Abstract
• Background and Aims Studying the genome structure of pines has been hindered by their large genomes and uniform karyotypes. Consequently our understanding of the genome organization and evolutionary changes in different groups of pines is extremely limited. However, techniques are now available that can surmount these difficulties. The purpose of this study was to exploit some of these techniques to characterize the genome differentiation between the two subgenera of Pinus: Pinus and Strobus.
• Methods Double-probe fluorescence in-situ hybridization (FISH) was used to localize the 5S and 18S rDNA loci on chromosomes of five species from the subgenus Strobus: P. bungeana, P. koraiensis, P. armandii, P. wallichiana and P. strobus.
• Key Results The rDNA FISH pattern varied considerably among the five species, with P. bungeana being the most distinct. By comparing the results obtained with those of previous rDNA FISH studies of members of the subgenus Pinus, several general features of rDNA loci distribution in the genus Pinus can be discerned: (a) species of subgenus Strobus generally have more rDNA loci than species of subgenus Pinus, correlating with their larger genomes in the subgenus Strobus; (b) there is a clear differentiation in 5S and 18S rDNA loci linkage patterns between the two subgenera; (c) variations in the rDNA FISH pattern correlate with phylogenetic relationships among species within the subgenus; (d) P. bungeana has fewer 18S rDNA sites than other pines investigated to date, but they give intense signals, and may reflect the primary distribution of the 18S–25S rDNA loci in the genus.
• Conclusions The stable differentiation in rDNA FISH pattern between the subgenera suggests that chromosomal rearrangements played a role in the splitting of the two subgenera, and transpositional events rather than major structural changes are likely responsible for the variable rDNA distribution patterns among species of the same subgenus with conserved karyotypes.
Keywords: 5S and 18S rDNA, FISH, karyotype, Pinus, subgenus Strobus
INTRODUCTION
The genus Pinus comprises more than 100 species, which are widely distributed in the northern hemisphere (Mirov, 1967). The genus is divided into two subgenera: Pinus and Strobus (Little and Critchfield, 1969). Morphological, anatomical and molecular data demonstrate that the two subgenera have considerably diverged (Mirov, 1967; Strauss and Doerksen, 1990; Liston et al., 1999; Wang et al., 1999). Fossil records indicate that the genus split into the two subgenera by the early Cretaceous (Miller, 1977). To date, the genome structure of pines is poorly understood, as is the differentiation between the two subgenera at the genome and chromosome levels.
The genome size of pines has been measured as the amount of DNA in their haploid genomes (C values) (Wakamiya et al., 1993; Hall et al., 2000; Joyner et al., 2001; Grotkopp et al., 2004). Investigations have revealed that members of the subgenus Strobus generally have larger genomes (27·36–37·68 pg C−1) than those in the subgenus Pinus (19·94–35·26 pg C−1). This variation has been difficult to relate to the karyotypic characters of pines in a phylogenetic framework, because pines show remarkable uniformity in karyotype. All the Pinus species have the same chromosome number, 2n = 24, composed of 11 pairs of metacentric chromosomes of similar length and a short heterobrachial chromosome (Pederick, 1970; Saylor, 1972, 1983). Fluorescent banding using the fluorochromes chromomycin A (CMA) and 4′,6-diamidino-2-phenylindole (DAPI) was applied to some Pinus species and was useful for chromosome identification (Hizume et al., 1983, 1989, 1990). However, investigations on the genome organization and structural rearrangements in pines have been limited due to the lack of resolution in the karyotypes. For these applications, in situ hybridization (ISH) or fluorescence in-situ hybridization (FISH) using an rDNA probe has proved to be an excellent tool in pines (Hizume et al., 1992, 2002; Lubaretz et al., 1996; Jacobs et al., 2000; Liu et al., 2003).
To date, the chromosomal localization of 18S–25S rDNA loci has been reported for 15 Pinus species, including P. elliotii (Doudrick et al., 1995), P. sylvestris (Karvonen et al., 1993; Lubaretz et al., 1996; Hizume et al., 2002), P. radiata (Gorman et al., 1992; Jacobs et al., 2000), P. taeda (Jacobs et al., 2000), P. densiflora, P. thunbergii (Hizume et al., 1992; 2002), P. nigra (Hizume et al., 2002), P. tabuliformis, P. yunanensis, P. densata, P. massoniana, P. merkusii (Liu et al., 2003), P. resinosa, P. banksiana and P. strobus (Nkongolo et al., 2004). Among these pines, only P. strobus belongs to the subgenus Strobus, the 14 other pines are all from the subgenus Pinus. Thus, there is a clear need to find out more about the localization of rDNA loci in subgenus Strobus to generalize the patterns of rDNA loci distribution in this subgenus and the differentiation between the two subgenera.
In the present study, the 5S and 18S rDNA loci localization patterns in five species (P. bungeana, P. koraiensis, P. armandii, P. wallichiana and P. strobus) of subgenus Strobus are investigated using DNA probes for the 5S rDNA and 18S rDNA simultaneously to address the following questions: (a) Do the number and location of rDNA loci vary between the two subgenera? (b) Does the difference in rDNA loci number correlate with difference in genome sizes between the two subgenera? (c) Can the variations in FISH patterns be explained within a phylogenetic framework of Pinus?
MATERIALS AND METHODS
Chromosome slide preparation
Five species representing three subsections of subgenus Strobus were selected for this study (Table 1). Seeds of each species were collected from natural stands or artificial plantations and germinated in Petri dishes on moist filter paper at room temperature. The primary root tips, grown to a length of 1–1·5 cm, were excised and pretreated with 0·05 % colchicine for 24 h at 25 °C, fixed in Carnoy's fixative (ethanol : acetic acid, 3 : 1, v/v) for 24 h, and macerated with a mixture of 1 % Y-23 pectolyase (Yakult) and 2 % R-10 cellulase (Yakult) at 37 °C for 45 min. Metaphase chromosome spreads were prepared by conventional squashing.
Table 1.
The five species of subgenus Strobus used in the study
| Species | Subsection* | Source |
|---|---|---|
| Pinus bungeana Zucc. | Gerardianae | Institute of Botany, Chinese Academy of Sciences, Beijing, China |
| Pinus koraiensis Sieb. et Zucc. | Cembrae | Changbai Mountain, Jilin, China |
| Pinus armandii Franch. | Strobi | Shengnongjia, Hubei, China |
| Pinus wallichiana Jackson. | Strobi | Yadong, Tibet, China |
| Pinus strobus L. | Strobi | North Carolina State University, NC, USA |
The taxonomic system of Little and Critchfield (1969) is followed.
Probe preparation
Genomic DNA of P. bungeana was isolated from seed megagametophyte and used as a template for PCR amplification. 18S rDNA sequence was amplified with the primers 5′-CTAGAGCTAATACGTGCAAC-3′ and 5′-GATAAGGTTCAGTGGACTTC-3′ (Troitsky et al., 1991). The PCR programme for 18S rDNA amplification consisted of 2 min at 95 °C for initial denaturation followed by 33 cycles of 30 s at 94 °C, 30 s at 55 °C and 2 min at 72 °C. In the last cycle the extension at 72 °C was extended to 8 min. The primers for 5S rDNA amplification were 5′-CGGTGCATTAATGCTGGTAT-3′ and 5′-CCCATCCGTGTACTACTCTC-3′ (Amarasinghe and Carlson, 1998). The PCR programme for 5S rDNA amplification was the same as for 18S rDNA, except that the annealing temperature was 60 °C. The fragment sizes of the 18S rDNA and 5S rDNA PCR product were approx. 1·8 kb and 600 bp, respectively. The purified rDNA PCR products were labelled by the random priming method using Klenow fragment (Promega). Biotin-16-dUTP (Roche) was used for labelling 5S rDNA and digoxigenin (DIG)-11-dUTP (Roche) for 18S rDNA labelling.
FISH
The chromosomal slides were treated with 100 μL RNase (100 µg mL−1 in 2× SSC) at 37 °C for 1 h and then soaked twice for 5 min in 2× SSC. The chromosomes were then digested with proteinase K (1 µg mL−1, Promega) at 37 °C for 10 min. The slides were dehydrated in a series of 70 %, 95 % and 100 % ethanol and then air-dried. A 20-μL hybridization solution, containing 10 % dextran sulfate, 50 % formamide, 10 ng μL−1 of sheared salmon sperm DNA, 2 ng μL−1 of denatured probes and 2× SSC, was added to each slide. The slides were placed in a moisture chamber, denatured at 85 °C for 13 min, then immediately placed in a hybridization oven and kept at 37 °C overnight for probe hybridization. The slides were then washed with 20 % formamide in 0·1× SSC at 42 °C for 10 min, followed by two washes with 2× SSC at 37 °C for 5 min, and 2× SSC at room temperature for 5 min. In-situ hybridization was repeated on three to five chromosome spreads for each species.
Before detection, the slides were soaked in detection buffer (5 % BSA, 100 mM Tris–HCl pH 7·5, 150 mM NaCl, 0·1 % Triton X-100) at 37 °C for 30 min. The 18S rDNA signals were detected using anti-DIG-rhodamine (2 ng mL−1; Roche) and 5S rDNA signals were detected using avidin–FITC (2 µg mL−1; Sigma, USA). The slides were counter-stained with 100 μL DAPI (2 µg mL−1), and observed under a fluorescence microscope (DMRBE, Leica) at 100 × 10 magnification. The FISH images were captured by a cooled CCD colour chromatic camera (Spot Enhanced; Diagnostic Instruments Inc.).
Chromosome length measurement
The arm lengths of chromosomes in the CCD-captured images of ten cells from each of two or three chromosome spreads of each species were measured. The arm ratio was calculated, and the average values were used to construct an ideogram of the karyotype for each species. The chromosomes were assigned consecutively according to their lengths, with the longest being assigned as I and the shortest as XII. For chromosomes of equal length, the chromosome with the longest short arm was assigned first. As well as the length and arm ratios of the chromosomes, secondary constrictions and FISH patterns are also informative characters for karyotyping. The location of rDNA loci on each chromosome is shown on the ideograms. The mapped positions are not precise quantitative representations, although the relative distances of each rDNA site to the centromere and telomere were considered while constructing the ideogram.
RESULTS
Clear, reproducible, unambiguous FISH signals from three to five FISH replicates of each species were registered. Weak signals in centromeric regions were not analysed. Examples of the 18S and 5S rDNA FISH pattern obtained for each species are shown in Fig. 1 and an ideogram of the karyotypes is presented in Fig. 2. The karyotype features of the five pine species are similar to those reported by Saylor (1983): chromosomes I–XI were found in all cases to be metacentric and to have similar lengths, while chromosome XII was always heterobrachial and short. However, the chromosomal distribution patterns of the 5S and 18S rDNA sites varied considerably among the five species.
Fig. 1.
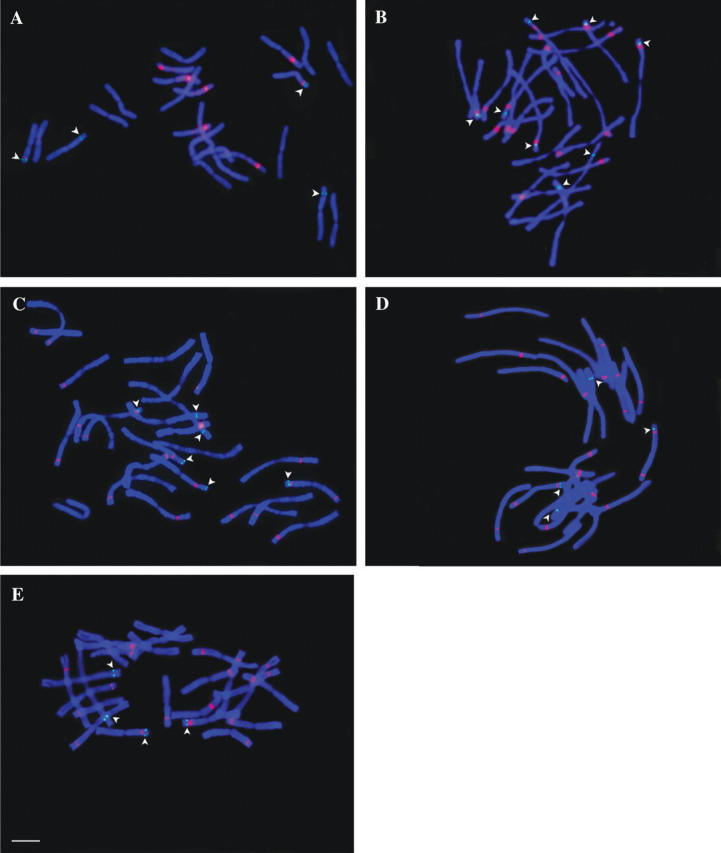
FISH localization of 18S rDNA (red) and 5S rDNA (green) loci on somatic chromosomes in (A) P. bungeana, (B) P. koraiensis, (C) P. armandii, (D) P. wallichiana and (E) P. strobus. Arrowheads indicate the 5S rDNA signals. Scale bar = 10 μm.
Fig. 2.
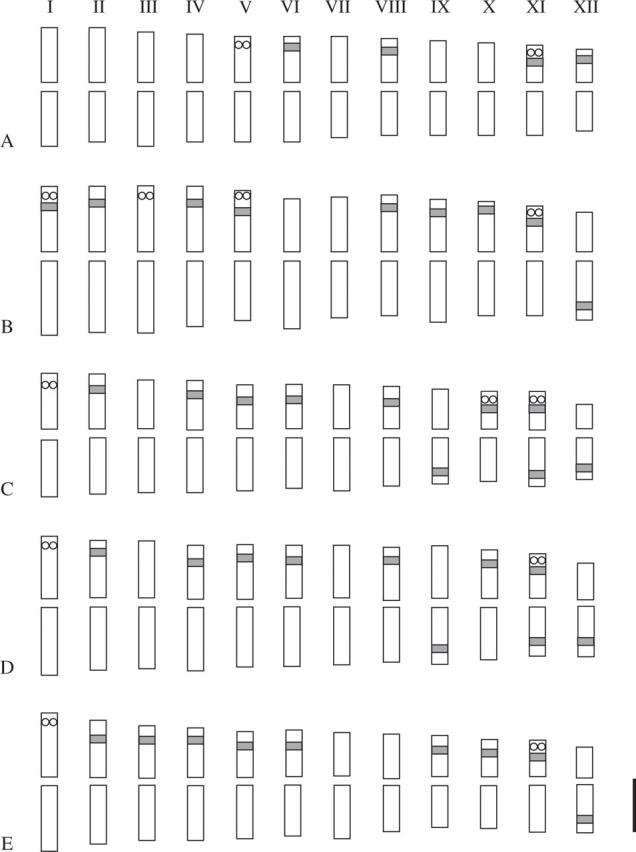
Ideogram of (A) P. bungeana, (B) P. koraiensis, (C) P. armandii, (D) P. wallichiana and (E) P. strobus karyotypes. The open dots represent signals of 5S rDNA and the solid bands represent strong 18S rDNA signals. Scale bar = 10 μm.
In P. bungeana (Figs 1A and 2A), four pairs of intense 18S rDNA site signals were detected in the interstitial regions of the short arms of chromosomes VI, VIII, XI and XII. Two pairs of 5S rDNA sites were detected, one localized on the short arm of chromosome V and the other together with an 18S rDNA site on the short arm of chromosome XI.
In P. koraiensis, nine pairs of 18S rDNA sites were detected on nine pairs of chromosomes, eight on the interstitial regions of the short arms of chromosomes I, II, IV, V, VIII, IX, X and XI and one on the long arm of chromosome XII. Four pairs of 5S rDNA sites were detected: one on the short arm of chromosome III, while the remaining three were localized with 18S rDNA sites on the short arms of chromosomes I, V and XI (Figs 1B and 2B).
In P. armandii, ten pairs of 18S rDNA and three pairs of 5S rDNA sites were detected. Seven pairs of 18S rDNA sites were localized in the interstitial regions of the short arms of chromosomes II, IV, V, VI, VIII, X and XI, while the other three were on the long arms of chromosomes IX, XI and XII. A notable feature was that both arms of chromosome XI hosted an 18S rDNA site. Two 5S rDNA sites were detected close to the 18S rDNA sites on the distal regions of the short arms of chromosomes X and XI, respectively. Another 5S rDNA site was situated alone on the short arm of chromosome I (Figs 1C and 2C).
In P. wallichiana, ten pairs of 18S rDNA and two pairs of 5S rDNA sites were detected. Similar to the rDNA distribution in P. armandii, seven pairs of 18S rDNA were found in the interstitial regions of the short arms of chromosomes II, IV, V, VI, VIII, X and XI and three pairs on the long arms of chromosomes IX, XI and XII. In addition, as in P. armandii, chromosome XI of P. wallichiana also hosted an 18S rDNA site on each of its arms and a 5S rDNA site in the distal region of the short arm. The other 5S rDNA site was found on the short arm of chromosome I (Figs 1D and 2D).
In P. strobus, nine pairs of 18S rDNA and two pairs of 5S rDNA sites were detected. Eight pairs of 18S rDNA sites were found in the interstitial regions of the short arms of chromosomes II, III, IV, V, VI, IX, X and XI and one on the long arm of chromosome XII. One 5S rDNA site was found close to the 18S rDNA site on the short arm of chromosome XI, and the other on chromosome I (Figs 1E and 2E).
In four of the species examined, except for P. bungeana, rDNA sites were detected on ten of the 12 chromosome pairs. Most of the 18S rDNA sites gave clear signals and were positioned in the interstitial regions corresponding to secondary constrictions. No clear, unambiguous 18S signals were observed at the centromere in any case, although weak signals were detected in centromeric regions of two or three pairs of chromosomes in each species. All the 5S rDNA sites were detected at distal positions. Based on the 5S and 18S rDNA distribution patterns and chromosomal morphology, chromosomes XI and XII can be unambiguously identified in all five species. Chromosome XI hosted a 5S and an 18S rDNA site on its short arm in all five species, and a second 18S rDNA site was hosted on the opposite arm of this chromosome in P. wallichiana and P. armandii. The rDNA FISH pattern of P. bungeana differed markedly from that of the other four species, since it contains only four pairs of 18S rDNA sites. The detected signal strength of these sites, however, was noticeably more intense than in the other four pines.
DISCUSSION
Genome research in Pinus has not progressed as rapidly as it has in model plants such as Oriza (Goff et al., 2002; Bowers et al., 2005; Cheng et al., 2005), Arabidopsis (Copenhaver et al., 1998; Arabidopsis Genome Initiative, 2000; Lysak et al., 2003) and Populus (Brunner et al., 2004; Sterky et al., 2004). Consequently, our understanding of the genome organization and chromosomal structure among different groups of pines is extremely limited. Karyotype analyses of Pinus species have revealed that chromosomal morphology is highly conserved within the genus, and that most of the chromosomes have similar lengths and arm ratios, making precise karyotyping difficult (Saylor, 1972, 1983). In the subgenus Strobus, for example, the correct ordering of chromosomes III–VIII has been ambiguous to date (Saylor, 1983). The use of double- or multiple-probe FISH, in combination with other chromosomal characters, has facilitated the identification of homologous chromosomes and the inference of genome evolution among plant species (Brown et al., 1999; Heslop-Harrison, 2000; Marcon et al., 2005; Taketa et al., 2005; Vaio et al., 2005). However, due to the uniformity of the karyotypes among pines, it is still not straightforward to identify precisely all the homologous chromosomes across species, even when the two rDNA markers are used. Among the five species analysed in this study, only chromosomes XI and XII can be aligned with certainty across species. The rDNA distribution patterns of the other ten similar-length chromosomes differed even among species of the same subsection, such as P. armandii, P. wallichiana and P. strobus. This finding is in accordance with the study by Saylor (1983), who found less similarity in karyotypic features among species of the same subsection in subgenus Strobus than in subgenus Pinus. This makes the alignment of homologous chromosomes among pines challenging. Other FISH markers need to be mapped along the chromosomes to facilitate precise identifications (Hizume et al., 2002).
This study reports, for the first time, the physical locations of 5S and 18S rDNA loci in species from the subgenus Strobus. Combined with the results of previous rDNA FISH analyses of members of the subgenus Pinus (Doudrick et al., 1995; Hizume et al., 2002; Liu et al., 2003), several general features of rDNA loci distribution in the genus Pinus can be discerned. The number of 5S and 18S rDNA loci vary from one to four pairs, and from seven to ten pairs, respectively (Table 2), significantly more than the one or two pairs of 5S rDNA loci and one to five pairs of 18S–25S rDNA loci in diploid angiosperms (Castilho and Heslop-Harrison, 1995; Linares et al., 1996; de Melo and Guerra, 2003; Vaio et al., 2005). The difference is, presumably, partly related to the evolutionary divergence between angiosperm and gymnosperm genomes, and partly to the much larger genomes of gymnosperms (Leitch et al., 2005). Pines have large genomes, with estimates of approx. 1010 bp for the haploid genome (Ohri and Khoshoo, 1986; Wakamiya et al., 1993; Murray, 1998; Elsik and Williams, 2000). These huge genomes are largely (approx. 90 %) composed of repetitive DNA, together with many high-copy retro-element families (Kamm et al., 1996; Kossack and Kinlaw, 1999; Elsik and Williams, 2000). 18S–25S rDNA is one of the major components of the repetitive DNA in the genome (Bobola et al., 1992). Thus, it is not surprising to discover a high number of rDNA sites in Pinus.
Table 2.
The number of 18S–25S and 5S rDNA sites in the diploid genome of Pinus
| Species | 18S–25S rDNA | 5S rDNA | Reference |
|---|---|---|---|
| Subgenus Strobus | |||
| Pinus bungeana Zucc. | 8 strong | 4 strong | Present study |
| Pinus koraiensis Sieb. et Zucc. | 18 strong | 8 strong | Present study |
| Pinus armandii Franch. | 20 strong | 6 strong | Present study |
| Pinus wallichiana Jackson | 20 strong | 4 strong | Present study |
| Pinus strobus L. | 18 strong | 4 strong | Present study |
| Pinus strobus L. | 10–14 | – | Nkongolo et al. (2004) |
| Subgenus Pinus | |||
| P. banksiana Lamb. | 10–14 | – | Nkongolo et al. (2004) |
| Pinus elliotii Englem. | 16 strong | 2 strong, 4 weak | Doudrick et al. (1995) |
| Pinus radiata D. Don | 12 strong, 8 weak | 2 strong, 2 weak | Jacobs et al. (2000) |
| Pinus sylvestris L. | >16, 14 strong | 2 strong, 2 weak | Hizume et al. (2002); Karvonen et al. (1993); Lubaretz et al. (1996) |
| Pinus densiflora Sieb. et Zucc. | 14 strong | 2 strong, 2 weak | Hizume et al. (1992, 2002) |
| Pinus thunbergii Parl. | 10 strong, 2 weak | 2 strong, 2 weak | Hizume et al. (1992, 2002) |
| Pinus nigra Arnold | 16 | 2 strong, 2 weak | Hizume et al. (2002) |
| Pinus taeda L. | 12 strong, 8 weak | 2 strong, 2 weak | Jacobs et al. (2000) |
| Pinus tabuliformis Carr. | 14 strong, 10 weak | 2 strong, 2 weak | Liu et al. (2003) |
| Pinus densata Mast. | 10 strong, 8 weak | 2 strong, 1 weak | Liu et al. (2003) |
| Pinus yunnanensis Franch. | 16 strong, 4 weak | 2 strong, 2 weak | Liu et al. (2003) |
| Pinus massoniana Lamb. | 20 strong | 2 strong | Liu et al. (2003) |
| Pinus merkusii Jungh. et de Vriese | 12 strong, 4 weak | 2 strong, 2 weak | Liu et al. (2003) |
Within the genus, there is a tendency for the genomes to be larger in subgenus Strobus than in subgenus Pinus (Hall et al., 2000; Joyner et al., 2001; Grotkopp et al., 2004). Genome size seems to have remained stable or increased from the ancestral genome size over evolutionary time in the subgenus Strobus, while it has decreased in most subsections of the subgenus Pinus (Grotkopp et al., 2004). Interestingly, more rDNA loci were observed in subgenus Strobus than in subgenus Pinus, except for P. bungeana (Table 2). rDNA FISH results can vary among studies, mainly due to differences in experimental procedures including the homology of the probes and the hybridization and washing stringency. The present results were compared with previous results on five species of subgenus Pinus obtained under the same experimental conditions (Liu et al., 2003). The comparison indicates that there are nine or ten pairs of 18S rDNA loci and two to four pairs of 5S rDNA loci in species of subgenus Strobus as compared with five to ten pairs of 18S rDNA loci, excluding the weak signals in centromeric regions, and one or two pairs of 5S rDNA loci in species of subgenus Pinus (Liu et al., 2003) (Table 2). Thus, more chromosomes contained rDNA loci in subgenus Strobus than in subgenus Pinus. Three to five pairs of chromosomes in subgenus Pinus have no rDNA loci, while only two pairs of chromosomes have no rDNA loci in all the species of subgenus Strobus examined except for P. bungeana. Another difference between the two subgenera is that weak 18S–25S rDNA signals have been detected from the pericentromeric regions of most chromosomes in subgenus Pinus (Hizume et al., 2001; Liu et al., 2003), while in subgenus Strobus such signals were detected in only two or three pairs of chromosomes, and nearly all the distinct 18S rDNA signals originated from the interstitial regions. The weak 18S–25S rDNA signals in the centromeric regions reflect the presence of homologous sequences. However, these sequences seem to be nonfunctional (Hizume et al., 2001). It has been suggested that these weak signals might be the remnants of primary sites of 18S–25S rDNA that once existed at the centromeres but were later moved to distal sites by chromosomal rearrangements (Liu et al., 2003). If this is true, the variation in the presence of weak 18S rDNA signals in centromeric regions between the subgenera should reflect, to some degree at least, genome rearrangements accompanying their divergence.
Double-rDNA FISH facilitates visualization of the relative positions of the two classes of rDNA loci on chromosomes. In subgenus Pinus, all the 5S sites are situated on chromosomes that contain one 18S–25S site. The two rDNA sites are either situated on the same arm, with the 5S site in the interstitial region and the 18S–25S site toward the terminal region, or on different arms of the same chromosome (Liu et al., 2003). However, in subgenus Strobus the relative positions of the two rDNA sites examined were reversed, with the 5S site toward the terminal region and the 18S site toward the interstitial region. In addition, one 5S site in subgenus Strobus was observed on a chromosome with no 18S–25S rDNA site. Among the pines that have been examined by double-rDNA FISH, the 5S and 18S–25S rDNA loci linkage pattern seems to be a stable character for each subgenus. Such a stable differentiation in rDNA distribution patterns between the subgenera is more likely caused by chromosome inversion and/or translocation, suggesting that chromosomal rearrangements played a role in the splitting of the two subgenera.
The variations in rDNA distribution have phylogenetic implications, since the closeness of taxa is correlated to the similarity of their rDNA FISH patterns (Hizume et al., 2002; Liu et al., 2003). Among the species of subgenus Strobus examined here, the 18S rDNA site patterns of the two Asian species of subsection Strobi, P. armandii and P. wallichiana, are most similar, while the North American P. strobus, which is in the same subsection, diverges from them less in this respect than P. koraiensis of subsection Cembrae (Fig. 2). These relationships agree well with the rDNA- and chloroplast DNA-based phylogeny (Liston et al., 1999; Wang et al., 1999). The most unique rDNA pattern was found in P. bungeana. It showed the fewest 18S rDNA sites, but the signal strength of these sites was noticeably more intense than those of other pines, indicating that high copy numbers of the 18S–25S rDNA repeats are present within each site. These sites may represent the primary sites of 18S–25S rDNA before some of them moved out and became dispersed among more chromosomal sites in the speciation of pines. Molecular phylogeny supports a close-to-basal position for P. bungeana in subgenus Strobus (Liston et al., 1999; Wang et al., 1999). It would be interesting to examine P. gerardiana, from the same subsection as P. bungeana, and pines of the subsection Balfourianae (which lies at the basal position of the subgenus Strobus) to test the hypothesis that the condensed pattern of 18S–25S rDNA sites found in P. bungeana reflects the primary distribution of these loci in the Pinus genome. Variations in rDNA FISH pattern among angiosperms of the same ploidy level have been attributed to chromosomal rearrangements, transpositional events and gene silencing (Moscone et al., 1999; de Melo and Guerra, 2003; Marcon et al., 2005) The structural similarity among the karyotypes of pines within each subgenus suggest that major chromosomal structural rearrangments (e.g. translocation/inversion) are not frequent among species of the same subgenus. Mechanisms such as transposition of rDNA and amplification of cryptic minor rDNA sites by unequal crossover have been proposed for the dispersed genomic organization of rDNA in angiosperms and fungi (Drouin and de Sá, 1995; Dubcovsky and Dvořák, 1995; Rooney and Ward, 2005). Such mechanisms are likely to be involved in the origin of the variation in the rDNA loci distribution among closely related Pinus species.
Acknowledgments
We thank Dr Bailian Li, North Carolina State University, for seeds of P. strobus, and Dr Alfred E. Szmidt, Kyushu University, for critically reading the manuscript and making valuable comments on it. This study was supported by grants from the Natural Science Foundation of China (NSFC 30325006, 30121003).
LITERATURE CITED
- Amarasinghe V, Carlson JE. 1998. Physical mapping and characterization of 5S rRNA genes in Douglas-fir. Journal of Heredity 89: 495–500. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bobola MS, Smith DE, Klein AS. 1992. Five major nuclear ribosomal repeats represent a large and variable fraction of the genomic DNA of Picea rubens and P. mariana. Molecular Biology and Evolution 9: 125–137. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Arias MA, Asher R, Avise JA, Ball RT, Brewer GA, et al.2005. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proceedings of the National Academy of Sciences of the USA 102: 13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SE, Stephens JL, Lapitan NL, Knudson DL. 1999. FISH landmarks for barley chromosomes (Hordeum vulgare L.). Genome 42: 274–281. [PubMed] [Google Scholar]
- Brunner AM, Busov VB, Strauss SH. 2004. Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends in Plant Science 9: 49–56. [DOI] [PubMed] [Google Scholar]
- Castilho A, Heslop-Harrison JS. 1995. Physical mapping of 5S and 18S-25S rDNA and repetitive DNA sequences in Aegilops umbellulata. Genome 38: 91–96. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Chung M-C, Liu S-M, Chen S-K, Kao F-Y, Lin S-J, et al.2005. A fine physical map of the rice chromosome 5. Molecular Genetics and Genomics 274: 337–345. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Browne WE, Preuss D. 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proceedings of the National Academy of Sciences of the USA 95: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudrick RL, Heslop-Harrison JS, Nelson CD, Schmidt T, Nance WL, Schwarzacher T. 1995. Karyotype of slash pine (Pinus elliottii var. elliottii) using patterns of fluorescence in situ hybridzation and fluorochrome banding. Journal of Heredity 86: 289–296. [Google Scholar]
- Drouin G, de Sá MM. 1995. The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Molecular Biology and Evolution 12: 481–493. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvořák J. 1995. Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Williams CG. 2000. Retroelements contribute to the excess low-copy-number DNA in pine. Molecular Genetics and Genomics 264: 47–55. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, et al.2002. A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Gorman SW, Teasdale RD, Cullis CA. 1992. Structure and organiztion of the 5S rRNA genes (5S DNA) in Pinus radiata (Pinaceae). Plant Systematics and Evolution 183: 223–234. [Google Scholar]
- Grotkopp E, Rejmanek M, Sanderson MJ, Rost TL. 2004. Evolution of genome size in pines (Pinus) and its life-history correlates: supertree analyses. Evolution 58: 1705–1729. [DOI] [PubMed] [Google Scholar]
- Hall SE, Dvorak WS, Johnston JS, Price HJ, Williams CG. 2000. Flow cytometric analysis of DNA content for tropical and temperate New World pines. Annals of Botany 86: 1081–1086. [Google Scholar]
- Heslop-Harrison JS. 2000. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12: 617–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizume M, Ohgiku A, Tanaka A. 1983. Chromosome banding in the genus Pinus. I. Identification of chromosomes in P. nigra by fluorescent banding method. Botanical Magazine 96: 273–276. [Google Scholar]
- Hizume M, Ohgiku A, Tanaka A. 1989. Chromosome banding in the genus Pinus. II. Interspecific variation of fluorescent banding patterns in P. densiflora and P. thunbergii. Botanical Magazine 102: 25–36. [Google Scholar]
- Hizume M, Arai M, Tanaka A. 1990. Chromosome banding in the genus Pinus. III. Fluorescent banding pattern of P. luchuensis and its relationships among the Japanese diploxylon pines. Botanical Magazine 103: 103–111. [Google Scholar]
- Hizume M, Ishida F, Murata M. 1992. Multiple locations of the rRNA genes in chromosomes of pines, Pinus densiflora and P. thunbergii. Japanese Journal of Genetics 67: 389–396. [Google Scholar]
- Hizume M, Shibata F, Maruyama Y, Kondo T. 2001. Cloning of DNA sequences localized on proximal fluorescent chromosome bands by microdissection in Pinus densiflora Sieb. & Zucc. Chromosoma 110: 345–351. [DOI] [PubMed] [Google Scholar]
- Hizume M, Shibata F, Matsusaki Y, Garajova Z. 2002. Chromosome identification and comparative karyotypic analyses of four Pinus species. Theoretical and Applied Genetics 105: 491–497. [DOI] [PubMed] [Google Scholar]
- Jacobs MD, Gardner RC, Murray BG. 2000. Cytological characterization of heterochromatin and rDNA in Pinus radiata and P. taeda. Plant Systematics and Evolution 223: 71–79. [Google Scholar]
- Joyner KL, Wang X-R, Johnston JS, Price HJ, Williams CG. 2001. DNA content for Asian pines parallels New World relatives. Canadian Journal of Botany 79: 192–196. [Google Scholar]
- Kamm A, Doudrick RL, Heslop-Harrison JS, Schmidt T. 1996. The genomic and physical organization of Ty1-copia-like sequences as a component of large genomes in Pinus elliottii var. elliottii and other gymnosperms. Proceedings of the National Academy of Sciences of the USA 93: 2708–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvonen P, Karjalainen M, Savolainen O. 1993. Ribosomal RNA genes in Scots pines (Pinus sylvestris L.): chromosomal organization and structure. Genetica 88: 59–68. [Google Scholar]
- Kossack DS, Kinlaw CS. 1999. IFG, a gypsy-like retrotransposon in Pinus (Pinaceae), has an extensive history in pines. Plant Molecular Biology 39: 417–426. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. 2005. Evolution of DNA amounts across land plants (Embryophyta). Annals of Botany 95: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares C, Gonzalez J, Ferrer E, Fominaya A. 1996. The use of double fluorescence in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S-5·8S-26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome 39: 535–542. [DOI] [PubMed] [Google Scholar]
- Liston A, Robinson WA, Pinero D, Alvarezbuylla ER. 1999. Phylogenetics of Pinus (Pinaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Molecular Phylogenetics and Evolution 11: 95–109. [DOI] [PubMed] [Google Scholar]
- Little Jr EL, Critchfield WB. 1969. Subdivisions of the genus Pinus (pines). USDA Forest Service, Washington D.C. Miscellaneous Publication No. 1144.
- Liu Z-L, Zhang D, Hong D-Y, Wang X-R. 2003. Chromosomal localization of 5S and 18S-5·8S-25S ribosomal DNA sites in five Asian Pinus species using fluorescence in situ hybridization. Theoretical and Applied Genetics 106: 198–204. [DOI] [PubMed] [Google Scholar]
- Lubaretz O, Fuchs J, Ahne R, Meister A, Schubert I. 1996. Karyotyping of three Pinaceae species via fluorescent in situ hybridization and computer-aided chromosome analysis. Theoretical and Applied Genetics 92: 411–416. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Pecinka A, Schubert I. 2003. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Research 11: 195–204. [DOI] [PubMed] [Google Scholar]
- Marcon AB, Barros ICL, Guerra M. 2005. Variation in chromosome numbers, CMA bands and 45S rDNA sites in species of Selaginella (Pteridophyta). Annals of Botany 95: 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo NF, Guerra M. 2003. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Annals of Botany 92: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CN. 1977. Mesozoic conifers. Botanical Review 43: 217–280. [Google Scholar]
- Mirov NT. 1967. The genus Pinus. New York: Ronald Press.
- Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D. 1999. Quantitative karyotyping and dual-color FISH mapping of 5S and 18S-25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42: 1224–1233. [DOI] [PubMed] [Google Scholar]
- Murray BG. 1998. Nuclear DNA amounts in gymnosperms. Annals of Botany 82 (Suppl. A): 3–15. [Google Scholar]
- Nkongolo KK, Kim NS, Michae P. 2004. Detection and physical mapping of the 18S-5·8S-26S rDNA and the pKFJ660 probe with microsatellite sequences derived from the rice blast fungus (Magnaporthe grisea) in conifer species. Hereditas 140: 70–78. [DOI] [PubMed] [Google Scholar]
- Ohri D, Khoshoo TN. 1986. Genome size in gymnosperms. Plant Systematics and Evolution 153: 119–132. [Google Scholar]
- Pederick LA. 1970. Chromosome relationships between Pinus species. Silvae Genetica 19: 171–180. [Google Scholar]
- Rooney AP, Ward TJ. 2005. Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proceedings of the National Academy of Sciences of the USA 102: 5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylor LC. 1972. Karyotype analysis of the genus Pinus—subgenus Pinus. Silvae Genetica 21: 155–163. [Google Scholar]
- Saylor LC. 1983. Karyotype analysis of the Genus Pinus—subgenus Strobus. Silvae Genetica 32: 119–124. [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, et al.2004. A Populus EST resource for plant functional genomics. Proceedings of the National Academy of Sciences of the USA 101: 13951–13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss SH, Doerksen AH. 1990. Restriction fragment analysis of pine phylogeny. Evolution 44: 1081–1096. [DOI] [PubMed] [Google Scholar]
- Taketa S, Ando H, Takeda K, Ichii M, von Bothmer R. 2005. Ancestry of American polyploid Hordeum species with the I genome inferred from 5S and 18S-25S rDNA. Annals of Botany 96: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troitsky AV, Melekhovets Y, Rakhimova GM, Bobrava VK, Valiejo-Roman KM, Autonov AS. 1991. Angiosperm origin and early stages of seed plant evolution deduced from rRNA sequence comparisons. Journal of Molecular Evolution 32: 253–261. [DOI] [PubMed] [Google Scholar]
- Vaio M, Speranza P, Valls JF, Guerra M, Mazzella C. 2005. Localization of the 5S and 45S rDNA sites and cpDNA sequence analysis in species of the Quadrifaria group of Paspalum (Poaceae, Paniceae). Annals of Botany 96: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamiya I, Newton RJ, Johnston JS, Price HJ. 1993. Genome size and environmental factors in the genus Pinus. American Journal of Botany 80: 1235–1241. [Google Scholar]
- Wang X-R, Tsumura Y, Yoshimaru H, Nagasaka K, Szmidt AE. 1999. Phylogenetic relationships of Eurasian pines (Pinus, Pinaceae) based on chloroplast rbcl, matK, rpl20-rps18 spacer, and trnV intron sequences. American Journal of Botany 86: 1742–1753. [PubMed] [Google Scholar]


