Abstract
• Background and Aims A possible role of host tree identity in the structuring of vascular epiphyte communities has attracted scientific attention for decades. Specifically, it has been suggested that each host tree species has a specific subset of the local species pool according to its own set of properties, e.g. physicochemical characteristics of the bark, tree architecture, or leaf phenology patterns.
• Methods A novel, quantitative approach to this question is presented, taking advantage of a complete census of the vascular epiphyte community in 0·4 ha of undisturbed lowland forest in Panama. For three locally common host-tree species (Socratea exorrhiza, Marila laxiflora, Perebea xanthochyma) null models were created of the expected epiphyte assemblages assuming that epiphyte colonization reflected random distribution of epiphytes in the forest.
• Key Results In all three tree species, abundances of the majority of epiphyte species (69–81 %) were indistinguishable from random, while the remaining species were about equally over- or under-represented compared with their occurrence in the entire forest plot. Permutations based on the number of colonized trees (reflecting observed spatial patchiness) yielded similar results. Finally, a third analysis (canonical correspondence analysis) also confirmed host-specific differences in epiphyte assemblages. In spite of pronounced preferences of some epiphytes for particular host trees, no epiphyte species was restricted to a single host.
• Conclusions The epiphytes on a given tree species are not simply a random sample of the local species pool, but there are no indications of host specificity either.
Keywords: Epiphytes, community assembly, null model, host preference, colonization, Panama, Orchidaceae, Bromeliaceae, Araceae
INTRODUCTION
Vascular epiphytes, i.e. non-parasitic plants using trees only as structural support, comprise a major proportion of tropical phytodiversity. While rarely exceeding 15 % of the vascular flora in lowland rainforests (e.g. Croat, 1978), their contribution may exceed 50 % in some montane forests with >120 species in 1·5 ha (Kelly et al., 2004). Individual trees may have >80 species growing on them (Kreft et al., 2004; Krömer et al., 2005). Many hypotheses have been put forward to account for the local co-existence of such a hyper-diverse group of plants, for example, frequent disturbance (bark defoliation, detached branches, tree falls) that prevents competitive exclusion (Benzing, 1981), vertical niche diversification (Johansson, 1974; Gentry and Dodson, 1987) or host tree specificity (Went, 1940).
Subsequent observational and descriptive studies have provided quantitative evidence for niche assembly along vertical abiotic gradients as well as for the importance of disturbance for epiphyte population and community processes (Griffiths and Smith, 1983; Hietz, 1997; Hietz and Briones, 1998; Zotz et al., 2005), while there is little support for the notion of strict host-specificity in vascular epiphytes (Benzing, 1990; Zimmerman and Olmsted, 1992). However, failure to find a one-to-one match between particular species pairs of host trees and epiphytes is not equivalent to ‘neutrality’ of host tree species identity in respect to the structuring of epiphyte communities. Went (1940) came close to the concept of species-specificity by proposing that the occurrence of certain epiphyte species was solely linked to host-tree identity since he could not explain their distribution with physical factors characterizing the host trees (e.g. bark roughness, age of host tree, humus accumulation and light availability). Rather than tree identity, the fact that each potential host tree species offers a different set of architectural traits (e.g. branch angles, diameters, etc.), chemical and morphological bark characteristics, phenological patterns, or microclimatic regimes suggests that there could be rather unique epiphyte assemblages on each host tree species: Zotz et al. (1999) called these assemblages ‘phorophyte-specific epiphyte spectra’. The existence of such spectra, in turn, would directly link the local tree diversity to local epiphyte diversity, albeit probably in a rather diffuse way. Comments on differences of tree species in their suitability for vascular epiphytes, either in general or for particular epiphyte taxa, abound in the literature, although most are rather anecdotal (e.g. Mesler, 1975; Cribb et al., 2002; Moran and Russell, 2004). Quantitative and experimental approaches, on the other hand, are rare (e.g. Benzing, 1978; Callaway et al., 2002), and all these studies tested either the preference of only one or two epiphyte species for a set of host trees (Benzing, 1978; Ackerman et al., 1989; Callaway et al., 2002) or host specificity in the strict sense (Zimmerman and Olmsted, 1992; Migenis and Ackerman, 1993). This paper presents a novel approach that constitutes a critical test of the notion of phorophyte-specific epiphyte spectra. In it the null hypothesis that the species assemblage found on a particular host tree species in a forest is just a random subset of the local species pool of epiphytes is examined. This was done taking advantage of a complete inventory of the vascular epiphytes in 0·4 ha of a tropical lowland forest in Panama (Zotz, 2004). A study on host specificity of vascular epiphytes in a tropical rainforest faces the prominent problem that tree species as well as epiphyte species are frequently very rare. In the study plot at San Lorenzo, only the three tree species Socratea exorrhiza, Marila laxiflora and Perebea xanthochyma were sufficiently common and frequently used by epiphytes to warrant their inclusion in this study allowing for appropriate statistical power.
MATERIALS AND METHODS
Study site and species
The data for this study were collected at the San Lorenzo Canopy Crane Site that is located near the Atlantic coast of the Republic of Panama. The forest around this facility has not experienced any severe human disturbance for at least 200 years (Condit et al., 2004). Average annual rainfall is approx. 3100 mm (Wright et al., 2003). A 52-m-tall construction crane covers approx. 9000 m2 of forest with its jib of 54 m. A small gondola allowed easy access to the vascular epiphytes in this area, although the use of binoculars was necessary in rare cases. Briefly, between mid-1999 and early 2002 each individual epiphyte in an area of 0·4 ha on all trees with a diameter at breast height of >1 cm was registered with species name, plant size and location on the tree (Zotz, 2004; G. Zotz, unpubl. res.). The census included 1358 trees, 389 of which had epiphytes growing on them. Although both species richness and individual abundance correlated with tree size (Laube and Zotz, 2006; G. Zotz, unpubl. res.), epiphytes were occasionally found on trees with even the smallest diameter at breast height. Plant names of angiosperms follow the Flora of Panama checklist (D'Arcy, 1987), while fern names are according to Lellinger (1989).
The first tree species included in the analysis was Socratea exorrhiza (Mart.) H. Wendl. (Arecaceae). This stilt-root palm, which occurs from Nicaragua to northern South America, reaches up to 30 m (Croat, 1978). Unlike most other palms its trunk diameter increases with height (Schatz et al., 1985). There were 31 individuals in the study plot. Marila laxiflora Rusby (Clusiaceae), which is known only from wetter forests in Panama and occurred with 40 individuals at San Lorenzo, reaches similar heights (Croat, 1978). The 38 individuals of the third species, Perebea xanthochyma H. Karst. (Moraceae), reached up to 35 m. The species occurs from Costa Rica to Peru.
Data analysis
The local epiphyte species pool (hemi-epiphytes were excluded) consisted of 103 species with 13 099 individuals. The species pool on Socratea exorrhiza comprised 39 epiphyte species with 354 individuals, Marila laxiflora hosted 47 species with 496 individuals and Perebea xanthochyma 32 species with 227 individuals. Null models of the epiphyte assemblage on a given host tree species were created with R (Version 2.2.1; R Development Core Team, 2005) as follows. To create, for example, the null model for Socratea exorrhiza, 354 individuals were randomly selected from the complete list of epiphyte individuals in the local species pool. Individuals were drawn from the list with replacement. This process was repeated 1000 times and 95 % confidence intervals were obtained for each species by discarding the 25 highest and the 25 lowest values. Ranges expected by chance were then compared with the actual species abundances for each species separately. Accordingly, null models were created for the epiphyte assemblages on Marila laxiflora and Perebea xanthochyma.
Similarly, null models were created for the epiphyte distributions on the tree species as a random sample based on the number of host trees a given epiphyte species inhabited in the study plot. For a given epiphyte species that was growing on a tree species, the number of trees that were inhabited by epiphytes was randomly selected from the complete list of trees found in the study plot. For example, the epiphytic orchid, Scaphyglottis longicaulis, was found on 44 trees, but was absent on 1314 trees. To create a null model for the distribution on Socratea exorrhiza, 31 trees, i.e. the number of Socratea exorrhiza trees in the plot, were randomly selected from this complete tree list with replacement. This process was repeated 1000 times and 95 % confidence intervals were obtained as described above. Accordingly, tree-based null models were created for the epiphyte species on the 40 Marila laxiflora and 38 Perebea xanthochyma trees growing in the study plot.
Also a completely different method was used to analyse epiphyte species preferences, i.e. canonical correspondence analysis (CCA) with CANOCO software (Version 4.5; ter Braak and Šmilauer, 1997). This ordination technique is designed to detect the variation in species composition that can be explained best by environmental variables (host tree identity in the present case), which is achieved by combining aspects of regular ordination and regression (ter Braak, 1995). The resulting ordination diagrams express both the variation in species composition and the principal relationships between species and environmental variables. Including tree species identity as dummy variables, differences in the epiphyte assemblages among species were analysed using Monte Carlo permutation tests (with manual forward selection). Two separate CCAs were run, one with abundance data, another with binary (presence/absence) data. The data sets used in these analyses were not completely identical to the ones described above. First, infrequent epiphyte species were excluded, reducing the species number to, respectively, 43 (abundance data) and 39 (binary data) and, secondly, only trees with at least three epiphyte individuals were included, leaving 70 trees of three tree species. Ordinations were optimized by species and Monte Carlo permutation tests run 499 times.
RESULTS
If epiphyte species showed no preference for particular host tree species, their relative abundances on a tree should simply reflect their relative abundance in the forest as a whole. In slightly more than half of all cases (57·3 %) the null hypothesis of a random sample could indeed not be rejected. However, almost 43 % of the epiphyte species in the study plot showed a higher or lower abundance, respectively, on at least one of the focal tree species than expected by chance (Table 1). Three epiphyte species (Dicranoglossum panamense, Trichomanes angustifrons and Tillandsia anceps) were more frequent than expected by chance on all three tree species, while five epiphyte species (Pleurothallis brighamii, Trichosalpinx orbicularis, Trichomanes nummularium, Maxillaria uncata and Scaphyglottis graminifolia) were invariably less frequent than expected by chance.
Table 1.
Overview on the occurrence of epiphyte species on three host tree species in 0·4 ha of the San Lorenzo crane plot in relation to a null assemblage deduced from the local epiphyte pool
| Species | Family | Socratea | Marila | Perebea |
|---|---|---|---|---|
| Aechmea tillandsioides | Bromeliaceae | 0 | + | + |
| Ananthacorus angustifolius | Vittariaceae | + | 0 | + |
| Anthurium acutangulum | Araceae | 0 | + | 0 |
| Anthurium clavigerum | Araceae | + | + | 0 |
| Anthurium friedrichsthalii | Araceae | 0 | + | 0 |
| Anthurium hacumense | Araceae | 0 | + | 0 |
| Asplenium juglandifolium | Aspleniaceae | − | − | 0 |
| Asplenium serratum | Aspleniaceae | + | 0 | 0 |
| Campylocentrum micranthum | Orchidaceae | 0 | + | + |
| Campyloneurum occultum | Polypodiaceae | − | 0 | 0 |
| Catasetum viridiflavum | Orchidaceae | 0 | + | 0 |
| Codonanthe macradenia | Gesneriaceae | 0 | + | + |
| Columnea billbergiana | Gesneriaceae | + | 0 | 0 |
| Dichaea panamensis | Orchidaceae | − | + | 0 |
| Dicranoglossum panamense | Polypodiaceae | + | + | + |
| Elaphoglossum herminieri | Lomariopsidaceae | − | − | 0 |
| Elaphoglossum latifolium | Lomariopsidaceae | 0 | − | 0 |
| Elaphoglossum sporadolepis | Lomariopsidaceae | − | 0 | − |
| Encyclia fragrans | Orchidaceae | + | 0 | 0 |
| Epidendrum imatophyllum | Orchidaceae | 0 | + | 0 |
| Epidendrum nocturnum | Orchidaceae | + | 0 | 0 |
| Gongora quinquenervis | Orchidaceae | 0 | + | + |
| Guzmania subcorymbosa | Bromeliaceae | + | 0 | 0 |
| Maxillaria uncata | Orchidaceae | − | − | − |
| Microgramma lycopodioides | Polypodiaceae | 0 | + | 0 |
| Microgramma reptans | Polypodiaceae | 0 | + | 0 |
| Niphidium crassifolium | Polypodiaceae | 0 | − | − |
| Ornithocephalus bicornis | Orchidaceae | 0 | + | 0 |
| Peperomia rotundifolia | Piperaceae | 0 | + | 0 |
| Pleurothallis brighamii | Orchidaceae | − | − | − |
| Polypodium percussum | Polypodiaceae | 0 | − | 0 |
| Scaphyglottis graminifolia | Orchidaceae | − | − | − |
| Scaphyglottis longicaulis | Orchidaceae | 0 | − | − |
| Scaphyglottis prolifera | Orchidaceae | − | 0 | − |
| Sobralia fragrans | Orchidaceae | + | 0 | 0 |
| Stelis crescentiicola | Orchidaceae | 0 | − | 0 |
| Tillandsia anceps | Bromeliaceae | + | + | + |
| Tillandsia bulbosa | Bromeliaceae | 0 | − | 0 |
| Trichomanes angustifrons | Hymenophyllaceae | + | + | + |
| Trichomanes ekmannii | Hymenophyllaceae | 0 | + | + |
| Trichomanes nummularium | Hymenophyllaceae | − | − | − |
| Trichomanes ovale | Hymenophyllaceae | 0 | 0 | + |
| Trichosalpinx orbicularis | Orchidaceae | − | − | − |
| Vriesea gladioliflora | Bromeliaceae | + | 0 | + |
Species more frequent on a given host tree species than expected by random distribution are indicated with ‘+’; species less frequent are indicated with ‘−’; species occurring in a frequency as expected by random distribution are indicated with ‘0’. Excluded are the 59 species occurring in all three focal tree species in a frequency as expected by random distribution.
For a detailed analysis of host preference compare Appendices 1–3.
Socratea exorrhiza
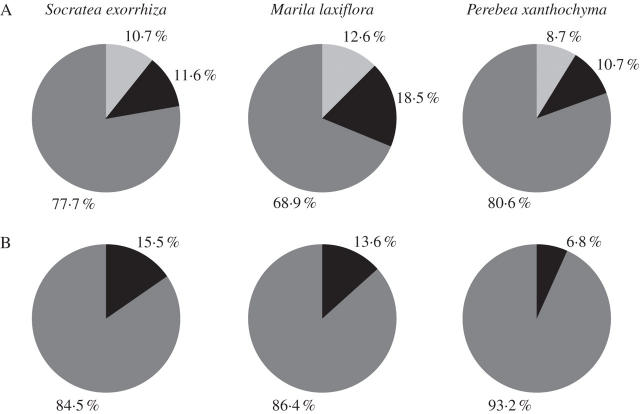
The abundance of the majority of epiphyte species (77·7 %) on Socratea exorrhiza could not be distinguished from random (Fig. 1A). A significantly higher abundance than expected was observed in 11·6 % of all cases, significantly lower abundance in 10·7 %. The most pronounced preference for Socratea exorrhiza was found in the bromeliad Guzmania subcorymbosa: 26 individuals inhabited the palm in the study plot whereas a maximum of only five individuals was expected by chance (Appendix 1). Substantial deviations from the expected abundances were also found in the aroid Anthurium clavigerum (17 individuals, 0–4 plants expected by chance), the gesneroid Columnea billbergiana (8 individuals, 0–2 expected) or the fern Ananthacorus angustifolius (51 individuals, 9–24 expected). On the other hand, otherwise locally rather common orchids were conspicuously absent (e.g. Scaphyglottis graminifolia, Maxillaria uncata and Trichosalpinx orbicularis). By chance alone, these species were expected to occur with up to 53 individuals on this palm.
Fig. 1.
Distribution of epiphyte species in the San Lorenzo forest plot according to (A) individual-based comparisons of the occurrence on host trees of a given species with that of a null model (white, species found with fewer individuals on a host tree species than expected by chance; black, species found with more individuals than expected by chance; grey, species found in a frequency indistinguishable from that expected by chance); (B) tree-based occurrence on host trees of a given species (black, species found on more trees than expected by chance; dark grey, species found on a number of trees indistinguishable from that expected by chance).
Marila laxiflora
On Marila laxiflora about two-thirds of the epiphyte species (68·9 %) showed an abundance indistinguishable from random (Fig. 1A). A portion of 18·5 % of the species that occurred in the San Lorenzo plot were significantly more abundant on this tree species than in the remaining plot, while 12·6 % showed a significantly lower abundance than expected. The strongest preference for this host tree was found in some ferns and aroids: Trichomanes angustifrons (13 individuals, 0–2 expected, Appendix 2), Dicranoglossum panamense (123 individuals, 13–30 expected) or Anthurium acutangulum (24 individuals, 1–8 expected). Among locally common orchid species only Pleurothallis brighamii was completely absent (20–40 individuals expected).
Perebea xanthochyma
The epiphyte assemblage of Perebea xanthochyma showed the smallest deviation from random among the three focal tree species: 80·6 % of all species showed an abundance indistinguishable from random (Fig. 1A). About equal proportions were more abundant (10·7 %) or less abundant (8·7 %) than expected. The strongest preference for this tree was observed in the fern Trichomanes angustifrons (24 individuals, 0–2 expected, Appendix 3) followed by the bromeliad Aechmea tillandsioides (12 individuals, 0–2 expected), and the orchid Campylocentrum micranthum (10 individuals, 0–2 expected). The orchids Scaphyglottis graminifolia, Pleurothallis brighamii and Trichosalpinx orbicularis were conspicuously absent. Under random distribution these species were expected to occur with up to 37 individuals.
Spatial patchiness
This first analysis has an obvious shortcoming because the null assemblages implicitly assume that individuals of a given epiphyte species are distributed evenly in the forest plot. As this is clearly not the case, a second series of null models that account for the patchiness of epiphyte spatial distributions was created: the number of trees for a given tree species, that an epiphyte species should inhabit, was determined based on the number of trees this species actually inhabited in the study plot as described in Materials and methods. The overall results were quite consistent with the first analysis. With few exceptions, species that were more common than expected by chance in the first analysis were also more common in the second (Table 2). Unambiguous exceptions were Vriesea gladioliflora (Socratea exorrhiza), Microgramma lycopodioides (Marila laxiflora) and Codonanthe macradenia (Perebea xanthochyma), while the tree-based occurrences of Tillandsia anceps and Trichomanes ovale on Perebea xanthochyma were marginally higher than expected. No species, however, could be shown to be less abundant than expected by chance in this tree-based analysis since the lower boundary of the null distribution almost always included zero (Fig. 1B).
Table 2.
Numbers of trees a given epiphyte species inhabited in 0·4 ha of the San Lorenzo Crane Plot (light and bold numbers) and generated numbers of a random distribution based on the distribution of a given species on all forest trees in the plot for the three host tree species Socratea exorrhiza, Marila laxiflora and Perebea xanthochyma
| Species | Socratea exorrhiza | Marila laxiflora | Perebea xanthochyma |
|---|---|---|---|
| Ananthacorus angustifolius | 11 (0; 4) | 5 (0; 5) | 7 (0; 5) |
| Anthrophyum lanceolatum | 0 (0; 2) | 1 (0; 2) | 1 (0; 2) |
| Anthurium acutangulum | 2 (0; 4) | 10 (0; 5) | 3 (0; 5) |
| Anthurium clavigerum | 12 (0; 3) | 8 (0; 4) | 0 (0; 3) |
| Anthurium friedrichsthallii | 1 (0; 3) | 8 (0; 4) | 1 (0; 4) |
| Anthurium hacumense | 1 (0; 3) | 7 (0; 3) | 0 (0; 3) |
| Asplenium serratum | 5 (0; 3) | 4 (0; 4) | 0 (0; 4) |
| Campyloneurum occultum | 0 (0; 2) | 3 (0; 3) | 1 (0; 3) |
| Campyloneurum phylliditis | 4 (0; 3) | 3 (0; 3) | 1 (0; 3) |
| Catasetum viridiflavum | 0 (0; 2) | 5 (0; 2) | 0 (0; 2) |
| Codonanthe macradenia | 4 (0; 4) | 8 (0; 4) | 1 (0; 4) |
| Dichaea panamensis | 0 (0; 3) | 5 (0; 3) | 1 (0; 3) |
| Dicranoglossum panamense | 8 (0; 6) | 17 (1; 8) | 17 (1; 7) |
| Elaphoglossum sporadolepis | 8 (0; 3) | 3 (0; 3) | 1 (0; 3) |
| Epidendrum nocturnum | 3 (0; 2) | 1 (0; 2) | 0 (0; 2) |
| Guzmania subcorymbosa | 3 (0; 2) | 2 (0; 3) | 1 (0; 2) |
| Microgramma lycopodioides | 3 (0; 2) | 1 (0; 3) | 1 (0; 3) |
| Niphidium crassifolium | 5 (0; 4) | 7 (0; 4) | 1 (0; 4) |
| Peperomia rotundifolia | 2 (0; 3) | 4 (0; 3) | 2 (0; 3) |
| Polypodium percussum | 3 (0; 3) | 1 (0; 3) | 1 (0; 3) |
| Scaphyglottis graminifolia | 0 (0; 3) | 2 (0; 3) | 0 (0; 3) |
| Scaphyglottis longicaulis | 4 (0; 3) | 2 (0; 4) | 1 (0; 4) |
| Scaphyglottis prolifera | 0 (0; 2) | 2 (0; 2) | 0 (0; 2) |
| Sobralia fragrans | 6 (0; 2) | 1 (0; 3) | 0 (0; 3) |
| Tillandsia anceps | 10 (0; 4) | 8 (0; 4) | 4 (0; 4) |
| Trichomanes angustifrons | 5 (0; 5) | 10 (0; 6) | 16 (0; 5) |
| Trichomanes ekmannii | 3 (0; 3) | 10 (0; 3) | 8 (0; 3) |
| Trichomanes ovale | 2 (0; 2) | 1 (0; 3) | 3 (0; 3) |
| Vriesea gladioliflora | 2 (0; 3) | 2 (0; 3) | 5 (0; 3) |
Numbers in brackets indicate the lower and upper boundary of the random generated tree numbers. Only species occurring on ≥5% of the host trees in the plot are shown. Bold numbers indicate species occurring on a higher number of trees than expected by random distribution.
Ordination
The ordination approach yielded similarly significant differences of epiphyte assemblages between tree species in the analyses of both the abundance data (Fig. 2) and the binary data (not shown). The explained variance, however, was very low in either case: only 5·4 % for abundance data and 4·7 % for binary data. Consistent with the very high proportion of species occurrences indistinguishable from random expectations in Perebea xanthochyma (approx. 80 %, Fig. 1A), the marginal effects of this species were not significant (Monte-Carlo permutation, P > 0·2), in contrast to the significant effects of the other two species (P < 0·05).
Fig. 2.
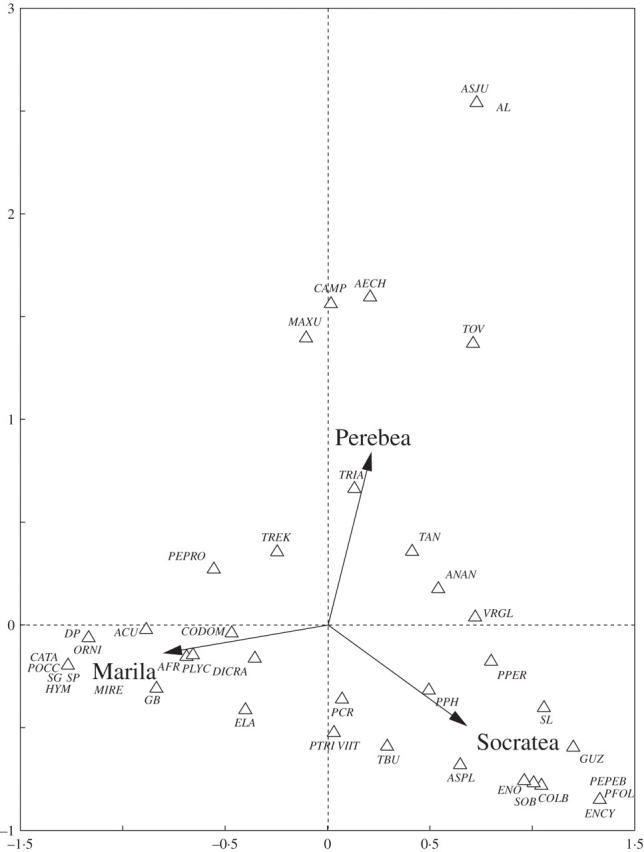
Biplot of a canonical correspondence analysis for epiphytes on the three host tree species Socratea exorrhiza, Marila laxiflora and Perebea xanthochyma. The plot is based on epiphyte abundance data, binary (presence/absence) data yield very similar results (not shown).
DISCUSSION
A preference of particular epiphyte species for particular host tree species has been reported repeatedly in the literature (Oliver, 1930; Mesler, 1975; Benzing, 1990; Male and Roberts, 2005). However, no study to date has tried to link the composition of the entire epiphyte assemblage occurring on a particular tree species to the local species pool in the quantitative manner of the present study. A majority of species (approx. 69–81 %, individual-based; approx. 85–93 %, tree-based) showed no bias in respect to the focal tree species (Fig. 1), their occurrence is thus consistent with the notion of a random assembly: individual trees are just redundant colonization opportunities for epiphytes irrespective of tree species identity. The remaining taxa were about equally over- or under-represented in abundance. This conclusion would change if the present analysis were confined to the more common species. If considering, for example, only the ten most common epiphyte species, the proportion of taxa deviating from a random sample would be much higher (cf. Appendices 1–3). However, as we were interested in the entire community and see no basis to distinguish unambiguously ‘common’ and ‘rare’ species, all species were included in the analysis.
The low proportion of taxa that were under-represented in abundance came as a surprise at least in the case of Socratea exorrhiza. A bias against a large proportion of species had been mostly expected considering the simple architecture of this palm that lacks features generally assumed to facilitate epiphyte establishment (Benzing, 1990), e.g. crotches, humus accumulations (Andrade and Nobel, 1996) or rough bark with a high water-holding capacity (Callaway et al., 2002). In addition, in contrast to other palm species that feature suitable horizontal growing sites for epiphytes in accumulated debris in persistent leaf bases (e.g. Copernicia tectorum; Holbrook and Putz, 1996), Socratea exorrhiza offers only vertical growing sites, which again is believed to hinder establishment (Benzing, 1990).
Among those taxa that were found more frequently than expected on Perebea xanthochyma, both in respect to the number of individuals and the number of inhabited trees, were filmy ferns of the genus Trichomanes, with the species T. ekmannii found on Perebea xanthochyma accounting for more than one-quarter of the entire plot population (Appendix 3; Zotz, 2004). Similarly, Socratea exorrhiza appeared to be a good host for all larger tank bromeliads growing in the study plot, the extreme case being Guzmania subcorymbosa, which was very rare on any other tree species. Individuals of rare epiphyte species that inhabited only one of the focal tree species were invariably found on other trees in the forest plot as well. Thus, no epiphyte species was restricted to a certain host tree species.
Due to an almost complete lack of information on the biology of most epiphytes it can only be speculated why particular epiphyte taxa may prefer a given tree species. The reasons why some species are not found on a palm, on the other hand, are less obscure. For example, there are a number of substrate specialists in the local epiphyte flora, for which particular requirements are known; there are so-called twig epiphytes such as Catopsis sessiliflora (Bromeliaceae; Zotz and Laube, 2005) or Notylia albida (Orchidaceae; Chase, 1987), or dead wood specialists such as Catasetum viridiflavum (Orchidaceae). In contrast to the crowns of larger dicotyledonous trees, which feature branches and twigs and frequently a large proportion of dead wood (Schulz and Wagner, 2002), these microhabitats do not exist on living palms and, not surprisingly, all these specialists were never observed there (Appendix 1). It is much less obvious, however, why Catopsis sessiliflora, for example, was not observed at all on the other two tree species either. Similar to the conspicuous absence of many locally common orchid species from Socratea exorrhiza, this absence may be more related to patchy species distributions than to real positive or negative substrate preferences. This is indicated, for example, by the regular occurrence of many (missing) orchids such as Scaphyglottis graminifolia or Dichaea panamensis on Socratea exorrhiza trees (compare Appendix 1) outside the study plot. Severe dispersal limitation in general, which probably causes this patchiness, is also suggested by the observation that orchids such as Maxillaria uncata (857 individuals in the 0·4 ha plot) and Trichosalpinx orbicularis (390 individuals; compare Zotz, 2004), although very abundant in individual numbers, are found only on a few trees in the plot (3·9 % of trees; G. Zotz, unpubl. res.). Finally, some of the positive and negative associations between host tree species and epiphyte species are likely to be false, considering the statistical methods used in the present study. The conclusions are not expected to be affected substantially by such possible artefacts, however, because (a) there are a large number of positive and negative associations; (b) the observed abundances of many species are very far from random expectations; and (c) three different approaches yielded qualitatively consistent results (Figs 1 and 2). Nevertheless, it is essential that future studies use descriptive data like the ones presented here as the basis for manipulative experiments to identify the mechanisms behind the observed deviations from random expectations.
In summary, comparing the actual epiphyte assemblages on a particular host tree with the ones expected by null models, no evidence was found for strict host specificity in any epiphyte. However, a significant positive or negative bias of individual epiphyte species was found in a large proportion of the local species pool. While Went's (1940) concept of species-specificity in the strict sense can thus be rejected, the extreme alternative can be dismissed as well; the epiphytes on the three focal tree species are not just a random subset of the local epiphyte community.
Acknowledgments
The University of Kaiserslautern, Germany provided a stipend for Stefan Laube during this work. We thank S. J. Wright, V. Horlyck, J. Herrera and E. Andrade (all Smithsonian Tropical Research Institute) for organizing work at the crane. Also we thank the Republic of Panama for research permits and for making their natural resources available for scientific study.
Appendix 1
Comparison of actual occurrence of epiphytes on Socratea exorrhiza and null assemblages created from the epiphyte pool in 0·4 ha of the San Lorenzo Crane Plot. The analysis is based on the individual number found on 31 Socratea exorrhiza trees. Shown are individual numbers of a given epiphyte species observed on Socratea exorrhiza and the expected range of individual numbers (lower and upper boundary of 95% confidence intervals). Bold names indicate species that were more common than expected; underlined names indicate species that are less common than expected.
Table 3.
| Individuals of null assemblage |
Rank |
|||||
|---|---|---|---|---|---|---|
| Species | Family | Individuals on Socratea | Lower boundary | Upper boundary | Null assemblage | Socratea |
| Scaphyglottis longicaulis | Orchidaceae | 41 | 30 | 54 | 1 | 3 |
| Scaphyglottis graminifolia | Orchidaceae | 0 | 30 | 53 | 2 | |
| Elaphoglossum sporadolepis | Lomariopsidaceae | 17 | 22 | 43 | 3 | 6 |
| Maxillaria uncata | Orchidaceae | 0 | 15 | 33 | 4 | |
| Pleurothallis brighamii | Orchidaceae | 0 | 13 | 31 | 5 | |
| Ananthacorus angustifolius | Vittariaceae | 51 | 9 | 24 | 6 | 1 |
| Dicranoglossum panamense | Polypodiaceae | 49 | 9 | 23 | 7 | 2 |
| Niphidium crassifolium | Polypodiaceae | 9 | 7 | 20 | 8 | 11 |
| Trichosalpinx orbicularis | Orchidaceae | 0 | 4 | 17 | 9 | |
| Scaphyglottis prolifera | Orchidaceae | 0 | 3 | 13 | 10 | |
| Dichaea panamensis | Orchidaceae | 0 | 3 | 14 | 11 | |
| Codonanthe macradenia | Gesneriaceae | 4 | 2 | 12 | 12 | 16 |
| Tillandsia anceps | Bromeliaceae | 22 | 2 | 12 | 13 | 5 |
| Trichomanes nummularium | Hymenophyllaceae | 0 | 1 | 10 | 14 | |
| Elaphoglossum herminieri | Lomariopsidaceae | 0 | 1 | 10 | 15 | |
| Tillandsia bulbosa | Bromeliaceae | 3 | 2 | 10 | 16 | 20 |
| Polypodium percussum | Polypodiaceae | 4 | 1 | 9 | 17 | 16 |
| Asplenium juglandifolium | Aspleniaceae | 0 | 1 | 8 | 18 | |
| Sobralia fragrans | Orchidaceae | 14 | 1 | 9 | 19 | 8 |
| Anthurium friedrichsthalii | Araceae | 3 | 1 | 9 | 20 | 20 |
| Peperomia rotundifolia | Piperaceae | 3 | 1 | 9 | 21 | 20 |
| Asplenium serratum | Aspleniaceae | 14 | 0 | 8 | 22 | 8 |
| Campyloneurum occultum | Polypodiaceae | 0 | 1 | 8 | 23 | |
| Campyloneurum phyllitidis | Polypodiaceae | 6 | 0 | 7 | 24 | 14 |
| Stelis crescentiicola | Orchidaceae | 0 | 0 | 7 | 25 | |
| Trichomanes ovale | Hymenophyllaceae | 3 | 0 | 7 | 26 | 20 |
| Elaphoglossum latifolium | Lomariopsidaceae | 0 | 0 | 6 | 27 | |
| Anthurium acutangulum | Araceae | 2 | 0 | 6 | 28 | 26 |
| Vriesea gladioliflora | Bromeliaceae | 13 | 0 | 7 | 29 | 10 |
| Peperomia ebingeri | Piperaceae | 4 | 0 | 6 | 30 | 16 |
| Microgramma lycopodioides | Polypodiaceae | 3 | 0 | 6 | 31 | 20 |
| Catopsis sessiliflora | Araceae | 0 | 0 | 6 | 32 | |
| Anthurium hacumense | Araceae | 1 | 0 | 5 | 33 | 30 |
| Guzmania subcorymbosa | Bromeliaceae | 26 | 0 | 5 | 34 | 4 |
| Catasetum viridiflavum | Orchidaceae | 0 | 0 | 5 | 35 | |
| Polystachya foliosa | Orchidaceae | 2 | 0 | 4 | 36 | 26 |
| Lockhartia acuta | Orchidaceae | 0 | 0 | 4 | 37 | |
| Anthrophyum lanceolatum | Vittariaceae | 0 | 0 | 5 | 38 | |
| Vittaria lineata | Vittariaceae | 1 | 0 | 4 | 39 | 30 |
| Trigonidium egertonianum | Orchidaceae | 0 | 0 | 4 | 40 | |
| Maxillaria discolor | Orchidaceae | 0 | 0 | 5 | 41 | |
| Aspasia principissa | Orchidaceae | 0 | 0 | 4 | 42 | |
| Microgramma reptans | Polypodiaceae | 1 | 0 | 3 | 43 | 30 |
| Masdevallia livingstoneana | Orchidaceae | 0 | 0 | 4 | 44 | |
| Epidendrum difforme | Orchidaceae | 0 | 0 | 3 | 45 | |
| Anthurium durandii | Araceae | 0 | 0 | 3 | 46 | |
| Anthurium clavigerum | Araceae | 17 | 0 | 4 | 47 | 6 |
| Trichomanes ekmannii | Hymenophyllaceae | 4 | 0 | 4 | 48 | 16 |
| Peperomia macrostachia | Piperaceae | 1 | 0 | 4 | 49 | 30 |
| Epidendrum nocturnum | Orchidaceae | 6 | 0 | 3 | 50 | 14 |
| Anthurium scandens | Araceae | 0 | 0 | 3 | 51 | |
| Anthurium brownii | Araceae | 2 | 0 | 3 | 52 | 26 |
| Campylocentrum micranthum | Orchidaceae | 0 | 0 | 3 | 53 | |
| Philodendron sagittifolium | Araceae | 0 | 0 | 3 | 54 | |
| Pecluma pectinata | Polypodiaceae | 0 | 0 | 3 | 55 | |
| Trichocentrum capistratum | Orchidaceae | 0 | 0 | 3 | 56 | |
| Hylocereus monacanthus | Cactaceae | 0 | 0 | 2 | 57 | |
| Stenospermation angustifolium | Araceae | 0 | 0 | 2 | 58 | |
| Aechmea tillandsioides | Bromeliaceae | 1 | 0 | 2 | 59 | 30 |
| Notylia albida | Orchidaceae | 0 | 0 | 2 | 60 | |
| Mormodes powellii | Orchidaceae | 0 | 0 | 2 | 61 | |
| Dimerandra emarginata | Orchidaceae | 1 | 0 | 2 | 62 | 30 |
| Polypodium triseriale | Polypodiaceae | 2 | 0 | 2 | 63 | 26 |
| Trichomanes angustifrons | Hymenophyllaceae | 8 | 0 | 2 | 64 | 12 |
| Trichopilia maculata | Orchidaceae | 0 | 0 | 2 | 65 | |
| Gongora quinquenervis | Orchidaceae | 0 | 0 | 1 | 66 | |
| Columnea billbergiana | Gesneriaceae | 8 | 0 | 2 | 67 | 12 |
| Oncidium ampliatum | Orchidaceae | 0 | 0 | 2 | 68 | |
| Philodendron radiatum | Araceae | 0 | 0 | 2 | 69 | |
| Hecistopteris pumila | Vittariaceae | 0 | 0 | 2 | 70 | |
| Ornithocephalus powellii | Orchidaceae | 0 | 0 | 1 | 71 | |
| Sobralia panamensis | Orchidaceae | 0 | 0 | 2 | 72 | |
| Epiphyllum phyllanthus | Cactaceae | 0 | 0 | 2 | 73 | |
| Pleurothallis verecunda | Orchidaceae | 1 | 0 | 1 | 74 | 30 |
| Ornithocephalus bicornis | Orchidaceae | 0 | 0 | 1 | 75 | |
| Polypodium costaricense | Polypodiaceae | 0 | 0 | 1 | 76 | |
| Guzmania musaica | Bromeliaceae | 0 | 0 | 1 | 77 | |
| Elleanthus longibracteatus | Orchidaceae | 1 | 0 | 1 | 78 | 30 |
| Huperzia dichotoma | Selaginellaceae | 0 | 0 | 1 | 79 | |
| Epidendrum imatophyllum | Orchidaceae | 0 | 0 | 1 | 80 | |
| Anetium citrifolium | Vittariaceae | 0 | 0 | 1 | 81 | |
| Encyclia fragrans | Orchidaceae | 3 | 0 | 1 | 82 | 20 |
| Peperomia obtusifolia | Piperaceae | 0 | 0 | 1 | 83 | |
| Epidendrum schlechterianum | Orchidaceae | 0 | 0 | 1 | 84 | |
| Encyclia aemula | Orchidaceae | 0 | 0 | 1 | 85 | |
| Maxillaria crassifolia | Orchidaceae | 0 | 0 | 1 | 86 | |
| Cochleanthes lipscombiae | Orchidaceae | 0 | 0 | 1 | 87 | |
| Trichomanes punctatum | Hymenophyllaceae | 1 | 0 | 1 | 88 | 30 |
| Trichomanes godmanii | Hymenophyllaceae | 0 | 0 | 1 | 89 | |
| Encyclia chimborazoensis | Orchidaceae | 0 | 0 | 1 | 90 | |
| Anthurium bakeri | Araceae | 0 | 0 | 1 | 91 | |
| Trichomanes anadromum | Hymenophyllaceae | 0 | 0 | 1 | 92 | |
| Lockhartia pittieri | Orchidaceae | 0 | 0 | 1 | 93 | |
| Kefersteinia sp. | Orchidaceae | 0 | 0 | 1 | 94 | |
| Hymenophyllum brevifrons | Hymenophyllaceae | 0 | 0 | 1 | 95 | |
| Jacquiniella pedunculata | Orchidaceae | 0 | 0 | 1 | 96 | |
| Pleurothallis grobyi | Orchidaceae | 0 | 0 | 1 | 97 | |
| Caularthron bilamellatum | Orchidaceae | 0 | 0 | 1 | 98 | |
| Werauhia sanguinolenta | Bromeliaceae | 0 | 0 | 1 | 99 | |
| Peperomia cordulata | Piperaceae | 0 | 0 | 1 | 100 | |
| Jacquinella sp. | Orchidaceae | 0 | 0 | 1 | 101 | |
| Drymonia serrulata | Gesneriaceae | 1 | 0 | 1 | 102 | 30 |
| Maxillaria variabilis | Orchidaceae | 0 | 0 | 1 | 103 | |
Appendix 2
Comparison of actual occurrence of epiphytes on Marila laxiflora and null assemblages created from the epiphyte pool in 0·4 ha of the San Lorenzo Crane Plot. The analysis is based on the individual number found on 40 Marila laxiflora trees. Shown are individual numbers of a given epiphyte species observed on Marila laxiflora and the expected range of individual numbers (lower and upper boundary of 95% confidence intervals). Bold names indicate species which were more common than expected; underlined names indicate species that are less common than expected.
Table 4.
| Individuals of null assemblage |
Rank |
|||||
|---|---|---|---|---|---|---|
| Species | Family | Individuals on Marila | Lower boundary | Upper boundary | Null assemblage | Marila |
| Scaphyglottis longicaulis | Orchidaceae | 4 | 46 | 73 | 1 | 26 |
| Scaphyglottis graminifolia | Orchidaceae | 25 | 44 | 71 | 2 | 4 |
| Elaphoglossum sporadolepis | Lomariopsidaceae | 34 | 32 | 58 | 3 | 2 |
| Maxillaria uncata | Orchidaceae | 5 | 23 | 44 | 4 | 22 |
| Pleurothallis brighamii | Orchidaceae | 0 | 20 | 40 | 5 | |
| Ananthacorus angustifolius | Vittariaceae | 25 | 14 | 32 | 6 | 4 |
| Dicranoglossum panamense | Polypodiaceae | 123 | 13 | 30 | 7 | 1 |
| Niphidium crassifolium | Polypodiaceae | 10 | 11 | 27 | 8 | 16 |
| Trichosalpinx orbicularis | Orchidaceae | 1 | 8 | 22 | 9 | 37 |
| Dichaea panamensis | Orchidaceae | 19 | 5 | 17 | 10 | 7 |
| Scaphyglottis prolifera | Orchidaceae | 9 | 5 | 18 | 11 | 18 |
| Tillandsia anceps | Bromeliaceae | 16 | 4 | 15 | 12 | 9 |
| Codonanthe macradenia | Gesneriaceae | 28 | 4 | 16 | 13 | 3 |
| Elaphoglossum herminieri | Lomariopsidaceae | 0 | 3 | 13 | 14 | |
| Trichomanes nummularium | Hymenophyllaceae | 0 | 3 | 13 | 15 | |
| Tillandsia bulbosa | Bromeliaceae | 2 | 3 | 12 | 16 | 30 |
| Polypodium percussum | Polypodiaceae | 1 | 2 | 12 | 17 | 37 |
| Sobralia fragrans | Orchidaceae | 2 | 2 | 12 | 18 | 30 |
| Anthurium friedrichsthalii | Araceae | 14 | 2 | 11 | 19 | 10 |
| Asplenium juglandifolium | Aspleniaceae | 0 | 2 | 11 | 20 | |
| Peperomia rotundifolia | Piperaceae | 18 | 2 | 11 | 21 | 8 |
| Asplenium serratum | Aspleniaceae | 7 | 2 | 11 | 22 | 20 |
| Campyloneurum occultum | Polypodiaceae | 6 | 1 | 10 | 23 | 21 |
| Stelis crescentiicola | Orchidaceae | 0 | 1 | 9 | 24 | |
| Campyloneurum phyllitidis | Polypodiaceae | 3 | 1 | 9 | 25 | 28 |
| Trichomanes ovale | Hymenophyllaceae | 1 | 1 | 9 | 26 | 37 |
| Vriesea gladioliflora | Bromeliaceae | 4 | 1 | 8 | 27 | 26 |
| Elaphoglossum latifolium | Lomariopsidaceae | 0 | 1 | 8 | 28 | |
| Peperomia ebingeri | Piperaceae | 0 | 0 | 8 | 29 | |
| Anthurium acutangulum | Araceae | 24 | 1 | 8 | 30 | 6 |
| Microgramma lycopodioides | Polypodiaceae | 12 | 0 | 8 | 31 | 12 |
| Catopsis sessiliflora | Araceae | 0 | 0 | 7 | 32 | |
| Anthurium hacumense | Araceae | 10 | 0 | 7 | 33 | 16 |
| Catasetum viridiflavum | Orchidaceae | 11 | 0 | 6 | 34 | 15 |
| Guzmania subcorymbosa | Bromeliaceae | 2 | 0 | 6 | 35 | 30 |
| Polystachya foliosa | Orchidaceae | 0 | 0 | 6 | 36 | |
| Anthrophyum lanceolatum | Vittariaceae | 2 | 0 | 5 | 37 | 30 |
| Lockhartia acuta | Orchidaceae | 0 | 0 | 5 | 38 | |
| Trigonidium egertonianum | Orchidaceae | 0 | 0 | 6 | 39 | |
| Aspasia principissa | Orchidaceae | 0 | 0 | 5 | 40 | |
| Maxillaria discolor | Orchidaceae | 0 | 0 | 5 | 41 | |
| Masdevallia livingstoneana | Orchidaceae | 0 | 0 | 5 | 42 | |
| Vittaria lineata | Vittariaceae | 1 | 0 | 5 | 43 | 37 |
| Trichomanes ekmannii | Hymenophyllaceae | 12 | 0 | 4 | 44 | 12 |
| Peperomia macrostachia | Piperaceae | 0 | 0 | 4 | 45 | |
| Anthurium clavigerum | Araceae | 9 | 0 | 5 | 46 | 18 |
| Microgramma reptans | Polypodiaceae | 12 | 0 | 4 | 47 | 12 |
| Anthurium durandii | Araceae | 0 | 0 | 4 | 48 | |
| Epidendrum nocturnum | Orchidaceae | 1 | 0 | 4 | 49 | 37 |
| Epidendrum difforme | Orchidaceae | 0 | 0 | 4 | 50 | |
| Anthurium scandens | Araceae | 0 | 0 | 4 | 51 | |
| Campylocentrum micranthum | Orchidaceae | 5 | 0 | 3 | 52 | 22 |
| Anthurium brownii | Araceae | 1 | 0 | 4 | 53 | 37 |
| Philodendron sagittifolium | Araceae | 0 | 0 | 4 | 54 | |
| Trichocentrum capistratum | Orchidaceae | 0 | 0 | 4 | 55 | |
| Pecluma pectinata | Polypodiaceae | 0 | 0 | 4 | 56 | |
| Notylia albida | Orchidaceae | 1 | 0 | 3 | 57 | 37 |
| Stenospermation angustifolium | Araceae | 0 | 0 | 3 | 58 | |
| Aechmea tillandsioides | Bromeliaceae | 5 | 0 | 3 | 59 | 22 |
| Oncidium ampliatum | Orchidaceae | 0 | 0 | 2 | 60 | |
| Hylocereus monacanthus | Cactaceae | 0 | 0 | 3 | 61 | |
| Trichomanes angustifrons | Hymenophyllaceae | 13 | 0 | 2 | 62 | 11 |
| Dimerandra emarginata | Orchidaceae | 0 | 0 | 3 | 63 | |
| Trichopilia maculata | Orchidaceae | 0 | 0 | 2 | 64 | |
| Columnea billbergiana | Gesneriaceae | 1 | 0 | 2 | 65 | 37 |
| Epiphyllum phyllanthus | Cactaceae | 0 | 0 | 2 | 66 | |
| Philodendron radiatum | Araceae | 0 | 0 | 2 | 67 | |
| Gongora quinquenervis | Orchidaceae | 3 | 0 | 2 | 68 | 28 |
| Mormodes powellii | Orchidaceae | 0 | 0 | 2 | 69 | |
| Hecistopteris pumila | Vittariaceae | 0 | 0 | 2 | 70 | |
| Polypodium triseriale | Polypodiaceae | 2 | 0 | 2 | 71 | 30 |
| Anetium citrifolium | Vittariaceae | 0 | 0 | 2 | 72 | |
| Ornithocephalus bicornis | Orchidaceae | 5 | 0 | 2 | 73 | 22 |
| Sobralia panamensis | Orchidaceae | 1 | 0 | 2 | 74 | 37 |
| Ornithocephalus powellii | Orchidaceae | 0 | 0 | 2 | 75 | |
| Pleurothallis verecunda | Orchidaceae | 2 | 0 | 2 | 76 | 30 |
| Epidendrum schlechterianum | Orchidaceae | 1 | 0 | 1 | 77 | 37 |
| Encyclia fragrans | Orchidaceae | 0 | 0 | 1 | 78 | |
| Polypodium costaricense | Polypodiaceae | 0 | 0 | 1 | 79 | |
| Epidendrum imatophyllum | Orchidaceae | 2 | 0 | 1 | 80 | 30 |
| Peperomia obtusifolia | Piperaceae | 0 | 0 | 1 | 81 | |
| Encyclia chimborazoensis | Orchidaceae | 0 | 0 | 1 | 82 | |
| Huperzia dichotoma | Selaginellaceae | 0 | 0 | 1 | 83 | |
| Elleanthus longibracteatus | Orchidaceae | 0 | 0 | 1 | 84 | |
| Encyclia aemula | Orchidaceae | 0 | 0 | 1 | 85 | |
| Guzmania musaica | Bromeliaceae | 0 | 0 | 1 | 86 | |
| Trichomanes punctatum | Hymenophyllaceae | 0 | 0 | 1 | 87 | |
| Trichomanes anadromum | Hymenophyllaceae | 1 | 0 | 1 | 88 | 37 |
| Pleurothallis grobyi | Orchidaceae | 0 | 0 | 1 | 89 | |
| Cochleanthes lipscombiae | Orchidaceae | 0 | 0 | 1 | 90 | |
| Jacquinella sp. | Orchidaceae | 0 | 0 | 1 | 91 | |
| Jacquiniella pedunculata | Orchidaceae | 0 | 0 | 1 | 92 | |
| Trichomanes godmanii | Hymenophyllaceae | 0 | 0 | 1 | 93 | |
| Maxillaria variabilis | Orchidaceae | 0 | 0 | 1 | 94 | |
| Lockhartia pittieri | Orchidaceae | 0 | 0 | 1 | 95 | |
| Kefersteinia sp. | Orchidaceae | 0 | 0 | 1 | 96 | |
| Peperomia cordulata | Piperaceae | 0 | 0 | 1 | 97 | |
| Drymonia serrulata | Gesneriaceae | 0 | 0 | 0 | 98 | |
| Werauhia sanguinolenta | Bromeliaceae | 0 | 0 | 1 | 99 | |
| Maxillaria crassifolia | Orchidaceae | 0 | 0 | 1 | 100 | |
| Caularthron bilamellatum | Orchidaceae | 0 | 0 | 1 | 101 | |
| Hymenophyllum brevifrons | Hymenophyllaceae | 0 | 0 | 1 | 102 | |
| Anthurium bakeri | Araceae | 0 | 0 | 1 | 103 | |
Appendix 3
Comparison of actual occurrence of epiphytes on Perebea xanthochyma and null assemblages created from the epiphyte pool in 0·4 ha of the San Lorenzo Crane Plot. The analysis is based on the individual number found on 38 Perebea xanthochyma trees. Shown are individual numbers of a given epiphyte species observed on Perebea xanthochyma and the expected range of individual numbers (lower and upper boundary of 95 % confidence intervals). Bold names indicate species which were more common than expected; underlined names indicate species that are less common than expected.
Table 5.
| Individuals of null assemblage |
Rank |
|||||
|---|---|---|---|---|---|---|
| Species | Family | Individuals on Perebea | Lower boundary | Upper boundary | Null assemblage | Perebea |
| Scaphyglottis longicaulis | Orchidaceae | 6 | 18 | 37 | 1 | 12 |
| Scaphyglottis graminifolia | Orchidaceae | 0 | 17 | 37 | 2 | |
| Elaphoglossum sporadolepis | Lomariopsidaceae | 1 | 13 | 30 | 3 | 18 |
| Maxillaria uncata | Orchidaceae | 7 | 8 | 23 | 4 | 11 |
| Pleurothallis brighamii | Orchidaceae | 0 | 7 | 20 | 5 | |
| Dicranoglossum panamense | Polypodiaceae | 43 | 4 | 16 | 6 | 1 |
| Ananthacorus angustifolius | Vittariaceae | 33 | 5 | 17 | 7 | 2 |
| Niphidium crassifolium | Polypodiaceae | 1 | 3 | 14 | 8 | 18 |
| Trichosalpinx orbicularis | Orchidaceae | 0 | 2 | 12 | 9 | |
| Dichaea panamensis | Orchidaceae | 1 | 1 | 9 | 10 | 18 |
| Tillandsia anceps | Bromeliaceae | 17 | 1 | 9 | 11 | 4 |
| Scaphyglottis prolifera | Orchidaceae | 0 | 1 | 9 | 12 | |
| Trichomanes nummularium | Hymenophyllaceae | 0 | 1 | 8 | 13 | |
| Codonanthe macradenia | Gesneriaceae | 11 | 1 | 9 | 14 | 7 |
| Elaphoglossum herminieri | Lomariopsidaceae | 0 | 0 | 7 | 15 | |
| Tillandsia bulbosa | Bromeliaceae | 0 | 0 | 7 | 16 | |
| Sobralia fragrans | Orchidaceae | 0 | 0 | 7 | 17 | |
| Polypodium percussum | Polypodiaceae | 1 | 0 | 7 | 18 | 18 |
| Peperomia rotundifolia | Piperaceae | 5 | 0 | 6 | 19 | 14 |
| Anthurium friedrichsthalii | Araceae | 1 | 0 | 6 | 20 | 18 |
| Asplenium juglandifolium | Aspleniaceae | 6 | 0 | 6 | 21 | 12 |
| Asplenium serratum | Aspleniaceae | 0 | 0 | 6 | 22 | |
| Campyloneurum occultum | Polypodiaceae | 1 | 0 | 6 | 23 | 18 |
| Stelis crescentiicola | Orchidaceae | 0 | 0 | 5 | 24 | |
| Trichomanes ovale | Hymenophyllaceae | 8 | 0 | 5 | 25 | 9 |
| Campyloneurum phyllitidis | Polypodiaceae | 1 | 0 | 5 | 26 | 18 |
| Peperomia ebingeri | Piperaceae | 1 | 0 | 5 | 27 | 18 |
| Vriesea gladioliflora | Bromeliaceae | 8 | 0 | 5 | 28 | 9 |
| Elaphoglossum latifolium | Lomariopsidaceae | 0 | 0 | 5 | 29 | |
| Anthurium acutangulum | Araceae | 4 | 0 | 5 | 30 | 15 |
| Catopsis sessiliflora | Araceae | 0 | 0 | 4 | 31 | |
| Microgramma lycopodioides | Polypodiaceae | 1 | 0 | 5 | 32 | 18 |
| Polystachya foliosa | Orchidaceae | 0 | 0 | 3 | 33 | |
| Catasetum viridiflavum | Orchidaceae | 0 | 0 | 4 | 34 | |
| Anthurium hacumense | Araceae | 0 | 0 | 4 | 35 | |
| Guzmania subcorymbosa | Bromeliaceae | 2 | 0 | 4 | 36 | 16 |
| Trigonidium egertonianum | Orchidaceae | 0 | 0 | 3 | 37 | |
| Lockhartia acuta | Orchidaceae | 0 | 0 | 3 | 38 | |
| Maxillaria discolor | Orchidaceae | 0 | 0 | 3 | 39 | |
| Anthrophyum lanceolatum | Vittariaceae | 1 | 0 | 3 | 40 | 18 |
| Aspasia principissa | Orchidaceae | 0 | 0 | 3 | 41 | |
| Anthurium clavigerum | Araceae | 0 | 0 | 3 | 42 | |
| Trichomanes ekmannii | Hymenophyllaceae | 12 | 0 | 3 | 43 | 5 |
| Vittaria lineata | Vittariaceae | 0 | 0 | 3 | 44 | |
| Masdevallia livingstoneana | Orchidaceae | 0 | 0 | 3 | 45 | |
| Epidendrum difforme | Orchidaceae | 0 | 0 | 3 | 46 | |
| Microgramma reptans | Polypodiaceae | 0 | 0 | 3 | 47 | |
| Epidendrum nocturnum | Orchidaceae | 0 | 0 | 3 | 48 | |
| Anthurium durandii | Araceae | 1 | 0 | 3 | 49 | 18 |
| Campylocentrum micranthum | Orchidaceae | 10 | 0 | 2 | 50 | 8 |
| Peperomia macrostachia | Piperaceae | 0 | 0 | 3 | 51 | |
| Anthurium scandens | Araceae | 0 | 0 | 3 | 52 | |
| Anthurium brownii | Araceae | 0 | 0 | 2 | 53 | |
| Philodendron sagittifolium | Araceae | 0 | 0 | 2 | 54 | |
| Pecluma pectinata | Polypodiaceae | 0 | 0 | 2 | 55 | |
| Trichocentrum capistratum | Orchidaceae | 1 | 0 | 2 | 56 | 18 |
| Aechmea tillandsioides | Bromeliaceae | 12 | 0 | 2 | 57 | 5 |
| Stenospermation angustifolium | Araceae | 0 | 0 | 2 | 58 | |
| Notylia albida | Orchidaceae | 0 | 0 | 2 | 59 | |
| Dimerandra emarginata | Orchidaceae | 0 | 0 | 2 | 60 | |
| Hylocereus monacanthus | Cactaceae | 0 | 0 | 2 | 61 | |
| Oncidium ampliatum | Orchidaceae | 1 | 0 | 1 | 62 | 18 |
| Trichomanes angustifrons | Hymenophyllaceae | 24 | 0 | 2 | 63 | 3 |
| Mormodes powellii | Orchidaceae | 0 | 0 | 2 | 64 | |
| Epiphyllum phyllanthus | Cactaceae | 1 | 0 | 1 | 65 | 18 |
| Columnea billbergiana | Gesneriaceae | 0 | 0 | 1 | 66 | |
| Trichopilia maculata | Orchidaceae | 0 | 0 | 1 | 67 | |
| Hecistopteris pumila | Vittariaceae | 0 | 0 | 1 | 68 | |
| Philodendron radiatum | Araceae | 0 | 0 | 1 | 69 | |
| Anetium citrifolium | Vittariaceae | 0 | 0 | 1 | 70 | |
| Ornithocephalus powellii | Orchidaceae | 0 | 0 | 1 | 71 | |
| Peperomia obtusifolia | Piperaceae | 0 | 0 | 1 | 72 | |
| Polypodium triseriale | Polypodiaceae | 0 | 0 | 1 | 73 | |
| Huperzia dichotoma | Selaginellaceae | 0 | 0 | 1 | 74 | |
| Encyclia fragrans | Orchidaceae | 0 | 0 | 1 | 75 | |
| Gongora quinquenervis | Orchidaceae | 2 | 0 | 1 | 76 | 16 |
| Pleurothallis verecunda | Orchidaceae | 1 | 0 | 1 | 77 | 18 |
| Guzmania musaica | Bromeliaceae | 0 | 0 | 1 | 78 | |
| Ornithocephalus bicornis | Orchidaceae | 0 | 0 | 1 | 79 | |
| Sobralia panamensis | Orchidaceae | 0 | 0 | 1 | 80 | |
| Epidendrum imatophyllum | Orchidaceae | 0 | 0 | 1 | 81 | |
| Trichomanes godmanii | Hymenophyllaceae | 0 | 0 | 1 | 82 | |
| Polypodium costaricense | Polypodiaceae | 0 | 0 | 1 | 83 | |
| Trichomanes punctatum | Hymenophyllaceae | 0 | 0 | 1 | 84 | |
| Elleanthus longibracteatus | Orchidaceae | 0 | 0 | 1 | 85 | |
| Encyclia chimborazoensis | Orchidaceae | 0 | 0 | 1 | 86 | |
| Pleurothallis grobyi | Orchidaceae | 0 | 0 | 1 | 87 | |
| Epidendrum schlechterianum | Orchidaceae | 0 | 0 | 1 | 88 | |
| Encyclia aemula | Orchidaceae | 0 | 0 | 1 | 89 | |
| Trichomanes anadromum | Hymenophyllaceae | 0 | 0 | 1 | 90 | |
| Cochleanthes lipscombiae | Orchidaceae | 0 | 0 | 1 | 91 | |
| Maxillaria crassifolia | Orchidaceae | 0 | 0 | 1 | 92 | |
| Jacquiniella pedunculata | Orchidaceae | 0 | 0 | 1 | 93 | |
| Caularthron bilamellatum | Orchidaceae | 0 | 0 | 0 | 94 | |
| Lockhartia pittieri | Orchidaceae | 0 | 0 | 1 | 95 | |
| Drymonia serrulata | Gesneriaceae | 0 | 0 | 0 | 96 | |
| Jacquinella sp. | Orchidaceae | 0 | 0 | 0 | 97 | |
| Kefersteinia sp. | Orchidaceae | 0 | 0 | 0 | 98 | |
| Hymenophyllum brevifrons | Hymenophyllaceae | 0 | 0 | 0 | 99 | |
| Peperomia cordulata | Piperaceae | 0 | 0 | 1 | 100 | |
| Maxillaria variabilis | Orchidaceae | 0 | 0 | 0 | 101 | |
| Werauhia sanguinolenta | Bromeliaceae | 0 | 0 | 0 | 102 | |
| Anthurium bakeri | Araceae | 0 | 0 | 0 | 103 | |
LITERATURE CITED
- Ackerman JD, Montalvo AM, Vera AM. 1989. Epiphyte host specificity of Encyclia krugii, a Puerto Rican endemic orchid. Lindleyana 4: 74–77. [Google Scholar]
- Andrade JL, Nobel PS. 1996. Habitat, CO2 uptake, and growth for the CAM epiphytic cactus Epiphyllum phyllanthus in a Panamanian tropical forest. Journal of Tropical Ecology 12: 291–306. [Google Scholar]
- Benzing DH. 1978. Germination and early establishment of Tillandsia circinnata Schlecht. (Bromeliaceae) on some of its hosts and other supports in Southern Florida. Selbyana 5: 95–106. [Google Scholar]
- Benzing DH. 1981. Bark surfaces and the origin and maintenance of diversity among angiosperm epiphytes: a hypothesis. Selbyana 5: 248–255. [Google Scholar]
- Benzing DH. 1990. Vascular epiphytes. General biology and related biota. Cambridge: Cambridge University Press.
- ter Braak CJF. 1995. Ordination. In: Jongman RHG, ter Braak CJF, van Tongeren OFR, eds. Data analysis in community and landscape ecology. Cambridge: Cambridge University Press, 91–173.
- ter Braak CJF, Šmilauer P. 1997. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community ordination, Version 4.5. Ithaca, NY: Microcomputer Power.
- Callaway R, Reinhart K, Moore G, Moore D, Pennings S. 2002. Epiphyte host preferences and host traits: mechanisms for species-specific interactions. Oecologia 132: 221–230. [DOI] [PubMed] [Google Scholar]
- Chase MW. 1987. Obligate twig epiphytism in the Oncidiinae and other neotropical orchids. Selbyana 10: 24–30. [Google Scholar]
- Condit R, Aguilar S, Hernandez A, Perez R, Lao S, Angehr G, et al. 2004. Tropical forest dynamics across a rainfall gradient and the impact of an El Niño dry season. Journal of Tropical Ecology 20: 51–72. [Google Scholar]
- Cribb PJ, Du Puy D, Bosser J. 2002. An unusual new epiphytic species of Eulophia (Orchidaceae) from southeastern Madagascar. Adansonia 24: 169–172. [Google Scholar]
- Croat T. 1978. Flora of Barro Colorado Island. Stanford, CA: Stanford University Press.
- D'Arcy WG. 1987. Flora of Panama: checklist and index. St Louis, MO: Missouri Botanical Garden.
- Gentry AH, Dodson CH. 1987. Diversity and biogeography of neotropical vascular epiphytes. Annals of the Missouri Botanical Garden 74: 205–233. [Google Scholar]
- Griffiths H, Smith JAC. 1983. Photosynthetic pathways in the Bromeliaceae of Trinidad: relations between life-forms, habitat preference and the occurrence of CAM. Oecologia 60: 176–184. [DOI] [PubMed] [Google Scholar]
- Hietz P. 1997. Population dynamics of epiphytes in a Mexican humid montane forest. Journal of Ecology 85: 767–777.
- Hietz P, Briones O. 1998. Correlation between water relations and within-canopy distribution of epiphytic ferns in a Mexican cloud forest. Oecologia 114: 305–316. [DOI] [PubMed] [Google Scholar]
- Holbrook NM, Putz FE. 1996. Water relations of epiphytic and terrestrially-rooted strangler figs in Venezuelan palm savanna. Oecologia 106: 424–431. [DOI] [PubMed] [Google Scholar]
- Johansson D. 1974. Ecology of vascular epiphytes in West African rain forest. Acta Phytogeographica Suecica 59: 1–136. [Google Scholar]
- Kelly DL, O'Donovan G, Feehan J, Murphy S, Drangeid SO, Marcano-Berti L. 2004. The epiphyte communities of a montane rain forest in the Andes of Venezuela: patterns in the distribution of the flora. Journal of Tropical Ecology 20: 643–666. [Google Scholar]
- Kreft H, Köster N, Küper W, Nieder J, Barthlott W. 2004. Diversity and biogeography of vascular epiphytes in Western Amazonia, Yasuní, Ecuador. Journal of Biogeography 31: 1463–1476. [Google Scholar]
- Krömer T, Kessler M, Gradstein SR, Acebey A. 2005. Diversity patterns of vascular epiphytes along an elevational gradient in the Andes. Journal of Biogeography 32: 1799–1809. [Google Scholar]
- Laube S, Zotz G. 2006. Long-term changes of the vascular epiphyte assemblage on the palm Socratea exorrhiza in a lowland forest in Panama. Journal of Vegetation Science (in press).
- Lellinger DB. 1989. The ferns and fern-allies of Costa Rica. Panama, and the Chocó (Part I: Psilotaceae through Dicksoniaceae). Washington, DC: American Fern Society.
- Male TD, Roberts GE. 2005. Host associations of the strangler fig Ficus watkinsiana in a subtropical Queensland rain forest. Austral Ecology 30: 229–236. [Google Scholar]
- Mesler MR. 1975. The gametophytes of Ophioglossum palmatum L. American Journal of Botany 62: 982–992. [Google Scholar]
- Migenis LE, Ackerman JD. 1993. Orchid–phorophyte relationships in a forest watershed in Puerto Rico. Journal of Tropical Ecology 9: 231–240. [Google Scholar]
- Moran R, Russell R. 2004. The occurrence of Trichomanes godmanii (Hymenophyllaceae) on Welfia georgii (Arecaceae) at the La Selva Biological Station, Costa Rica. American Fern Journal 94: 70–76. [Google Scholar]
- Oliver WRB. 1930. New Zealand epiphytes. Journal of Ecology 18: 1–50. [Google Scholar]
- R Development Core Team. 2005. R: A Language and Environment for Statistical Computing, V 2.2.1. Vienna, Austria: R Foundation for Statistical Computing (http:\\www.R-project.org).
- Schatz GE, Williamson GB, Cogswell CM, Stam AM. 1985. Stilt roots and growth of arboreal palms. Biotropica 17: 206–209. [Google Scholar]
- Schulz A, Wagner T. 2002. Influence of forest type and tree species on canopy ants (Hymenoptera: Formicidae) in Budongo Forest, Uganda. Oecologia 133: 224–232. [DOI] [PubMed] [Google Scholar]
- Went FW. 1940. Soziologie der Epiphyten eines tropischen Regenwaldes. Annales du Jardin Botanique de Buitenzorg 50: 1–98. [Google Scholar]
- Wright SJ, Horlyck V, Basset Y. 2003. Tropical Canopy Biology Program, Republic of Panama. In: Basset Y, Horlyck V, Wright SJ, eds. Studying forest canopies from above: the International Canopy Crane Network. Smithsonian Tropical Research Institute (Panama) and United Nations Environmental Programme (UNEP), 137–139.
- Zimmerman JK, Olmsted IC. 1992. Host tree utilization by vascular epiphytes in a seasonally inundated forest (Tintal) in Mexico. Biotropica 24: 402–407. [Google Scholar]
- Zotz G. 2004. How prevalent is crassulaceaen acid metabolism among vascular epiphytes? Oecologia 138: 184–192. [DOI] [PubMed] [Google Scholar]
- Zotz G, Laube S. 2005. Tank function in the epiphytic bromeliad, Catopsis sessiliflora. Ecotropica 11: 63–68. [Google Scholar]
- Zotz G, Bermejo P, Dietz H. 1999. The epiphyte vegetation of Annona glabra on Barro Colorado Island, Panama. Journal of Biogeography 26: 761–776. [Google Scholar]
- Zotz G, Laube S, Schmidt G. 2005. Long-term population dynamics of the epiphytic bromeliad, Werauhia sanguinolenta. Ecography 28: 806–814. [Google Scholar]