Abstract
• Background Many plant species can modify their root architecture to enable them to forage for heterogeneously distributed nutrients in the soil. The foraging response normally involves increased proliferation of lateral roots within nutrient-rich soil patches, but much remains to be understood about the signalling mechanisms that enable roots to sense variations in the external concentrations of different mineral nutrients and to modify their patterns of growth and development accordingly.
• Scope In this review we consider different aspects of the way in which the nitrogen supply can modify root branching, focusing on Arabidopsis thaliana. Our current understanding of the mechanism of nitrate stimulation of lateral root growth and the role of the ANR1 gene are summarized. In addition, evidence supporting the possible role of auxin in regulating the systemic inhibition of early lateral root development by high rates of nitrate supply is presented. Finally, we examine recent evidence that an amino acid, l-glutamate, can act as an external signal to elicit complex changes in root growth and development.
• Conclusions It is clear that plants have evolved sophisticated pathways for sensing and responding to changes in different components of the external nitrogen supply as well as their own internal nitrogen status. We speculate on the possibility that the effects elicited by external l-glutamate represent a novel form of foraging response that could potentially enhance a plant's ability to compete with its neighbours and micro-organisms for localized sources of organic nitrogen.
Keywords: Arabidopsis thaliana, auxin, dissolved organic nitrogen, foraging, glutamate, lateral roots, MADS box transcription factor, nitrate, nitrogen, root architecture, root development, roots, signalling, Thlaspi caerulescens
INTRODUCTION
The growth and development of a root system is highly sensitive to modification by both intrinsic and extrinsic factors (Forde and Lorenzo, 2001; Bloom et al., 2002; Porterfield, 2002). Intrinsic factors that influence root growth and development include the supply of photosynthate from the shoot and the nutrient status of the plant; extrinsic factors include the supply and distribution of nutrients in the soil, soil compaction and gradients of water potential. One important aspect of root plasticity is the proliferation of lateral roots that occurs within soil patches enriched in certain nutrients, including 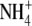 ,
, 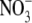 , Pi (Robinson, 1994; Forde and Lorenzo, 2001) and even Zn2+ (Haines, 2002). This form of plant behaviour is commonly referred to as foraging, as it represents the preferential utilization of habitat patches that are rich in essential resources (Hutchings and de Kroon, 1994).
, Pi (Robinson, 1994; Forde and Lorenzo, 2001) and even Zn2+ (Haines, 2002). This form of plant behaviour is commonly referred to as foraging, as it represents the preferential utilization of habitat patches that are rich in essential resources (Hutchings and de Kroon, 1994).
Increased branching (of shoots or roots) in resource-rich conditions serves to enhance the precision with which the resource-capturing structures (leaves or roots) are placed within the environment (Sutherland and Stillman, 1988). Thus, an understanding of the mechanisms underlying root foraging is very much dependent on understanding how intrinsic and extrinsic nutritional factors influence root branching. In this brief review we will consider recent progress towards elucidating the signalling mechanisms by which different forms of nitrogen regulate root branching in Arabidopsis thaliana.
EFFECT OF EXTERNAL NITRATE ON LATERAL ROOT GROWTH AND THE ROLE OF THE ANR1 GENE
The ability to respond to localized nitrate supplies by proliferating lateral roots within the nitrate-rich zone is a property common to many species of plants (Robinson, 1994; Hodge, 2004). In barley, this ability is due to a combination of increased numbers of lateral roots and increased rates of lateral root elongation (Drew and Saker, 1975). In Arabidopsis, the primary effect of a localized nitrate treatment was to stimulate lateral root elongation (Zhang and Forde, 1998; Linkohr et al., 2002), with one report indicating a small localized increase in lateral root numbers (Linkohr et al., 2002). This stimulation of lateral root elongation appears to be attributable to a signalling effect from the 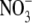 ion itself rather than to a downstream metabolite (Zhang and Forde, 1998; Zhang et al., 1999). Nitrate stimulates lateral root elongation by increasing rates of cell production in the root tips directly exposed to the signal (rather than through any effect on cell elongation) (Zhang et al., 1999).
ion itself rather than to a downstream metabolite (Zhang and Forde, 1998; Zhang et al., 1999). Nitrate stimulates lateral root elongation by increasing rates of cell production in the root tips directly exposed to the signal (rather than through any effect on cell elongation) (Zhang et al., 1999).
How the nitrate signal is converted into an increase in meristematic activity in the root tip is an intriguing question that as yet has no clear answer. One component of the 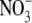 signalling pathway has been identified in the form of the product of the ANR1 gene, which is a member of the MADS box family of transcription factors (Zhang and Forde, 1998). Using a reverse genetic approach it was shown that lateral roots of Arabidopsis lines in which ANR1 was down-regulated were defective in their response to a localized supply of
signalling pathway has been identified in the form of the product of the ANR1 gene, which is a member of the MADS box family of transcription factors (Zhang and Forde, 1998). Using a reverse genetic approach it was shown that lateral roots of Arabidopsis lines in which ANR1 was down-regulated were defective in their response to a localized supply of 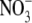 . We have obtained additional evidence that ANR1 is a positive regulator of lateral root growth using transgenic Arabidopsis lines in which ANR1 can be rapidly post-translationally activated by a treatment with the synthetic steroid dexamethasone (DEX). These transgenic lines carry a construct constitutively expressing a translational fusion between ANR1 and the ligand-binding domain of the rat glucocorticoid receptor (rGR). The ANR1-rGR fusion protein is held inactive in a cytoplasmic complex with the HSP90 protein until addition of DEX, when it is released and is able to enter the nucleus where it can activate or repress its target genes (Picard et al., 1988). When seedlings of these ANR1-rGR lines were treated with 1 μm DEX, lateral root growth was strongly stimulated (Filleur et al., 2005). Remarkably, even though the ANR1-rGR gene was expressed under the control of a strong constitutive promoter (CaMV 35S), the effect of the DEX treatment was specific to lateral root growth, suggesting that one or more components of the regulatory pathway of which ANR1 is a part must be absent in the primary root tip.
. We have obtained additional evidence that ANR1 is a positive regulator of lateral root growth using transgenic Arabidopsis lines in which ANR1 can be rapidly post-translationally activated by a treatment with the synthetic steroid dexamethasone (DEX). These transgenic lines carry a construct constitutively expressing a translational fusion between ANR1 and the ligand-binding domain of the rat glucocorticoid receptor (rGR). The ANR1-rGR fusion protein is held inactive in a cytoplasmic complex with the HSP90 protein until addition of DEX, when it is released and is able to enter the nucleus where it can activate or repress its target genes (Picard et al., 1988). When seedlings of these ANR1-rGR lines were treated with 1 μm DEX, lateral root growth was strongly stimulated (Filleur et al., 2005). Remarkably, even though the ANR1-rGR gene was expressed under the control of a strong constitutive promoter (CaMV 35S), the effect of the DEX treatment was specific to lateral root growth, suggesting that one or more components of the regulatory pathway of which ANR1 is a part must be absent in the primary root tip.
Evidence has been obtained using transgenic lines constitutively overexpressing ANR1 that there is an 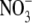 -dependent component of the ANR1 signalling pathway (Y. Gan and B.G. Forde, unpubl. obs.). These lines showed increased rates of lateral root growth, but the effect was dependent on the presence of
-dependent component of the ANR1 signalling pathway (Y. Gan and B.G. Forde, unpubl. obs.). These lines showed increased rates of lateral root growth, but the effect was dependent on the presence of 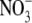 in the medium. Thus, ANR1 overexpression appears to be necessary but not sufficient for stimulating lateral root growth. One possible model to explain this observation is that ANR1 is post-translationally regulated and that an
in the medium. Thus, ANR1 overexpression appears to be necessary but not sufficient for stimulating lateral root growth. One possible model to explain this observation is that ANR1 is post-translationally regulated and that an 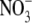 signal is required to convert it to its active form. Alternatively,
signal is required to convert it to its active form. Alternatively, 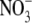 could be required to induce another essential component of the signalling pathway (Walch-Liu et al., 2005).
could be required to induce another essential component of the signalling pathway (Walch-Liu et al., 2005).
Initial studies using Arabidopsis root cultures had indicated that ANR1 itself was 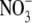 -inducible (Zhang and Forde, 1998). However, recent results obtained with mature hydroponically grown Arabidopsis plants have established that ANR1 expression in roots of intact plants is up-regulated approx. two-fold after 2.5 d of N starvation and rapidly down-regulated when
-inducible (Zhang and Forde, 1998). However, recent results obtained with mature hydroponically grown Arabidopsis plants have established that ANR1 expression in roots of intact plants is up-regulated approx. two-fold after 2.5 d of N starvation and rapidly down-regulated when 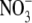 or another N source is re-supplied (Gan et al., 2005). Consistent with these data, an earlier study had shown that ANR1 was down-regulated when
or another N source is re-supplied (Gan et al., 2005). Consistent with these data, an earlier study had shown that ANR1 was down-regulated when 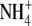 -grown Arabidopsis seedlings were treated with
-grown Arabidopsis seedlings were treated with 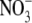 (Wang et al., 2000).
(Wang et al., 2000).
Because ANR1 is a positive regulator of lateral root growth, its down-regulation under conditions of N sufficiency suggests a possible mechanism for feedback regulation of lateral root growth rates by the N status of the plant. It is well established that lateral root growth in one part of the root system is not only dependent on the immediate external 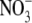 concentration but also on the amount of
concentration but also on the amount of 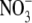 supplied to the remainder of the root system (Drew et al., 1973). The inhibition of early lateral root development by high shoot
supplied to the remainder of the root system (Drew et al., 1973). The inhibition of early lateral root development by high shoot 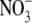 concentrations has already been identified as one mechanism for feedback regulation of root branching (Stitt and Feil, 1999; Zhang et al., 1999). It has therefore been suggested that the down-regulation of ANR1 under conditions of high N status (not necessarily
concentrations has already been identified as one mechanism for feedback regulation of root branching (Stitt and Feil, 1999; Zhang et al., 1999). It has therefore been suggested that the down-regulation of ANR1 under conditions of high N status (not necessarily 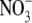 status) could provide an effective means of feedback regulating elongation of the mature lateral root (Gan et al., 2005). In this case, the inverse relationship between ANR1 expression and N status can be viewed as a mechanism for modulating the intensity of the lateral root response to a localized
status) could provide an effective means of feedback regulating elongation of the mature lateral root (Gan et al., 2005). In this case, the inverse relationship between ANR1 expression and N status can be viewed as a mechanism for modulating the intensity of the lateral root response to a localized 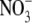 supply to take into account the plant's demand for N.
supply to take into account the plant's demand for N.
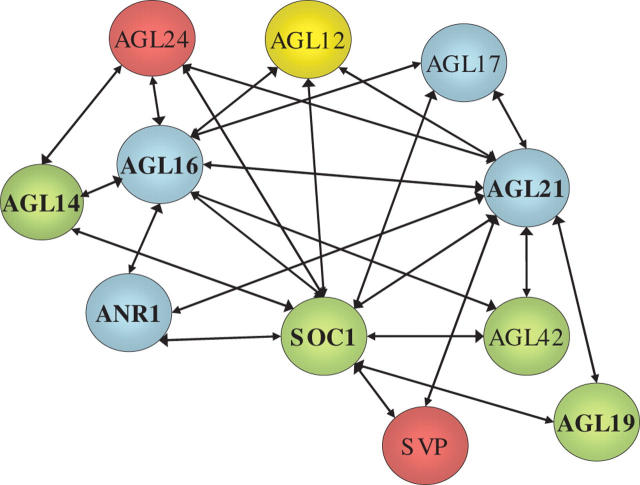
There is evidence for widespread interactions between MADS box proteins at both the post-translational and the transcriptional levels. MADS box factors bind to DNA as dimers, which can be either homodimers or heterodimers with other MADS box proteins (Theissen et al., 2000). There are also numerous cases in which one MADS box protein has been found to regulate the transcription of another MADS box gene, either directly or indirectly (Riechmann and Meyerowitz, 1997; Jack, 2004). With at least half the >100 members of the MADS box gene family in Arabidopsis shown to be transcribed in roots (Rounsley et al., 1995; Alvarez-Buylla et al., 2000; Burgeff et al., 2002; Parenicova et al., 2003), this gives ample scope for possible regulatory interactions between ANR1 and other MADS box genes and also for functional redundancy between different members of the gene family, as is the case among some MADS box genes involved in flower development (Jack, 2004). It has been suggested that the AGL21 gene, which is a member of the same clade as ANR1 and appears to have a similar spatial pattern of expression in roots, may be functionally redundant with ANR1 (Burgeff et al., 2002).
In an attempt to throw light on these issues, a recent study has used quantitative real-time PCR (qRT-PCR) to compare the responsiveness of ANR1 and 11 other root-expressed MADS box genes to fluctuations in the supply of N, P and S (Gan et al., 2005). Four MADS box genes (AGL12, AGL17, AGL18 and AGL79) were unresponsive to a 2.5-d period of N starvation or to a 3-h period of 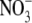 resupply. However, the other seven MADS box genes responded in a similar way to ANR1, although less strongly. These included two members of the ANR1 clade, AGL16 and AGL21, with only one member of this clade (AGL17) failing to respond. Three of the other N-regulated genes belong to the SOC1-like clade (AGL14, AGL19 and SOC1), and the other two (AGL26 and AGL56) belong to the poorly characterized type I lineage of MADS box genes. SOC1 was the only gene of those tested that was responsive to changes in P and S supply. Any of the five type II MADS box genes, particularly the two members of the ANR1 clade (AGL16 and AGL21) could be candidates for having roles that at least partially overlap with those of ANR1.
resupply. However, the other seven MADS box genes responded in a similar way to ANR1, although less strongly. These included two members of the ANR1 clade, AGL16 and AGL21, with only one member of this clade (AGL17) failing to respond. Three of the other N-regulated genes belong to the SOC1-like clade (AGL14, AGL19 and SOC1), and the other two (AGL26 and AGL56) belong to the poorly characterized type I lineage of MADS box genes. SOC1 was the only gene of those tested that was responsive to changes in P and S supply. Any of the five type II MADS box genes, particularly the two members of the ANR1 clade (AGL16 and AGL21) could be candidates for having roles that at least partially overlap with those of ANR1.
In the same study, the expression of these 11 MADS box genes was examined in an ANR1 knock-out mutant to test for possible regulatory interactions (Gan et al., 2005). It was found that inactivation of ANR1 had no discernible effect on the expression of any of the genes, indicating that these genes are not under either direct or indirect control of ANR1, at least at the transcriptional level.
Recent evidence, however, has suggested the possibility of regulatory interactions between some of the root-expressed MADS box genes at the protein level. A comprehensive map of protein–protein interactions among the Arabidopsis MADS box family has been compiled using a matrix-based yeast two-hybrid screen (de Folter et al., 2005). Figure 1 shows one part of this ‘interactome’ map, representing only those type II MADS box proteins whose expression has been reported in roots (Parenicova et al., 2003) and which interacted either with ANR1 or with MADS box proteins that themselves interacted with ANR1. The complexity of the network of potential interactions is remarkably high, with each of the 11 proteins interacting with 2–10 other MADS box proteins. However, these findings must be treated with caution because the interactions have not been confirmed in planta and in any case can only be biologically meaningful if the relevant proteins are expressed in the same cells at the same time. Nevertheless, it is noteworthy that ANR1 was found to interact with only three other proteins (AGL16, AGL21 and SOC1), all of which are regulated by the N supply in a similar manner to ANR1 (Gan et al., 2005).
Fig. 1.
Network of potential protein–protein interactions involving ANR1 and other root-expressed members of the type II lineage of MADS box proteins. Those proteins for which interactions were detected in the matrix-based yeast two-hybrid system (de Folter et al., 2005) are connected by a line. Different clades are colour-coded: blue, ANR1-like; red, SVP-like; green: SOC1-like. AGL12 is an orphan MADS box protein. Those proteins encoded by genes that are co-regulated by changes in the N supply (Gan et al., 2005) are indicated by bold type.
SYSTEMIC INHIBITION OF LATERAL ROOT DEVELOPMENT AT HIGH RATES OF NITRATE SUPPLY
When Arabidopsis seedlings were grown at external 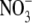 concentrations ≥10 mm, early lateral root development was inhibited, resulting in the accumulation of short laterals that were blocked just after emergence from the primary root (Zhang et al., 1999). The finding that this effect was enhanced in a nitrate reductase-deficient mutant suggested that the internal
concentrations ≥10 mm, early lateral root development was inhibited, resulting in the accumulation of short laterals that were blocked just after emergence from the primary root (Zhang et al., 1999). The finding that this effect was enhanced in a nitrate reductase-deficient mutant suggested that the internal 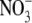 concentration rather than accumulation of the products of
concentration rather than accumulation of the products of 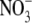 assimilation was the key factor triggering the developmental response (Zhang et al., 1999). It has been proposed that the accumulation of high tissue concentrations of
assimilation was the key factor triggering the developmental response (Zhang et al., 1999). It has been proposed that the accumulation of high tissue concentrations of 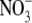 in the leaf are responsible for generating a long-distance signal that regulates lateral root development (Scheible et al., 1997; Zhang et al., 1999). The nature of this long-distance signal is unknown, but an obvious candidate is auxin because it has been shown that shoot-derived auxin is important for stimulating lateral root emergence (but not initiation) in Arabidopsis seedlings (Bhalerao et al., 2002). It has been hypothesized that
in the leaf are responsible for generating a long-distance signal that regulates lateral root development (Scheible et al., 1997; Zhang et al., 1999). The nature of this long-distance signal is unknown, but an obvious candidate is auxin because it has been shown that shoot-derived auxin is important for stimulating lateral root emergence (but not initiation) in Arabidopsis seedlings (Bhalerao et al., 2002). It has been hypothesized that 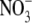 accumulation in the shoot may inhibit the flux of auxin to the root, leading to a failure of the lateral roots to pass an auxin-requiring checkpoint in lateral root development (Forde, 2002).
accumulation in the shoot may inhibit the flux of auxin to the root, leading to a failure of the lateral roots to pass an auxin-requiring checkpoint in lateral root development (Forde, 2002).
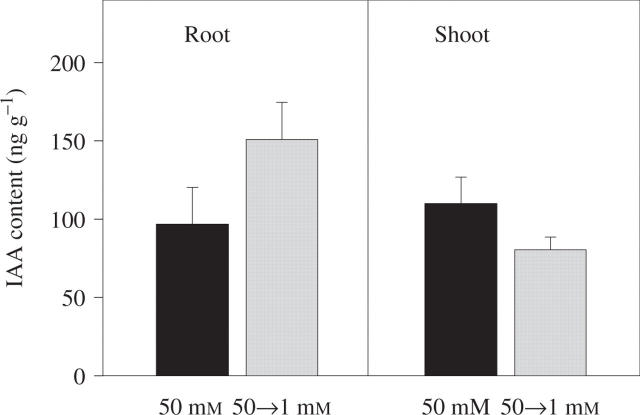
We have tested this hypothesis by measuring tissue auxin concentrations in the root 24 h after transferring Arabidopsis seedlings from 50 mm 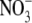 to 1 mm
to 1 mm 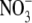 . It has been found that lateral roots whose development is arrested by growth on 50 mm
. It has been found that lateral roots whose development is arrested by growth on 50 mm 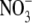 begin to develop normally 24–48 h after removal of the high
begin to develop normally 24–48 h after removal of the high 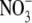 concentration (unpublished results). If auxin is involved in regulating this process, its concentration in the root might therefore be expected to increase in the period preceding the release of the lateral roots from their inhibition. In agreement with this prediction, we observed a 50 % increase in the IAA (indole 3-acetic acid) content of the roots of seedlings transferred to 1 mm
concentration (unpublished results). If auxin is involved in regulating this process, its concentration in the root might therefore be expected to increase in the period preceding the release of the lateral roots from their inhibition. In agreement with this prediction, we observed a 50 % increase in the IAA (indole 3-acetic acid) content of the roots of seedlings transferred to 1 mm 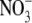 compared with those that were maintained on 50 mm
compared with those that were maintained on 50 mm 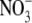 (Fig. 2). A small decrease seen in shoot IAA content was not statistically significant. In soybean it has similarly been found that plants grown on 8 mm
(Fig. 2). A small decrease seen in shoot IAA content was not statistically significant. In soybean it has similarly been found that plants grown on 8 mm 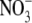 had a four-fold lower concentration of IAA in their roots than those grown on 1 mm
had a four-fold lower concentration of IAA in their roots than those grown on 1 mm 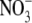 (Caba et al., 2000), suggesting that down-regulation of the root auxin content by high rates of
(Caba et al., 2000), suggesting that down-regulation of the root auxin content by high rates of 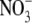 supply may be a widespread phenomenon in plants. The mechanism by which this effect of
supply may be a widespread phenomenon in plants. The mechanism by which this effect of 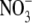 is achieved remains to be established, but could involve, for example, an inhibition of IAA biosynthesis in the shoot or some form of restriction of IAA transport from the shoot to the root.
is achieved remains to be established, but could involve, for example, an inhibition of IAA biosynthesis in the shoot or some form of restriction of IAA transport from the shoot to the root.
Fig. 2.
Effect of short-term changes in the external 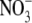 supply on the IAA (indole 3-acetic acid) content of Arabidopsis roots and shoots. Arabidopsis seedlings were germinated and grown at 25 °C on vertical agar plates containing 50 mm KNO3. The composition of the medium and the growth conditions were as previously described (Zhang et al., 1999). After 10 d, when lateral roots had initiated but had arrested at around the time of emergence, half of the seedlings were transferred to fresh plates containing 50 mm
supply on the IAA (indole 3-acetic acid) content of Arabidopsis roots and shoots. Arabidopsis seedlings were germinated and grown at 25 °C on vertical agar plates containing 50 mm KNO3. The composition of the medium and the growth conditions were as previously described (Zhang et al., 1999). After 10 d, when lateral roots had initiated but had arrested at around the time of emergence, half of the seedlings were transferred to fresh plates containing 50 mm 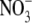 as controls and the other half were transferred to plates containing 1 mm KNO3. In other experiments, lateral root out-growth could be observed during the second day after transfer in seedlings transferred to 1 mm KNO3, but not in controls maintained on 50 mm KNO3 (data not shown). Twenty-four hours after transfer the roots and shoots were harvested separately and their IAA contents determined immunologically using an enzyme-linked immunoadsorbent assay (ELISA) (Veselov et al., 1992). Data are expressed per g fresh weight and are the means obtained from two independent experiments each having two replicates (± standard error; n = 4).
as controls and the other half were transferred to plates containing 1 mm KNO3. In other experiments, lateral root out-growth could be observed during the second day after transfer in seedlings transferred to 1 mm KNO3, but not in controls maintained on 50 mm KNO3 (data not shown). Twenty-four hours after transfer the roots and shoots were harvested separately and their IAA contents determined immunologically using an enzyme-linked immunoadsorbent assay (ELISA) (Veselov et al., 1992). Data are expressed per g fresh weight and are the means obtained from two independent experiments each having two replicates (± standard error; n = 4).
There is also evidence that a second plant hormone, abscisic acid (ABA), may be involved in regulating the systemic effect of endogenous 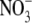 pools on lateral root development. Three ABA-insensitive mutants (abi4-1, abi4-2 and abi5-1) were insensitive to the inhibitory effect of
pools on lateral root development. Three ABA-insensitive mutants (abi4-1, abi4-2 and abi5-1) were insensitive to the inhibitory effect of 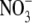 and four ABA synthesis mutants (aba1-1, aba2-3, aba2-4 and aba3-2) showed reduced sensitivity (Signora et al., 2001). To explain these data the authors suggested that there are two regulatory pathways mediating the inhibitory effects of
and four ABA synthesis mutants (aba1-1, aba2-3, aba2-4 and aba3-2) showed reduced sensitivity (Signora et al., 2001). To explain these data the authors suggested that there are two regulatory pathways mediating the inhibitory effects of 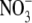 , one that is ABA-dependent and involves ABI4 and ABI5, and another that is ABA-independent. One possibility is that auxin is involved in long-distance signalling from shoot to root, while the ABA-dependent signalling pathway operates within the developing lateral root primordium.
, one that is ABA-dependent and involves ABI4 and ABI5, and another that is ABA-independent. One possibility is that auxin is involved in long-distance signalling from shoot to root, while the ABA-dependent signalling pathway operates within the developing lateral root primordium.
EFFECT OF EXTERNAL l-GLUTAMATE ON PRIMARY ROOT GROWTH AND ROOT BRANCHING
While investigating the effect of different N sources on plant development we observed that primary root growth in aseptically grown seedlings of Arabidopsis was markedly inhibited in the presence of even low concentrations of l-glutamate (0.05–0.5 mm) (Filleur et al., 2005; P.W.-L. et al., submitted for publication). This response was highly specific for l-glutamate as similar concentrations of the related amino acids aspartate, glutamine, γ-aminobutyric acid (GABA) and d-glutamate had no effect. The only other amino acid to affect root growth was tryptophan, which at 50 μm inhibited primary root growth by about 25 % (P. Walch-Liu and B. G. Forde, unpublished results). Tryptophan is a precursor of IAA (Muller et al., 1998) and its overall effects on root architecture (including stimulation of lateral root initiation) differed from those of l-glutamate but resembled those classically seen when Arabidopsis roots are treated with IAA (Evans et al., 1994; Celenza et al., 1995). Although these experiments were performed in the presence of 0.5 mm glutamine as background N source, l-glutamate had the same effect in the absence of any other form of N.
The effects of l-glutamate on root architecture are complex because, although inhibition of primary root growth and reduction in mitotic activity are detectable within 24 h of transfer to l-glutamate, lateral roots only acquire sensitivity to l-glutamate some time after emergence. It appears that l-glutamate sensitivity in lateral roots is developmentally regulated, with the result that they continue growth on glutamate until they reach a fairly uniform average length of 5–7 mm. A third effect, which is probably a consequence of the inhibition of primary root growth, is that lateral root outgrowth behind the primary root tip is stimulated. The net result is that l-glutamate-treated seedlings have a shorter, more branched root system not dissimilar to the phenotype seen when Arabidopsis seedlings were grown on a limiting supply of P (Williamson et al., 2001).
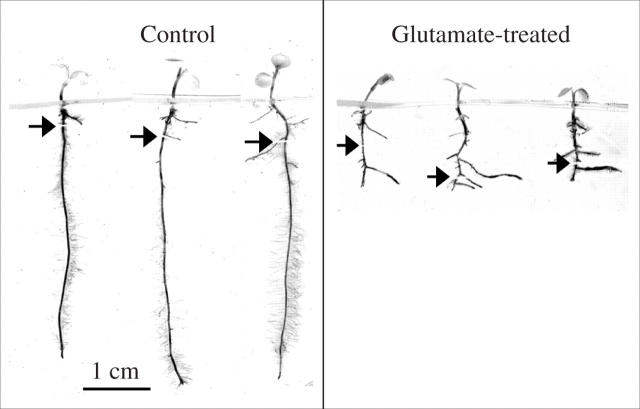
The effect of l-glutamate on Arabidopsis root growth was unexpected, but it is a phenomenon not restricted to this genus. Figure 3 shows the effect of l-glutamate on seedlings of Thlaspi caerulescens, a metal hyperacumulator related to Arabidopsis, illustrating how the l-glutamate treatment dramatically alters root architecture in a similar way to that seen in Arabidopsis (Filleur et al., 2005). Other taxa found to be sensitive to 1 mm glutamate were tomato, Icelandic poppy and another Arabidopsis relative, Thellungiella halophila (P. Walch-Liu and B. G. Forde, unpublished results). However, a survey of 19 different ecotypes of Arabidopsis has shown that within a single species a high degree of natural variation in l-glutamate sensitivity can exist. Among these ecotypes, from diverse geographical locations and habitats, the inhibition of primary root growth by 50 μm l-glutamate, measured over a 6-d period of treatment, varied from 0 to 80 % (P.W.-L. et al., submitted for publication). Nevertheless, even the least sensitive ecotype (RLD1) was sensitive to 1 mm l-glutamate, being inhibited by 26 %.
Fig. 3.
Effect of l-glutamate on root architecture in Thlaspi caerulescens. Seed of T. caerulescens was surface-sterilized with Na hypochlorite and germinated on vertical agar plates as previously described for Arabidopsis (Zhang et al., 1999) except that Phytagel™ (1 %, w/v) was used instead of agar and the medium contained 1 mm MgCl2 and 1 mm CaCl2 to aid solidification. The background N source was 0.5 mm glutamine. The temperature was 18–23 °C and the photon irradiance was approx. 120 μmol m2 s−1 with a 16 : 8-h light–dark regime. After 6 d growth, and before lateral roots had appeared, the seedlings were transferred to fresh plates containing the same medium plus either 1 mm KCl (control) or 1 mm potassium l-glutamate (treated) and incubated for a further 9 d before image capture. Arrows indicate the locations of the primary root tips at the time of transfer.
Arabidopsis roots have a high-affinity uptake system for l-glutamate which has a Km for l-glutamate of 14–15 μm (W. Koch and W. Frommer, personal communication). However, it seems unlikely that the effect of l-glutamate on root growth arises from its contribution to N metabolism. We have found that 50 μm l-glutamate has the same inhibitory effect on root growth whether it is applied with or without a ten-fold excess of glutamine, which is supplied as a N source. Furthermore, if 1 mm l-glutamate is supplied to the root system while avoiding contact with the primary root tip, primary root growth is unaffected. By contrast, if 50 μm l-glutamate is applied only to the root tip it has the same effect on primary root growth as 50 μm l-glutamate applied to the whole root system. We conclude that l-glutamate is detected specifically at the root tip and that the localized nature of this effect is most consistent with the l-glutamate signal being sensed extracellularly, presumably in the apoplast close to the root apex.
What is the mechanism of l-glutamate sensing? A study by Sivaguru and colleagues found that millimolar concentrations of l-glutamate inhibited Arabidopsis root growth and replicated the effects of Al3+ in rapidly depolymerizing cortical microtubules and depolarizing the plasma membrane (Sivaguru et al., 2003). Because this effect could be blocked with AP-5, an antagonist of mammalian ionotropic glutamate receptors (iGluRs), the authors concluded that plant homologues of these iGluRs may be involved in sensing the glutamate signal. The existence in plants of genes encoding homologues of mammalian iGluRs was first reported by Lam et al. (1998). It has since been established that Arabidopsis has 20 genes (AtGLR genes) belonging to the family of glutamate receptor-like proteins (Lacombe et al., 2001; Chiu et al., 2002). However, although there is physiological evidence for the existence of glutamate-gated cation channels in roots (Dennison and Spalding, 2000; Dubos et al., 2003; Demidchik et al., 2004), the ion transport properties of the AtGLR gene products and the identity of their ligand(s) are still unclear (Davenport, 2002; Demidchik et al., 2004).
We were unable to confirm the ability of AP-5 or other antagonists of mammalian iGluRs (DNQX or MK801) to block the inhibitory effect of l-glutamate on root growth. However, the phenomenon we observe appears to differ from that studied by Sivaguru and colleagues. They obtained a 60 % inhibition of root growth within 2.5 min of l-glutamate treatment, indicating rapid effect on cell elongation. By contrast, we find that the onset of inhibition of root growth is delayed, with only a 20 % decline measurable over the first 24 h, and that a decrease in mitotic activity, not cell elongation, accounts for this decline. Therefore, there may be a rapid, transient effect of l-glutamate on cell elongation that is mechanistically distinct from the longer term effect we observe. Nevertheless, the AtGLR genes at present are the likeliest candidates for a role in glutamate sensing. Expression studies have shown that 15 of the 20 AtGLR genes are expressed most strongly in roots, and five of these are root-specific (Chiu et al., 2002). We are currently screening T-DNA insertion mutants in individual members of this gene family to establish whether any are altered in the l-glutamate sensitivity of their root growth.
Our observations have led us to speculate on the possible physiological and ecological significance of the l-glutamate effect. Amino acids are present in soils as the largest component of the low-molecular-weight fraction of dissolved organic N (DON) (Lipson et al., 2001; Jones et al., 2005a, b). Traditionally it has been thought that microbial competition for this highly labile pool of organic N would be too intense for it to represent a significant source of N for plants. However, this view is now changing and there is strong evidence to suggest that both mycorrhizal and non-mycorrhizal plants in a variety of ecosystems directly absorb amino acids from the soil (Lipson and Nasholm, 2001; Neff et al., 2003). It has been pointed out that plants that were able to access the organic N pool directly would be able to release themselves from reliance on microbial mineralization to produce inorganic N, which is generally considered to be a bottleneck in the N cycle in soils (Neff et al., 2003). On this basis, there would have been strong selective pressure for plants to acquire mechanisms that enhanced their ability to compete with other plants and micro-organisms for organic N. One physiological adaptation that could have emerged as a consequence of this selective pressure is the range of high-affinity amino acid uptake systems with differing substrate specificities that exists in the roots of many plant species (Fischer et al., 1998; Lipson and Nasholm, 2001).
We propose that changes in root architecture in response to the presence of significant accumulations of l-glutamate in the soil may represent a second (morphological) adaptation that enhances a plant's ability to compete for organic N. It has been suggested that plants are most likely to be able to compete effectively with micro-organisms for soil amino acids within organic N-rich soil patches where the concentrations of these amino acids are highest (Raab et al., 1996; Jones et al., 2005b). The slowing of primary root growth, the increased root branching behind the root tip and the developmentally delayed inhibition of lateral root elongation, which are the responses observed when an Arabidopsis root system encounters a source of l-glutamate, can be seen as a potential foraging mechanism because they would serve to increase the precision of root placement within the soil (Sutherland and Stillman, 1988). Although the concentrations of l-glutamate normally found in the bulk soil solution may be too low to affect root growth (Jones et al., 2005b), within regions of decomposing organic matter its concentration can be expected to frequently exceed that needed to elicit a growth response in roots of sensitive genotypes. Plant and animal tissues contain free glutamate at millimolar concentrations (Joy et al., 1992; Young and Ajami, 2000) and an even larger pool of glutamate is available for proteolytic release in the protein fraction (Tapiero et al., 2002).
Glutamate appears to be widely used by both unicellular and multicellular organisms as a chemoattractant and foraging signal. It has previously been identified as an important cue for foraging behaviour in organisms as diverse as bacteria (Brown and Berg, 1974), protozoa (Van Houten et al., 2000), cnidarians (Bellis et al., 1991) and crustaceans (Trott et al., 1997). Future studies should be directed towards examining the relationship between the ability of a plant species or genotype to modify its root architecture in response to l-glutamate signals and its ability to compete for soil organic N, particularly when the organic N supply is spatially heterogeneous.
Acknowledgments
We thank Professor Volker Römheld (University of Hohenheim, Germany) for the gift of T. caerulescens seed. This work was supported by grants from the Biotechnology and Biological Sciences Research Council of the UK and Yara International ASA and by the Research Training Network PLUSN in the framework of the Human Potential Programme of the European Commission (contract no. HPRN-CT-2002-00247). I.I.I. was the recipient of an International Incoming Short Visit grant from the Royal Society of London.
LITERATURE CITED
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, et al. 2000. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant Journal 24: 457–466. [DOI] [PubMed] [Google Scholar]
- Bellis SL, Grosvenor W, Kasssimon G, Rhoads DE. 1991. Chemoreception in Hydra vulgaris (attenuata): initial characterization of two distinct binding sites for L-glutamic acid. Biochimica et Biophysica Acta 1061: 89–94. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant Journal 29: 325–332. [DOI] [PubMed] [Google Scholar]
- Bloom AJ, Meyerhoff PA, Taylor AR, Rost TL. 2002. Root development and absorption of ammonium and nitrate from the rhizosphere. Journal of Plant Growth Regulation 21: 416–431. [Google Scholar]
- Brown DA, Berg HC. 1974. Temporal stimulation of chemotaxis in Escherichia coli. Proceedings of the National Academy of Sciences of the USA 71: 1388–1392. [DOI] [PMC free article] [PubMed]
- Burgeff C, Liljegren SJ, Tapia-Lopez R, Yanofsky MF, Alvarez-Buylla ER. 2002. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214: 365–372. [DOI] [PubMed] [Google Scholar]
- Caba JM, Centeno ML, Fernandez B, Gresshoff PM, Ligero F. 2000. Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211: 98–104. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR. 1995. A pathway for lateral root formation in Arabidopsis thaliana. Genes and Development 9: 2131–2142. [DOI] [PubMed] [Google Scholar]
- Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, Coruzzi GM. 2002. Phylogenetic and expression analysis of the glutamate receptor-like gene family in Arabidopsis thaliana. Molecular Biology and Evolution 19: 1066–1082. [DOI] [PubMed] [Google Scholar]
- Davenport R. 2002. Glutamate receptors in plants. Annals of Botany 90: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Essah PA, Tester M. 2004. Glutamate activates cation currents in the plasma membrane of Arabidopsis root cells. Planta 219: 167–175. [DOI] [PubMed] [Google Scholar]
- Dennison KL, Spalding EP. 2000. Glutamate-gated calcium fluxes in Arabidopsis. Plant Physiology 124: 1511–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Saker LR. 1975. Nutrient supply and the growth of the seminal root system of barley. II. Localized, compensatory increases in lateral root growth and rates of nitrate uptake when nitrate supply is restricted to only part of the root system. Journal of Experimental Botany 26: 79–90. [Google Scholar]
- Drew MC, Saker LR, Ashley TW. 1973. Nutrient supply and the growth of the seminal root system in barley. I. The effect of nitrate concentration on the growth of axes and laterals. Journal of Experimental Botany 24: 1189–1202. [Google Scholar]
- Dubos C, Huggins D, Grant GH, Knight MR, Campbell MM. 2003. A role for glycine in the gating of plant NMDA-like receptors. Plant Journal 35: 800–810. [DOI] [PubMed] [Google Scholar]
- Evans ML, Ishikawa H, Estelle MA. 1994. Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild-type and auxin-response mutants. Planta 194: 215–222. [Google Scholar]
- Filleur S, Walch-Liu P, Gan Y, Forde BG. 2005. Nitrate and glutamate sensing by plant roots. Biochemical Transactions 33: 283–286. [DOI] [PubMed] [Google Scholar]
- Fischer WN, Andre B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, et al. 1998. Amino acid transport in plants. Trends in Plant Science 3: 188–195. [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, Parenicova L, Henz SR, Weigel D, et al. 2005. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. 2002. Local and long-range signalling pathways regulating plant responses to nitrate. Annual Review of Plant Biology 53: 203–224. [DOI] [PubMed] [Google Scholar]
- Forde BG, Lorenzo H. 2001. The nutritional control of root development. Plant and Soil 232: 51–68. [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG. 2005. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742. [DOI] [PubMed] [Google Scholar]
- Haines BJ. 2002. Zincophilic root foraging in Thlaspi caerulescens. New Phytologist 155: 363–372. [DOI] [PubMed] [Google Scholar]
- Hodge A. 2004. The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytologist 162: 9–24. [Google Scholar]
- Hutchings MJ, de Kroon H. 1994. Foraging in plants: the role of morphological plasticity in resource acquisition. Advances in Ecological Research 25: 159–238. [Google Scholar]
- Jack T. 2004. Molecular and genetic mechanisms of floral control. Plant Cell 16: S1–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Healey JR, Willett VB, Farrar JF, Hodge A. 2005a. Dissolved organic nitrogen uptake by plants—an important N uptake pathway? Soil Biology and Biochemistry 37: 413–423.
- Jones DL, Shannon D, Janvee-Fortune T, Farrar JF 2005b. Plant capture of free amino acids is maximized under high soil amino acid concentrations. Soil Biology and Biochemistry 37: 179–181.
- Joy KW, Blackwell RD, Lea PJ. 1992. Assimilation of nitrogen in mutants lacking enzymes of the glutamate synthase cycle. Journal of Experimental Botany 43: 139–145. [Google Scholar]
- Lacombe B, Becker D, Hedrich R, DeSalle R, Hollmann M, Kwak JM, et al. 2001. The identity of plant glutamate receptors. Science 292: 1486–1487. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Chiu J, Hsieh M-H, Meisel L, Oliveira IC, Shin M, et al. 1998. Glutamate receptor genes in plants. Nature 396: 125–126. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant Journal 29: 751–760. [DOI] [PubMed] [Google Scholar]
- Lipson D, Nasholm T. 2001. The unexpected versatility of plants: organic nitrogen use and availability in terrestrial ecosystems. Oecologia 128: 305–316. [DOI] [PubMed] [Google Scholar]
- Lipson DA, Raab TK, Schmidt SK, Monson RK. 2001. An empirical model of amino acid transformations in an alpine soil. Soil Biology and Biochemistry 33: 189–198. [Google Scholar]
- Muller A, Hillebrand H, Weiler EW. 1998. Indole-3-acetic acid is synthesized from L-tryptophan in roots of Arabidopsis thaliana. Planta 206: 362–369. [DOI] [PubMed] [Google Scholar]
- Neff JC, Chapin FS, Vitousek PM. 2003. Breaks in the cycle: dissolved organic nitrogen in terrestrial ecosystems. Frontiers in Ecology and the Environment 1: 205–211. [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, et al. 2003. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Salser SJ, Yamamoto KR. 1988. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell 54: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Porterfield DM. 2002. Environmental sensing and directional growth of plant roots. In: Waisel Y, Eshel A, Kafkafi U, eds. Plant roots: the hidden half. New York: Marcel Dekker, 471–487.
- Raab TK, Lipson DA, Monson RK. 1996. Non-mycorrhizal uptake of amino acids by roots of the alpine sedge Kobresia myosuroides: implications for the alpine nitrogen cycle. Oecologia 108: 488–494. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. 1997. MADS domain proteins in plant development. Biological Chemistry 378: 1079–1101. [PubMed] [Google Scholar]
- Robinson D. 1994. The responses of plants to non-uniform supplies of nutrients. New Phytologist 127: 635–674. [DOI] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. 1995. Diverse roles for MADS-box genes in Arabidopsis development. Plant Cell 7: 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Lauerer M, Schulze ED, Caboche M, Stitt M. 1997. Accumulation of nitrate in the shoot acts as a signal to regulate shoot–root allocation in tobacco. Plant Journal 11: 671–691. [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang HM. 2001. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant Journal 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Pike S, Gassmann W, Baskin TI. 2003. Aluminum rapidly depolymerizes cortical microtubules and depolarizes the plasma membrane: evidence that these responses are mediated by a glutamate receptor. Plant and Cell Physiology 44: 667–675. [DOI] [PubMed] [Google Scholar]
- Stitt M, Feil R. 1999. Lateral root frequency decreases when nitrate accumulates in tobacco transformants with low nitrate reductase activity: consequences for the regulation of biomass partitioning between shoots and root. Plant and Soil 215: 143–153. [Google Scholar]
- Sutherland WJ, Stillman RA. 1988. The foraging tactics of plants. Oikos 52: 239–244. [Google Scholar]
- Tapiero H, Mathe G, Couvreur P, Tew KD. 2002. Dossier: free amino acids in human health and pathologies. II. Glutamine and glutamate. Biomedicine and Pharmacotherapy 56: 446–457. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, et al. 2000. A short history of MADS-box genes in plants. Plant Molecular Biology 42: 115–149. [PubMed]
- Trott TJ, Voigt R, Atema J. 1997. Chemoreception by the red-jointed fiddler crab Uca minax (LeConte): spectral tuning properties of the walking legs. Marine and Freshwater Behaviour and Physiology 30: 239–249. [Google Scholar]
- Van Houten JL, Yang WQ, Bergeron A. 2000. Chemosensory signal transduction in Paramecium. Journal of Nutrition 130: 946S–949S. [DOI] [PubMed] [Google Scholar]
- Veselov SY, Kudoyarova GR, Egutkin NL, Gyulizade VZ, Mustafina AR, Kof EM. 1992. Modified solvent partitioning scheme providing increased specificity and rapidity of immunoassay for indole-3-acetic-acid. Physiologia Plantarum 86: 93–96. [Google Scholar]
- Walch-Liu P, Gan Y, Filleur S, Forde BG. 2005. Nitrogen signalling and the regulation of root development. Aspects of Applied Biology (in press).
- Wang RC, Guegler K, LaBrie ST, Crawford NM. 2000. Genomic analysis of a nutrient response in arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux S, Fitter AH, Leyser HMO. 2001. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiology 126: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young VR, Ajami AM. 2000. Glutamate: an amino acid of particular distinction. Journal of Nutrition 130: 892S–900S. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences of the USA 96: 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]