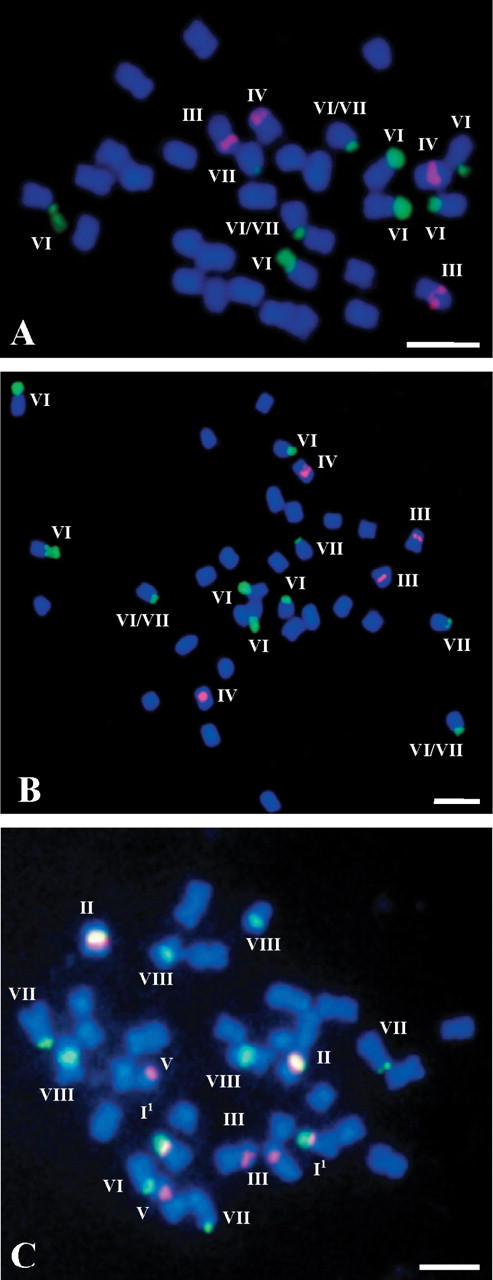
FISH of 5S and 25S rDNA probes to somatic metaphase chromosomes of (A and B) B. carinata ‘Yellow Dodolla’ and (C) B. juncea ‘685’.
Fig. 3.
FISH of 5S and 25S rDNA probes to somatic metaphase chromosomes of (A and B) Brassica napus ‘429’, (C) B. napus ‘Brukiew Wilhelmburska’ and (D) B. napus ‘Bpl 5-1’ (rapid-cycling form).
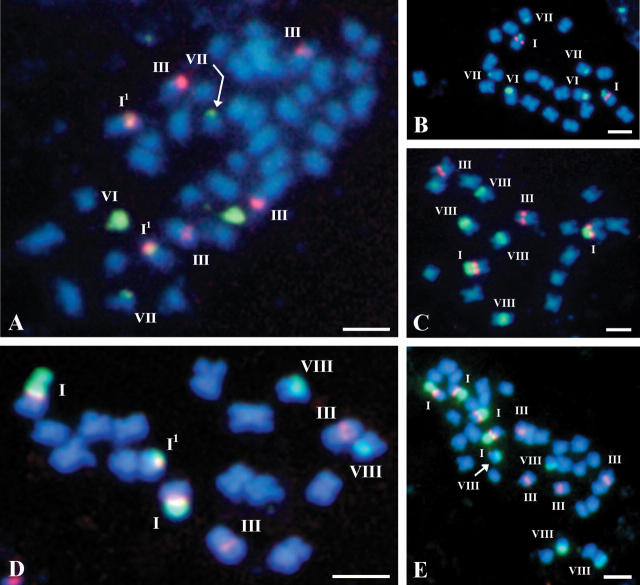
Fig. 4.
FISH of 5S and 25S rDNA probes to somatic metaphase chromosomes of (A) Diplotaxis muralis ‘690’, (B) Eruca vesicaria, (C) Raphanus sativus ‘186’, (D) R. sativus ‘Opolanka’ and (E) R. sativus ‘Agata’ (autotetraploid).
Fig. 5.
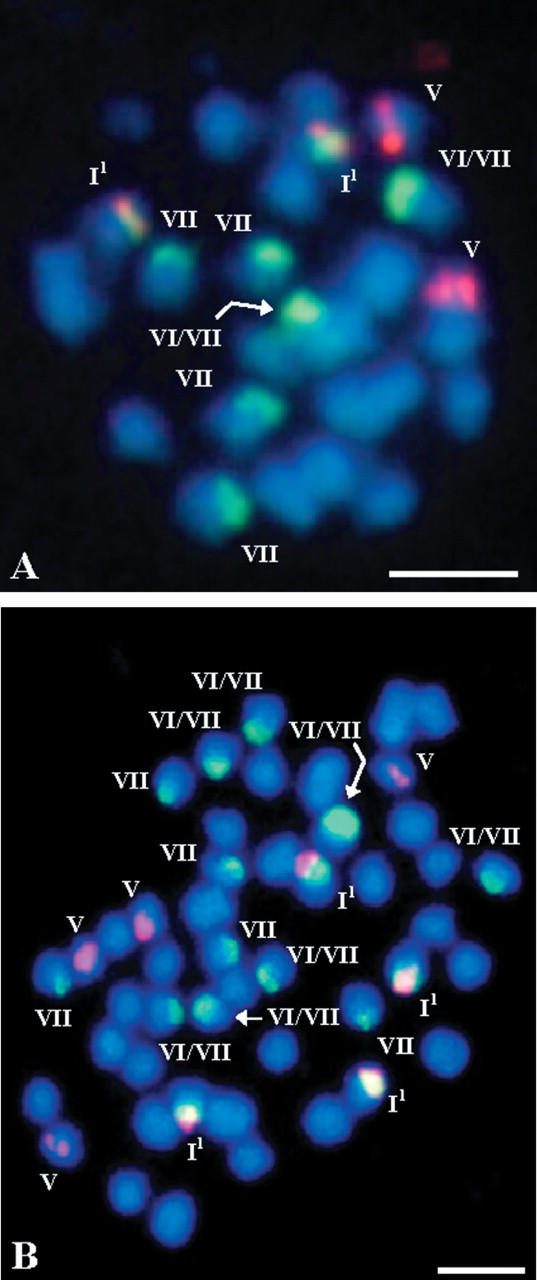
FISH of 5S and 25S rDNA probes to somatic metaphase chromosomes of (A) S. alba subsp. dissecta, (B) S. alba ‘774’ (autotetraploid).
Table 2.
Selected unreported distributions of rDNA chromosomal sites observed in 18 accessions of the 13 Brassicaceae species studied
| rDNA-bearing chromosomal types |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Taxon (genome) | 2n | I | II | III | IV | V | VI | VII | (VI/VII)* | VIII | Fig. |
| Brassica fruticulosa (F) subsp. fruticulosa | 16 | 21† | – | 2 | – | – | – | – | 2 | – | 1A |
| B. cretica (Cr) | 18 | – | – | – | 21 | – | – | 2 | – | – | 1B |
| 18 | – | – | – | 21 | – | 2 | – | 1 | – | 1C | |
| B. oleracea (C) ‘No. 4003’ | 18 | – | – | – | 21 | – | – | – | 2 | – | 1D |
| B. oxyrrhina (O) | 18 | 2 | – | 2 | – | – | – | – | 2 | – | 1E |
| B. rapa (A) ‘Schneeball’ | 20 | 2 | 2 | – | – | 2 | – | – | – | 2 | 1F |
| B. tournefortii (T) ‘1412’ | 20 | 2 | – | – | – | 2 | – | – | – | – | 1G |
| B. carinata (BC) ‘Yellow Dodolla’ | 34 | – | – | 2 | 2 | – | 6 | 1 | 2 | – | 2A |
| 34 | – | – | 2 | 2 | – | 6 | 2 | 2 | – | 2B | |
| B. juncea (AB) ‘685’ | 36 | 21 | 2 | 2 | – | 2 | 1 | 3 | – | 4 | 2C |
| B. napus (AC) ‘429’ | 38 | 2 | 3 | – | 2 | 2 | – | 2 | – | 3 | 3A |
| 38 | 2 | 5 | – | 2 | 2 | – | 2 | – | 2 | 3B | |
| ‘Brukiew Wilhelmburska’ | 38 | 2 | 4 | – | 2 | 2 | – | 2 | 2 | 4 | 3C |
| ‘Bpl 5-1’ (rapid-cycling form) | 38 | 2 + 21‡ | 4 | – | – | 2 | – | 2 | 2 | 2 | 3D |
| Diplotaxis muralis (Dm) ‘690’ | 42 | 21 | – | 2 + 2 | – | – | 2 | 2 | – | – | 4A |
| Eruca vesicaria (E) subsp. sativa | 22 | 2 | – | – | – | – | 2 | 3 | – | – | 4B |
| Raphanus sativus (R) ‘186’ | 18 | 2 | – | 2 | – | – | – | – | – | 4 | 4C |
| ‘Opolanka’ | 18 | 2 + 11 | – | 2 | – | – | – | – | – | 2 | 4D |
| ‘Agata’ (autotetraploid) | 36 | 4 | – | 4 | – | – | – | – | – | 4 | 4E |
| Sinapis alba (Sal) subsp. dissecta | 24 | 21 | – | – | – | 1 + 1 | – | 4 | 2 | – | 5A |
| ‘774’ (autotetraploid) | 48 | 41 | – | – | – | 2 + 2 | – | 5 | 7 | – | 5B |
Types VI and VII are considered as one group when not distinguishable.
Superscript numbers indicate the variant types I1, I2 and IV1.
Two values are given when distinct and consistent differences in size of signal occur on chromosomes of the same type.
Table 3.
The base and variant types of chromosomes bearing or lacking 5S rDNA (red) and 25S rDNA (green) loci in the genomes of Brassicaceae species
Differences in size of these chromosome types and in size of rDNA signals were observed between chromosomes of the same type within and between accessions; thus a type can refer to different chromosomes.
The size of the satellite in types I and VI may differ between accessions.
Non-distension of the secondary constriction and/or smaller size or loss of the satellite possibly produce type I1 from type I. Alternatively, type I1 may represent a different chromosome. Type I2, probably a variant of type I or I1, represents a chromosome variant observed so far only in the genome Sar.
Arrows indicate possible variation in the appearance of a certain chromosome type that leads to its manifestation as another type.
As far as is known, localization of 5S and 25S rDNA has been determined for the first time in six taxa: Brassica fruticulosa, B. cretica, B. oxyrrhina, B. tournefortii, Diplotaxis muralis and Eruca vesicaria (Figs 1A–C, E and G and 4A and B and Tables 1 and 2). In addition to the occurrence of the eight base types and possible loss of rDNA loci in types II, VI, VII and VIII, the presence of three new types of chromosomes with rDNA sites is documented from the Brassicaceae material studied (Table 3). The new types are treated as variants of types I (I1 and I2) and IV (IV1). Thus, type I might appear as type I1 by having a non-distended secondary constriction and/or a minor satellite, or by lacking the satellite. Alternatively, type I1 may represent a different intact or structurally altered chromosome, especially in those cases that have the expected number of type I chromosomes (B. napus ‘Bpl 5-1’ and Raphanus sativus ‘Opolanka’; Figs 3D and 4D, respectively). The variant type I2 was identified in the chromosome complement of Sinapis arvensis (data not shown). This type is similar to the variant type I1 except that the 5S rDNA site is localized proximally in the long arm and not distally on the short arm. The distension/non-distension of the secondary constriction, the presence/absence of the satellite or difference between satellite size resulting in variants I1 and I2 could well explain the difficulty in distinguishing types VI and VII in certain cases (Table 2), exemplified by B. carinata (Fig. 2A and B).
Type IV1, a variant of type IV specific for the C genome, is characterized by the presence of an intercalary site of 25S rDNA on the short arm and one proximal site of 5S rDNA, instead of two, on the long arm. This chromosomal type, present in the genome of the wild species B. cretica (Fig. 1B and C), was occasionally observed in some forms of B. oleracea (e.g. in some individuals of ‘No. 4003’; Fig. 1D).
Possibly interrelated base and variant chromosome types, occasionally indistinguishable types and loss of rDNA from certain types are indicated by arrows in Table 3. The prevalence of these features in many Brassicaceae species is the basis of the polymorphism in number, appearance and chromosomal distribution of both 5S and 25S ribosomal DNA loci.
Intergenomic variation in number of ribosomal DNA loci
The genomes of the materials studied are indicated in Table 1. For simplification the genome of Diplotaxis muralis (2n = 42) has been marked as Dm, although this species is considered to be an allotetraploid, most likely from D. viminea (2n = 2x = 20) and D. tenuifolia (2n = 2x = 22) (Martinez-Laborde, 1997). The number of 5S and 25S rDNA loci differs significantly between different genomes. The lowest somatic number of 25S rDNA sites (2) among the diploids was observed in the T genome of the wild species B. tournefortii (Fig. 1G), while the highest (10) was found in most accessions of B. rapa with the same chromosome number but containing the A genome (Table 1). Also, all three accessions of Sinapis alba invariably had eight sites of 25S rDNA (Fig. 5A). However, the most commonly observed number of 25S rDNA sites in the diploids was four (found in B. fruticulosa, B. oxyrrhina, the majority of individuals in B. cretica and almost all accessions of B. oleracea studied) and six (B. nigra, one accession of B. rapa, D. muralis, one accession of R. sativus and in S. arvensis).
Similarly, in the case of 5S rDNA there were several diploids, i.e. three accessions of B. nigra, one accession of B. cretica (Fig. 1B and C), all accessions of B. oleracea (Table 1 and Fig. 1D) and one accession of E. vesicaria (Fig. 4B) that possessed the minimum number of two sites per somatic chromosome complement. A large group of diploid species comprising B. fruticulosa (Fig. 1A), B. oxyrrhina (Fig. 1E), one accession of R. sativus (Fig. 4C), S. arvensis and three accessions of S. alba (Fig. 5A) has four sites. The highest number of 5S rDNA sites was again observed in B. rapa, from six (Hasterok et al., 2001) up to ten (Hasterok et al., 2005).
The two kinds of rDNA sites appeared on the same chromosome in 12 out of the 15 species studied. In these cases the sites were either co-localized on the same arm or were on different arms. It can be concluded from the above that despite having the same chromosome number, different genomes of Brassicaceae may differ up to 5-fold in the number of rDNA loci. A correlation between the increase in the number of rDNA sites and the increase of chromosome number in the genome, if existing at all, is very weak.
Variation in chromosomal distribution of rDNA loci
Comparative analyses revealed that polymorphism in the number and chromosomal distribution of 5S and 25S rDNA occurs both among different subspecies and varieties of a given species and within a population of individuals that belong to one variety or cultivar. However, no polymorphism was encountered within an individual. Inter- and/or intravarietal polymorphism was observed in seven out of 15 species, i.e. in B. cretica, B. oleracea, B. rapa, B. carinata, B. juncea, B. napus and R. sativus (Table 1). Noticeably, all species that contain the A genome in their chromosome complement, i.e. B. rapa, B. juncea and B. napus, displayed high polymorphism in number and chromosomal localization of rDNA sites. The chromosomes of this genome carry extraordinary high numbers of both 5S rDNA and 25S rDNA sites (Hasterok et al., 2001), a feature which is possibly the cause of their polymorphism in number and distribution.
Detailed analysis of rDNA distribution in the six accessions of B. rapa revealed that only the chromosomes with proximally and/or pericentromerically distributed ribosomal rRNA genes were polymorphic. These chromosomes represented types II and VIII. It seems that the same chromosome may occur in different materials as type II, type VIII or as an unidentified chromosome without any rDNA landmark, depending upon the presence or absence of proximal and pericentromeric 5S and 25S rDNA sites (Table 3). It seems that the proximal 5S rDNA site in the A genome is more liable to change than its pericentromeric 25S rDNA counterpart, as absence of the 5S rDNA site was more common than the lack of the 25S rDNA site. This was observed in three out of six accessions of B. rapa (Table 1) leading to the more frequent occurrence of type VIII compared with type II chromosomes.
In B. rapa ‘Schneeball’, two chromosomes of type I and only two chromosomes of each of types II and VIII constitute a total of six sites of 25S rDNA (Fig. 1F), which is the lowest so far observed for B. rapa (Table 1). All potential sites lacking rDNA were invariably proximal and pericentromeric in the five individuals studied. Absence of such loci from type II and VIII chromosomes makes these chromosomes unidentifiable. At the other extreme, the presence of both 5S and 25S rDNA sites in all potential sites leads to over-representation of type II chromosomes, raising the total number of each of the 5S and 25S rDNA sites to ten (subsp. trilocularis ‘K-151’) (Hasterok et al., 2005). Interestingly, no accession of A genome-containing Brassica species had only the 5S rDNA site retained at the proximal site of a putative type II chromosome (Table 3). Variation in numbers of chromosome types II and VIII also partially contributed to the polymorphism of rDNA sites in the two allopolyploid Brassica species that contain the A genome, i.e. B. juncea (Fig. 2C) and B. napus (Fig. 3A–D). On the other hand chromosome types V and I were not prone to such variation. Chromosome type V with a solitary 5S rDNA locus in the distal part of the short arm was never involved. Also chromosome type I, carrying 5S rRNA genes in the short arm and 25S rDNA in the secondary constriction and its adjacent regions, was almost constant in this genome. Cytochemical analysis of 18S–5.8S–25S rRNA gene activity by silver staining revealed that chromosome type I is the only type in the A genome that carries transcriptionally active clusters of these genes, while those in loci near centromeres are not expressed (Hasterok and Maluszynska, 2000). Possibly, the sites that contain such inactive 45S rDNA are more prone to loss from the chromosome. It is more difficult, however, to speculate if the same applies to 5S rDNA sites, as there is no cytochemical method available that could distinguish active and inactive loci of these genes.
In contrast to the A genome, genome B in B. nigra and genome C in B. oleracea contain more than one pair of chromosomes with distal sites of 25S rDNA (types VI and VII) which are either actually (Hasterok and Maluszynska, 2000) or potentially (Hasterok and Maluszynska, 1999) transcriptionally active. That the type I pair of chromosomes is the only one in the A genome that carries active 25S rDNA genes, possibly relates to the stability of this site. Apart from the occurrence of type I1 in some of the A genome-containing allotetraploids (i.e. in B. juncea ‘685’; Fig. 2C), the distal rDNA sites of type I in the A genome never undergo any marked change in number, implying the loss of the entire locus. This is not the case in type VI and VII chromosomes of the B, C and Cr genomes, where such distal loci may be polymorphic through their presence or absence both in diploids (Fig. 1B vs C) and more commonly in allopolyploid species of Brassica (e.g. Fig. 2A vs B, and 3A and B vs C and D).
The results of the present analyses based on several forms of both diploid and allopolyploid B or/and C genome-containing species suggest that probably the same chromosome can be either type VI or type VII, depending upon the appearance of its short arm (Table 3). As the phenomena of distended/non-distended secondary constriction, presence/absence of satellite, or variation in satellite size were consistent and in many cases form-specific, it is supposed that this may largely reflect some differences in the activity of individual loci (Flavell, 1986). An analogous situation might pertain to chromosome type I, where in some forms of B. juncea it possibly appeared as type I1 (e.g. Fig. 2C and Table 3).
Interindividual polymorphism was investigated in detail in two accessions of B. carinata, ‘s-67’ and ‘Yellow Dodolla’. The number of 45S rDNA sites was recorded for 20 individuals in each accession and ranged from eight to ten, while four sites of 5S rDNA were consistently found in both materials (Fig. 2A and B). Regardless of accession, the individuals with eight or ten sites of 45S rDNA in total represented 90 % of the population, while the remaining 10 % were the individuals with nine sites. Interindividual polymorphism in the number and/or distribution of 45S rDNA sites was also occasionally observed in B. cretica, some accessions of B. oleracea, B. juncea and B. napus (Table 1), but no detailed investigations were made.
An interesting case of chromosome polymorphism concerns type IV of the C genome. This type is characterized by the presence of two adjacent 5S rDNA sites near the centromere on the long arm and was recorded in earlier investigations of B. oleracea, B. napus and B. carinata (Armstrong et al., 1998; Hasterok et al., 2001; Kulak et al., 2002; Snowdon et al., 2002). That the 5S rDNA genes in this chromosome may be detectable as one signal instead of two in B. oleracea ‘No. 4003’ (Hasterok et al., 2005) was further encountered in the B. oleracea accessions: ‘Delikates Biala’ and ‘Kissendrup Swe’. Analysis of both metaphase and prometaphase chromosome spreads suggests that this phenomenon is more likely to be caused by a significant decrease in copy number, or complete loss, of 5S rDNA at one locus, rather than differences in the extent of chromatin condensation influencing mapping resolution.
The origin of chromosome type IV1 (Table 3), found in some individuals of B. oleracea ‘No. 4003’ (Fig. 1D) and ‘Delikates Biala’, is debatable. Its origin is connected with the presence of a highly polymorphic additional site of 25S rDNA which is localized interstitially in the short arm. In most accessions of B. oleracea studied so far, this locus is absent. The existence of such an interstitial locus of 25S rDNA on the 5S rDNA-bearing chromosome of the C genome has been reported by Howell et al. (2002) in another cultivar of B. oleracea var. alboglabra, but in this case the locus was additional to those on chromosomes types VI and VII. In the present study on the other hand, the existence of type IV1 chromosomes was always linked with an absence of a 25S locus on type VI or VII chromosomes. Two chromosomes type IV1 were also observed in all analysed individuals of B. cretica (Fig. 1B and C), the wild species that contains the Cr genome, which in number and morphometric characteristics of its chromosomes, as well as distribution of rDNA sites, closely resembles the C genome. This seems to confirm the hypothesis that B. cretica could be the putative ancestor of B. oleracea (Prakash and Hinata, 1980).
Further comparisons between the rDNA distribution patterns documented in the diploid species studied (Table 1) may be considered in a phylogenetical context. Based on traditional taxonomy and molecular analyses, primarily on ribosomal internal transcribed spacer (ITS) sequences, it has been shown that B. oleracea and B. rapa belong to the same clade, while B. nigra and Sinapis arvensis belong to another clade, and Raphanus is a ‘hybrid’ between these two clades (Yang et al., 1999; Koch et al., 2003; Warwick and Sauder, 2005). The present FISH results highlight the low number of sites of a less variable rDNA in B. oleracea in contrast to the high number of sites of a more variable rDNA in B. rapa, considered to be in the same clade. This may necessitate a re-evaluation of the interrelationships between these two species. Whether the deviation or divergence of rDNA patterns reflects possible differences in the evolutionary ages of these taxa or not remains to be substantiated. The number and variation in rDNA patterns of R. sativus represents an intermediate position between the invariable B. nigra and the variable B. rapa. Brassica fruticulosa and B. oxyrrhina with their nearly identical rDNA FISH patterns, and B. tournefortii with a slightly deviating pattern, are all closely related according to the ITS analysis (Warwick and Sauder, 2005). It is to be remembered that one should consider the ribosomal DNA FISH and molecular ITS data with caution when inferring phylogenies, due to limitations such as possible lack of homology, difficulties in defining orthologues from paralogues, and prevalence of non-functional rDNA pseudogenes (Alvarez and Wendel, 2003; Bailey et al., 2003; Dobigny et al., 2004).
Number of rDNA sites and polyploidization
It was shown by Maluszynska and Heslop-Harrison (1993) that the number of 45S ribosomal DNA sites in the genomes of the three allotetraploid crop species of Brassica was lower than the sum of the sites observed in their putative diploid ancestral species. The number of loci that were absent from chromosome complements of the allotetraploids was either two (B. carinata and B. juncea) or four (B. napus). In a later study, Snowdon (1997) reported an identical situation for B. juncea and B. napus, while the number of rDNA sites in the B. carinata genome equalled the sum of loci observed in its diploid ancestors. The decrease in the expected number of both 5S and 45S ribosomal DNA sites during polyploid evolution was also observed in other plant species, such as Scilla autumnalis (Vaughan et al., 1993), Nicotiana tabacum (Volkov et al., 1999) and in species of Sanguisorba (Mishima et al., 2002). This phenomenon is usually explained as a possible consequence of diploidization, i.e. a consequence of transient genome imbalance following hybridization. The ribosomal RNA genes contributed from the two ancestral genomes could constitute an excess of these genes and, as a consequence, precipitate the elimination of some dispensable loci (Dvorak, 1990).
The current observations do not support the above conclusion. It seems that there is no clear tendency to reduce or increase the number of rDNA loci in the allotetraploid species of Brassica. This was the case in the resynthesized B. napus ‘K-151’ × ‘No. 4003’ in which both numbers of 5S rDNA and 25S rDNA sites were exactly the sum of what was observed in the parental types (Table 1). The variation in rDNA patterns in the current material is most likely caused by distinct intervarietal and interindividual polymorphism in the number of ribosomal DNA sites observed in both diploid and allotetraploid species of Brassica.
Of interest is the comparison of the number and chromosomal localization of rDNA sites between the diploid (Fig. 4C and D) and autotetraploid (Fig. 4E) accessions of R. sativus (Tables 1 and 2). A clearly visible deficit of 25S rDNA loci in the autotetraploid is caused by a lower than expected number of type VIII chromosomes, or an absence of extra type I1 chromosomes. It can be quite difficult, however, to unambiguously interpret this observation. One possible explanation could be the absence of two pairs of type VIII chromosomes in the autotetraploid. On the other hand, the presence of two such pairs in the diploid which contributed to the tetraploid cannot be ruled out. The number and distribution of ribosomal DNA sites observed earlier in diploid R. sativus (Schrader et al., 2000) seem to suggest that the latter explanation is more likely.
In contrast to R. sativus, the numbers of both kinds of ribosomal DNA sites observed in the autotetraploid accession of S. alba (Fig. 5B) were exactly twice the numbers in the three diploid accessions, which may imply lack of highly polymorphic rDNA sites in the Sal genome. Also, this may suggest that in spite of both the higher numbers of chromosomes in the complement in general and 25S rDNA sites in particular, no detectable chromosome rearrangements involving ribosomal DNA-containing regions have occurred in the autotetraploid form of S. alba. Noticeably, changes in chromosome structure were reported for some other species, e.g. in the colchicine-induced autotetraploid of A. thaliana, which was explained in terms of a diploidization feature (Weiss and Maluszynska, 2000).
Acknowledgments
We are indebted to Dr Glyn Jenkins (University of Wales Aberystwyth, UK) and the two referees for their valuable comments on the manuscript. The authors acknowledge financial support from Polish National Committee of Scientific Research (grants 3 PO4C 013 22 and 6 PO4C 006 28) and by International Atomic Energy Agency grant. This study was also partially supported by the Royal Swedish Academy of Agriculture and Forestry (award to R.H. and W.K.H. in 2002) as well as by Einar and Inga Nilssons Foundation (award to R.H. and W.K.H. in 2003–2004).
LITERATURE CITED
- Ali HBM, Lysak A, Schubert I. 2005. Chromosomal localization of rDNA in the Brassicaceae. Genome 48: 341–346. [DOI] [PubMed] [Google Scholar]
- Alvarez I, Wendel JF. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29: 417–434. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Fransz P, Marshall DF, Jones GH. 1998. Physical mapping of DNA repetitive sequences to mitotic and meiotic chromosomes of Brassica oleracea var. alboglabra by fluorescence in situ hybridisation. Heredity 81: 666–673. [Google Scholar]
- Bailey CD, Carr TG, Harris SA, Hughes CE. 2003. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution 29: 435–455. [DOI] [PubMed] [Google Scholar]
- Cheng BF, Heneen WK, Pedersen C. 1995. Ribosomal RNA gene loci and their nucleolar activity in Brassica alboglabra Bailey. Hereditas 123: 169–173. [Google Scholar]
- Delseny M, McGrath JM, This P, Chevre AM, Quiros CF. 1990. Ribosomal RNA genes in diploid and amphidiploid Brassica and related species: organization, polymorphism, and evolution. Genome 33: 733–744. [Google Scholar]
- Dobigny G, Ducroz J-F, Robinson TJ, Volobouev V. 2004. Cytogenetics and cladistics. Systematic Biology 53: 470–484. [DOI] [PubMed] [Google Scholar]
- Dvorak J. 1990. Evolution of multigene families: the ribosomal RNA loci of wheat and related species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer Associates.
- Flavell RB. 1986. The structure and control of expression of ribosomal RNA genes. Oxford Surveys of Plant Molecular and Cell Biology 3: 251–274. [Google Scholar]
- Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G. 1998. Cytogenetics for the model system Arabidopsis thaliana. The Plant Journal 13: 867–876. [DOI] [PubMed] [Google Scholar]
- Fukui K, Ohmido N, Khush GS. 1994. Variability in rDNA loci in the genus Oryza detected through fluorescence in situ hybridization. Theoretical and Applied Genetics 87: 893–899. [DOI] [PubMed] [Google Scholar]
- Fukui K, Nakayama S, Ohmido N, Yoshiaki H, Yamabe M. 1998. Quantitative karyotyping of three diploid Brassica species by imaging methods and localization of 45S rDNA loci on the identified chromosomes. Theoretical and Applied Genetics 96: 325–330. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Dyer TA. 1980. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Research 8: 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R, Maluszynska J. 1999. Activation of silent rRNA genes in Brassica species. In: Abstracts of the 2nd European Cytogenetics Conference in Vienna, Austria. Cytogenetics and Cell Genetics 85: 135. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Maluszynska J. 2000. Nucleolar dominance does not occur in root tip cells of allotetraploid Brassica species. Genome 43: 574–579. [PubMed] [Google Scholar]
- Hasterok R, Jenkins G, Langdon T, Jones RN, Maluszynska J. 2001. Ribosomal DNA is an effective marker of Brassica chromosomes. Theoretical and Applied Genetics 103: 486–490. [Google Scholar]
- Hasterok R, Langdon T, Taylor S, Jenkins G. 2002. Combinatorial labelling of DNA probes enables multicolour fluorescence in situ hybridisation in plants. Folia Histochemica et Cytobiologica 40: 319–323. [PubMed] [Google Scholar]
- Hasterok R, Wolny E, Kulak S, Zdziechiewicz A, Maluszynska J, Heneen WK. 2005. Molecular cytogenetic analysis of Brassica rapa-Brassica oleracea var. alboglabra monosomic addition lines. Theoretical and Applied Genetics 111: 196–205. [DOI] [PubMed] [Google Scholar]
- Howell EC, Barker GC, Jones GH, Kearsey MJ, King GJ, Kop EP, et al. 2002. Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y, Nakayama S, O'Neil CM, Mathias RJ, Trick M, Fukui K. 1998. Visualization of the Brassica self-incompatibility S-locus on identified oilseed rape chromosomes. Plant Molecular Biology 38: 1081–1087. [DOI] [PubMed] [Google Scholar]
- Kianian SF, Quiros CF. 1992. Genetic analysis of major multigene families in Brassica oleracea and related species. Genome 35: 516–527. [Google Scholar]
- Koch M, Al-Shehbaz A, Mummenhoff K. 2003. Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae). Annals of the Missouri Botanical Garden 90: 151–171. [Google Scholar]
- Kulak S, Hasterok R, Maluszynska J. 2002. Karyotyping of Brassica amphidiploids using 5S and 25S rDNA as chromosome markers. Hereditas 136: 144–150 (erratum in Hereditas 137: 79–80). [DOI] [PubMed] [Google Scholar]
- McGrath JM, Quiros CF, Harada JJ, Landry BS. 1990. Identification of Brassica oleracea monosomic alien chromosome addition lines with molecular markers reveals extensive gene duplication. Molecular and General Genetics 223: 198–204. [DOI] [PubMed] [Google Scholar]
- Maluszynska J. 2002. In situ hybridization in plants—methods and applications. In: Jain SM, Brar DS, Ahloowalia BS, eds. Molecular techniques in crop improvement. Dordrecht: Kluwer Academic Publishers.
- Maluszynska J, Heslop-Harrison JS. 1993. Physical mapping of rDNA loci in Brassica species. Genome 36: 774–781. [DOI] [PubMed] [Google Scholar]
- Martinez-Laborde JB. 1997. A brief account of the genus Diplotaxis. In: Padulosi S, Pignone D, eds. Rocket: a Mediterranean crop of the world. Rome: International Plant Genetic Resources Institute.
- Mishima M, Ohmido, N, Fukui K, Yahara T. 2002. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110: 550–558. [DOI] [PubMed] [Google Scholar]
- Olin-Fatih M, Heneen WK. 1992. C-banded karyotypes of Brassica campestris, B. oleracea, and B. napus. Genome 35: 583–589. [DOI] [PubMed] [Google Scholar]
- Pedrosa A, Vallejos CE, Bachmair A, Schweizer D. 2003. Integration of common bean (Phaseolus vulgaris L.) linkage and chromosomal maps. Theoretical and Applied Genetics 106: 205–212. [DOI] [PubMed] [Google Scholar]
- Prakash S, Hinata K. 1980. Taxonomy, cytogenetics and origin of crop Brassicas: a review. Opera Botanica 55: 1–57. [Google Scholar]
- Schrader O, Budahn H, Ahne R. 2000. Detection of 5S and 25S rRNA genes in Sinapis alba, Raphanus sativus and Brassica napus by double fluorescence in situ hybridization. Theoretical and Applied Genetics 100: 665–669. [Google Scholar]
- Snowdon RJ. 1997. Fluorescence in situ hybridization techniques for Brassica: methodological development and practical applications. Giessen: Justus-Liebig-Universität.
- Snowdon RJ, Friedrich T, Friedt W, Kohler W. 2002. Identifying the chromosomes of the A- and C-genome diploid Brassica species B. rapa (syn. campestris) and B. oleracea in their amphidiploid B. napus. Theoretical and Applied Genetics 104: 533–538. [DOI] [PubMed] [Google Scholar]
- Snowdon R, Friedt W, Kohler A, Kohler W. 2000. Molecular cytogenetic localization and characterization of 5S and 25S rDNA loci for chromosome identication in oilseed rape (Brassica napus L.). Annals of Botany 86: 201–204. [Google Scholar]
- Snowdon RJ, Köhler W, Köhler A. 1997. Chromosomal localization and characterization of rDNA loci in the Brassica A and C genomes. Genome 40: 582–587. [DOI] [PubMed] [Google Scholar]
- Unfried I, Gruendler P. 1990. Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Research 18: 4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan HE, Jamilena M, Ruiz Rejón Ramos C, Parker JS, Garrido-Ramos MA. 1993. Loss of nucleolar-organizer regions during polyploid evolution in Scilla autumnalis. Heredity 71: 574–580. [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V. 1999. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Molecular Biology and Evolution 16: 311–320. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Sauder CA. 2005. Phylogeny of tribe Brassiceae (Brassicaceae) based on chloroplast restriction site polymorphisms and nuclear ribosomal internal transcribed spacer and chloroplast trnL intron sequences. Canadian Journal of Botany 83: 467–483. [Google Scholar]
- Weiss H, Maluszynska J. 2000. Chromosomal rearrangement in autotetraploid plants of Arabidopsis thaliana. Hereditas 133: 255–261. [DOI] [PubMed] [Google Scholar]
- Yang Y-W, Lai K-N, Tai P-Y, Ma T-W, Li W-H. 1999. Molecular phylogenetic studies of Brassica, Rorippa, Arabidopsis and allied genera based on the internal transcribed spacer region of 18S-25S rDNA. Molecular Phylogenetics and Evolution 13: 455–462. [DOI] [PubMed] [Google Scholar]