Abstract
• Background and Aims Dehydrins, or group 2 late embryogenic abundant proteins (LEA), are hydrophilic Gly-rich proteins that are induced in vegetative tissues in response to dehydration, elevated salt, and low temperature, in addition to being expressed during the late stages of seed maturation. With the aim of characterizing and studying genes involved in osmotic stress tolerance in coffee, several full-length cDNA-encoding dehydrins (CcDH1, CcDH2 and CcDH3) and an LEA protein (CcLEA1) from Coffea canephora (robusta) were isolated and characterized.
• Methods The protein sequences deduced from the full-length cDNA were analysed to classify each dehydrin/LEA gene product and RT–PCR was used to determine the expression pattern of all four genes during pericarp and grain development, and in several other tissues of C. arabica and C. canephora. Primer-assisted genome walking was used to isolate the promoter region of the grain specific dehydrin gene (CcDH2).
• Key Results The CcDH1 and CcDH2 genes encode Y3SK2 dehydrins and the CcDH3 gene encodes an SK3 dehydrin. CcDH1 and CcDH2 are expressed during the final stages of arabica and robusta grain development, but only the CcDH1 transcripts are clearly detected in other tissues such as pericarp, leaves and flowers. CcDH3 transcripts are also found in developing arabica and robusta grain, in addition to being detected in pericarp, stem, leaves and flowers. CcLEA1 transcripts were only detected during a brief period of grain development. Finally, over 1 kb of genomic sequence potentially encoding the entire grain-specific promoter region of the CcDH2 gene was isolated and characterized.
• Conclusions cDNA sequences for three dehydrins and one LEA protein have been obtained and the expression of the associated genes has been determined in various tissues of arabica and robusta coffees. Because induction of dehydrin gene expression is associated with osmotic stress in other plants, the dehydrin sequences presented here will facilitate future studies on the induction and control of the osmotic stress response in coffee. The unique expression pattern observed for CcLEA1, and the expression of a related gene in other plants, suggests that this gene may play an important role in the development of grain endosperm tissue. Genomic DNA containing the grain-specific CcDH2 promoter region has been cloned. Sequence analysis indicates that this promoter contains several putative regulatory sites implicated in the control of both seed- and osmotic stress-specific gene expression. Thus, the CcDH2 promoter is likely to be a useful tool for basic studies on the control of gene expression during both grain maturation and osmotic stress in coffee.
Keywords: Dehydrins, late embryogenic abundant protein (LEA), seed development, Coffea, C. canephora, C. arabica, Rubiaceae
INTRODUCTION
A group of proteins, called the late embryogenesis abundant proteins (LEA), have been shown to accumulate in a co-ordinated fashion during the latter stages of cotton seed development (Dure et al., 1981). Dehydrin proteins (DHN) are a sub-group of the LEA proteins that have also been called the ‘LEA D-11 family’ or LEA type 2 proteins (Dure, 1993; Close, 1996; Ingram and Bartels, 1996). Expression of the DHN proteins have been associated with the protection of various types of plant cells from osmotic stresses, such as those caused by desiccation, salt, and low temperatures (Skriver and Mundy, 1990; Close, 1996; Ingram and Bartels, 1996; Allagulova et al., 2003).
Over the last few years, direct experimental evidence linking higher expression of dehydrins and protection from osmotic stress has begun to appear in the literature. For example, arabidopsis plants engineered to overexpress a dehydrin fusion protein were found to have improved survival to low temperature exposures (Puhakainen et al., 2004). Similarly, expression of a citrus dehydrin protein in transgenic tobacco has been shown to give increased tolerance to low temperatures (Hara et al., 2003). Other supporting evidence for the linkage of dehydrins and tolerance to low temperature-induced stress are the observations that QTL (quantitative trait loci) for freezing tolerance and winter-hardiness map very closely to dehydrins (Close, 1996; Zhu et al., 2000). DHN genes are also expressed significantly in seeds towards the end of maturation, a period when the seed undergoes a developmentally programmed reduction in water content (Choi and Close, 2000; Nylander et al., 2001). The LEA/dehydrin proteins have been estimated to comprise up to 4 % of the total seed protein, and are thought to be involved in protecting the embryo and/or other seed tissues from the osmotic stresses associated with the low water content of mature seed (Roberts et al., 1993; Wise and Tunnacliffe, 2004).
Although a considerable number of dehydrin proteins have been isolated and studied, the precise physiochemical and/or structural mechanism(s) whereby these proteins function to protect cells from osmotic stress in vivo is unknown. The dehydrins are very hydrophilic proteins and exhibit an unusually low level of recognizable structure (Close, 1996; Soulages et al., 2003). A key characteristic of the dehydrins is the presence of one or more lysine-rich stretches of 15 amino acids, called the K motifs, that are predicted to form class A amphipathic alpha-helices (Dure, 1993; Close, 1996, 1997). Dehydrins can also contain two other motifs, an N-terminal Y segment (consensus V/TDE/QYGNP) and a serine-rich S segment which can be phosphorylated and is thought to participate in nuclear localization (Godoy et al., 1994; Close, 1997). It has been proposed that the short amphipathic K segments of dehydrin polypeptides interact with solvent-exposed hydrophobic patches on proteins undergoing partial denaturation, and thereby interfere with protein aggregate formation (Close, 1996). Amphipathic K helixes could also be involved in binding membrane lipids and thus could play a more specific role in protecting lipoproteins, proteins located in membranes, and/or the membrane structure itself (Close, 1996; Koag et al., 2003). An alternative proposal for at least part of the protective effect of dehydrins is the ability of these very stable, but relatively unstructured proteins to tightly bind and organize water molecules (Soulages et al., 2003). This latter effect could help slow the water loss from cells under dehydration conditions, and also possibly improve the stability of certain macromolecules by the development of dehydrin-based regions of more tightly bound ‘ordered’ water around these molecules.
The quality of a coffee beverage is directly related to the biochemical composition and the structural features of the mature grain. However, currently little is known concerning the influence of important grain components like the polysaccharides, proteins and lipids, on coffee quality. One approach to addressing this question is to study the expression of genes encoding these major components, or enzymes involved in their synthesis, and to determine if any correlation can be established between the expression of these genes and specific coffee qualities. Towards this goal, the work presented here describes the isolation and characterization of cDNA encoding a group of proteins that are relatively highly expressed during late grain development, the dehydrins.
The coffee dehydrin cDNAs were identified in the recently established coffee EST (expressed sequence tag) database which contains more than 47 000 ESTs generated from different EST libraries, including several grain-specific and pericarp-specific libraries (Lin et al., 2005). Here, five full length cDNAs isolated from these libraries, representing three distinct dehydrin genes that are strongly expressed in coffee grain, are described. In addition, a cDNA is presented which encodes an LEA transcript that is highly expressed during a very limited phase of grain maturation and may be associated with tissues which undergo programmed cell death. Using RT–PCR, the expression of these coffee genes have been studied in several tissues, and during several stages of development in both the grain and pericarp tissues of the coffee cherry. A genomic fragment encoding the putative promoter region of the coffee dehydrin CcDH2, which is exclusively expressed during grain development, has also been isolated and sequenced. The potential uses of this promoter for future studies of coffee grain development and osmotic stress tolerance, and how the dehydrin proteins could influence coffee flavour generation during the coffee roasting process are discussed.
MATERIALS AND METHODS
Plant materials
Freshly harvested roots, young leaves, stem, flowers and fruit at different stages of development [small green fruit (SG), large green fruit (LG), yellow fruit (Y) and red fruit (R)] were obtained from Coffea arabica ‘Catura T-2308’ grown under greenhouse conditions (25 °C, 70 % RH) in Tours, France. Freshly harvested young leaves were obtained from Coffea canephora ‘BP409’ grown under greenhouse conditions (25 °C, 70 % RH) in Tours, France, and roots, stems, flowers and fruit from Coffea canephora ‘BP409’ were obtained from plants field grown in Indonesia (Indonesian Coffee and Cacao Research Institute, ICCRI). After harvesting, the fresh tissues were frozen immediately in liquid nitrogen until RNA extraction.
RNA extraction and synthesis of cDNA
RNA was extracted from frozen tissues and treated with DNase I as described elsewhere (Simkin et al., 2006). cDNA was then prepared from 4 μg of DNase I-treated RNA using oligo (dT20) according to the protocol of the Superscripttm II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). The absence of contaminating genomic DNA was verified using primers for a ubiquitously expressed coffee chalcone isomerase gene that spanned an intron in this gene (Simkin et al., 2006).
RT–PCR analysis
PCR reactions were carried out using the Coffea arabica and Coffea canephora cDNA samples described above. The primers used are described in Table 1. PCR reactions (50 μL) were set up containing 10 μL of a 1/100-fold dilution of the cDNAs (except for CcDH2 where 10 μL of a 1/1000-fold dilution of the cDNA set was used), 1 μM of each primer, 5 μL of 10× ThermoPol Buffer (New England Biolabs, Beverly, MA, USA), 1 μL of DMSO, 200 μM of dNTPs and 2 units of Taq polymerase (New England Biolabs). The cycling conditions were 2 min at 94 °C, 35 cycles (except for CcDH1 where 40 cycles was used) of 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1.5 min. The final extension step was for 7 min at 72 °C. The RT–PCR products were resolved on 2 % (w/v) agarose gels and stained with ethidium bromide. The CcRPL39 gene encodes the constitutively expressed coffee L39 protein (a 60S ribosomal large subunit protein). The expression of this gene was used as a semi-quantitative control to verify that each RNA sample was transcribed into cDNA at relatively similar efficiencies.
Table 1.
List of primers used for RT–PCR and quantitative RT–PCR experiments
| Oligo name | 5′→3′ Sequence | Oligo name | 5′→3′ Sequence |
|---|---|---|---|
| rpl39_forward | TGGCGAAGAAGCAGAGGCAGA | DH2_reverse | TCAAGCGCACAACAAGGTCC |
| rpl39_reverse | TTGAGGGGGAGGGTAAAAAG | DH3_forward | AGGTGGTGGTCAGAAGAAGAC |
| DH1_forward | GAAGAAGGGGATGAAGGAG | DH3_reverse | GACACACTGGAAAGCTGCTA |
| DH1_reverse | TACGGACAAACACACTACAG | LEA1_forward | CCAATAACAGCTCAAGAATCA |
| DH2_forward | CCTCCAACAACCACCACTG | LEA1_reverse | TTCCCTTCCATCCCACTCT |
Isolation and characterization of the DH2 promoter region
The promoter sequence of CcDH2 was isolated using the method described in the Genome Walker® kit (BD Sciences-Clontech). The method used to isolate the genomic DNA from Coffea canephora ‘BP409’ was described by Crouzillat et al. (1996). The CcDH2 specific forward Genome Walker® primer used was: DH2aprimer1–5′-TGTGCTCCTGATGCTCTCTGTCCTTGTGC-3′. An approx. 2.1-kB fragment was isolated using HindIII-digested Coffea canephora ‘BP409’ genomic DNA ligated to the Genome Walker® adaptor sequence. The PCR was carried out in a 50-μL reaction using the Clontech Advantage® 2 PCR kit as described by the supplier using 0.5-μm final concentrations of DH2aprimer1 and the Genome Walker® AP1 primer. The PCR reaction was carried out using the following conditions: 94 °C for 2 s and 72 °C 3 min (seven cycles), 94 °C for 2 s and 67 °C 3 min (32 cycles), followed by 4 min at 67 °C. The major PCR fragment obtained was then cloned into the plasmid pCR4-TOPO® (Invitrogen). A plasmid (pJMc1) containing the appropriate insert was purified and its insert was completely sequenced. To verify that the inserts in pJMc1 and pcccs30w8a4 were from the same gene, the corresponding overlapping sequence of these two clones was re-amplified from genomic DNA using the primers DH2a geneup 5′-ATAGTGACCTTAATAGCGATCTTGTTGC-3′ and DH2agenelow 5′-CCAAATCAAATCAAACCAAGCAAATC-3′.The PCR reaction was performed with Coffea canephora ‘BP409’ genomic DNA and using Taq (New England Biolabs) as described by the supplier except 1 μm of the specific primers was used (DH2a geneup and DH2a genelow). The PCR conditions were 94 °C for 1 min, then 35 cycles of 94 °C for 1 min, 58 °C for 1.5 min and 72 °C for 3 min, followed by 7 min at 72 °C. The main PCR fragment produced was then cloned into pCR4-TOPO®. The resulting plasmid containing the appropriate insert (pVC1) was purified and its insert was completely sequenced.
Southern blot analysis
Genomic DNA was prepared as described previously (Crouzillat et al., 1996). Five micrograms of genomic DNA from C. canephora BP 409 DNA was digested overnight with the appropriate enzymes (10 U μg−1) according to the supplier's recommendations and the products were separated on a 0.8 % agarose gel. Southern blotting and hybridizations were carried out as described previously (Crouzillat et al., 1996). The probe was generated by first PCR amplifying the insert of the CcDH2 clone cccs30w8a4 with the primers T3 + T7. This PCR product was then labelled with [32P]dCTP using the ‘rediprime™ II random prime labelling system’ kit (Amersham).
RESULTS
Identification and characterization of coffee dehydrin cDNA
An EST database of >47 000 C. canephora EST sequences has recently been generated using RNA isolated from young leaves and from the grain and pericarp tissues of cherries harvested at different stages of development (Lin et al., 2005). An annotated ‘unigene’ set (i.e. contig set) of 13 175 sequences was then obtained by clustering the most highly similar, overlapping ESTs and annotating the unigenes thus generated. The unigenes were screened for dehydrin sequences using various approaches, including a search of the unigene annotations with the keyword ‘dehydrin’ and by using various arabidopsis and tomato dehydrin protein sequences to do a tBlastn search of the coffee unigene set. The various search protocols yielded several candidate dehydrin unigenes, and the longest cDNA clone for each of these unigenes was isolated from the library and completely sequenced.
DNA sequence analysis of the selected full-length cDNA clones indicated that there were essentially four unique sequences representing three different dehydrin genes. The corresponding genes have been named CcDH1, CcDH2 and CcDH3. Two apparently allelic sequences of the gene CcDH1 were identified by sequencing two of the longest cDNAs in unigene #121870. The cDNA clones CcDH1a (pcccl26i7) and CcDH1b (pcccs30w27m8) were 880 and 901 bp long, respectively. The two ORF (open reading frame) sequences exhibited five single base changes, and CcDH1b had an insertion of nine bases, which together resulted in six amino acid changes. CcDH1a and CcDH1b encode proteins of 172 amino acids and 175 amino acids having predicted molecular weights of approx. 17.8 kDa and 18.1 kDa, respectively. Several other sequence differences also exist in the untranslated regions, including a 12-bp deletion in CcDH1a. Two distinct poly (A)-containing unigenes, #123406 and #123405, were found to encode the ORF for CcDH2. Unigene #123405 (pcccs46w30p1; CcDH2b) is composed of two ESTs and differs from the unigene sequence #123406 (pcccs30w8a4; CcDH2a) by the presence of an intron sequence. Interestingly, when the intron/exon borders of the cDNA for DH2b (cccs46w30p1; accession number DQ323990) were examined in detail, it was observed that the 3′ junction had the sequence ttatgg/TCG while other genomic sequences obtained by genome walking (see below) had the sequence ttatag/(T or A)CG. The 761-bp cDNA pcccs30w8a4 (CcDH2a) encodes a protein 162 amino acids long with the predicted molecular weight of 17.4 kDa. Finally, a third coffee gene, CcDH3, was encoded by a single unigene #123385. This gene is represented by the 835-bp insert of plasmid pcccwc22w11a5, which encodes a protein of 227 amino acids with the predicted approximate molecular weight of 25.1 kDa.
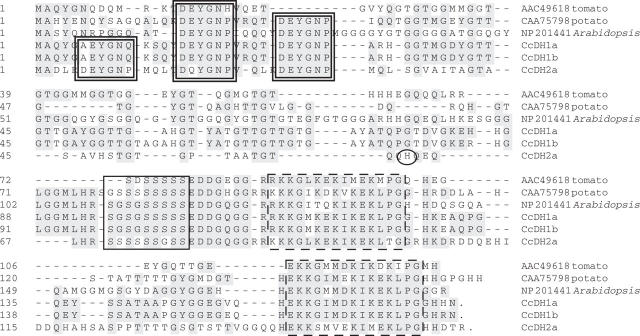
Alignments of the coffee dehydrins with the most homologous protein sequences found in the non-redundant protein database, and the analysis of the presence of the dehydrin-specific amino acid motifs Y, S and K (Close, 1996), are shown in Figs 1 and 2. The dehydrins fall into two classes, with CcCDH1 and CcDH2 having the structure Y3SK2 and CcDH3 having the structure SK3. Of those protein sequences with the Y3SK2 structure, CcDH1a and CcDH1b showed absolute conservation in each of the three motifs, as well as in the two conserved regions that precede each of the two K motifs, indicating that these two sequences are allelic. In contrast, CcDH2 is clearly different from CcDH1. While CcDH2 has the structure Y3SK2, it exhibits punctual differences from CcDH1 in all but one of the Y, S and K motifs, as well as more significant differences outside these dehydrin-specific motifs. The CcDH1 and CcDH2 encode proteins with calculated pIs that are near neutral, and their hydrophilicity plots indicate that these proteins are very hydrophilic throughout (data not shown). The calculated pI of the protein encoded by CcDH3 is slightly acidic (5.47), and this protein is also very hydrophilic.
Fig. 1.
The optimized alignment of C. canephora Y3SK2-type dehydrins with several of the closest plant homologues. This alignment was generated using the Clustal W program in the MegAlign software (DNASTAR) and then further optimized manually. Shaded blocks indicate identical amino acids. The doubled rectangles demarcate the Y-segments, the single dark rectangles demarcate the S-segments, and the rectangles with broken lines demarcate the K-segments. The black circle represents the single amino acid difference between CcDH2a and CcDH2b. Accession numbers: CcDH1a, DQ323987; CcDH1b, DQ323988; CCDH2a, DQ323989; Lycopersicon esculentum dehydrin TAS14, AAC49618; Solanum commersonii dehydrin Dhn1, CAA75798; Arabidopsis thaliana dehydrin RAB18, NP_201441.
Fig. 2.
The optimized alignment of the C. canephora SK3-type dehydrin with several of the closest plant homologues. This alignment was generated as described for Fig. 1. Shaded blocks indicate identical amino acids. The single dark rectangles demarcate the S-segments, and the rectangles with broken lines demarcate the K-segments. Accession numbers: CcDH3, DQ333960; Nicotiana tabacum dehydrin, BAD13499; Solanum tuberosum dehydrin homologue CI7, T07779; Arabidopsis thaliana dehydrin, CAA62449.
Characterization of a cDNA encoding a coffee LEA protein
Using a cDNA library constructed from RNA prepared from coffee grain 30 weeks after fertilization (M. Ben Amor et al., unpubl. res.), a full-length cDNA clone (pDav1-59; 1232 bp) encoding an LEA protein was isolated and sequenced and renamed CcLEA1. Also the EST database was searched for ‘unigenes’ annotated as LEA proteins. This search produced nine gene sequences, one of which (unigene #119994) corresponded to the previously isolated sequence of CcLEA1 (Table 2). It was decided to focus on CcLEA1 in more detail in this work because of its unusual expression pattern. Based on its EST representation, this gene appears to be strongly and exclusively expressed during only one period of grain development (30 weeks after flowering). The protein sequence encoded by CcLEA1 is 357 amino acids and has a predicted molecular weight of 39.5 kDa. The calculated pI for CcLEA1 is slightly basic (8.17), and hydrophilicity plotting shows that, although this protein does not have large regions of hydrophobicity, it is clearly less hydrophilic than the three coffee dehydrins. It is noted that the first 30 N-terminal residues of CcLEA1 form one of the two small hydrophobic regions found in this protein. Figure 3 shows the alignment of CcLEA1 with the three most homologous sequences found in the non-redundant Genbank protein database. While the overall identity values of these aligned sequences only ranged from 34.7 % for the arabidopsis sequence to 47.9 % for the Picea (white spruce) sequence, a closer examination of the alignment shows that there are several short, but highly conserved regions in these proteins. It is also noted that all the related protein sequences had relatively similar hydrophilicity profiles to CcLEA1 and each had it's most significant hydrophobic patch located in the N-terminal 1–25 amino acids, possibly indicating this protein has a leader peptide sequence to direct synthesis into the endoplasmic reticulum (data not shown). Interestingly, CcLEA1 has a rather striking proline-rich segment in the N-terminal region that is clearly absent from the other proteins in Fig. 3. However, CcLEA1 does have the conserved set of cysteine residues that were previously defined in the maize root cap proteins.
Table 2.
Coffea canephora unigene sequences annotated as late embryogenesis abundant proteins (the corresponding unigene sequences can be found at http://www.sgn.cornell.edu)
|
In-silico expression (no. of ESTs in libraries) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Unigene name | No. of ESTs | Whole cherries 22 weeks | Seed 18 weeks | Seed 30 weeks | Seed 42 weeks | Seed 46 weeks | Pericarp | Leaf |
| 124784 | 26 | 17 | 9 | |||||
| 124426 | 26 | 9 | 17 | |||||
| 119994 | 22 | 22 | ||||||
| 124428 | 10 | 1 | 3 | 4 | 2 | |||
| 121426 | 8 | 1 | 5 | 2 | ||||
| 124123 | 5 | 3 | 1 | 1 | ||||
| 120926 | 2 | 2 | ||||||
| 131122 | 1 | 1 | ||||||
| 127677 | 1 | 1 | ||||||
Fig. 3.
The optimized alignment of the C. canephora late embryogenesis abundant protein CcLEA1 with several of the closest plant homologues. This alignment was generated as described for Fig. 1. Shaded blocks indicate identical amino acids. Conserved cysteines are marked by an asterisk, and the positions of two less highly conserved cysteines are marked by a circle. Accession numbers: CcLEA1, DQ333961; Arabidopsis thaliana late embryogenesis abundant related protein, NP_200248; Picea glauca late embryogenesis abundant protein EMB7, T09288; Zea mays root cap protein 2, BAA75477.
RT–PCR expression analysis of CcDH1, CcDH2, CcDH3 and CcLEA-1 genes in different coffee tissues and during coffee grain development
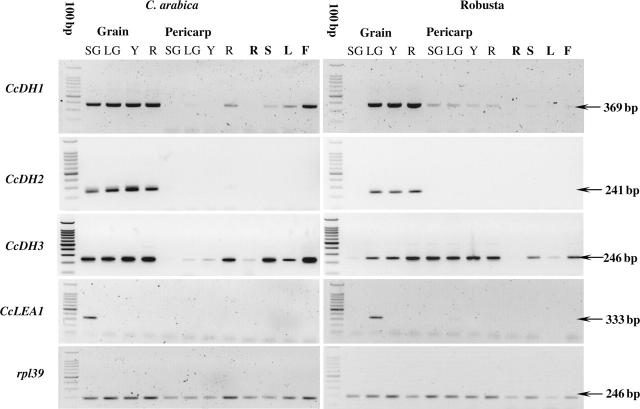
RT–PCR analysis was carried out for each of the three coffee dehydrin genes, and the results of this experiment are shown in Fig. 4. CcDH1 was expressed significantly in arabica grain at all the stages examined, and in the three last stages examined for robusta. It has previously been shown that there are significant differences between the gene expression profiles observed between the small green arabica and small green robusta grain samples used here. For example, small green grain from arabica, but not from robusta, have detectable transcripts of genes associated with endosperm expansion such as the oleosins and the 11S storage protein (Simkin et al., 2006). Close examination of the original gels indicated that very low levels of CcDH1 expression could also be detected in several of the other tissues tested, although no signal was detected for arabica in the small green pericarp and yellow pericarp samples, or for robusta in the root or young leaf samples. Among the tissues having expression of CcDH1, the arabica flower sample appeared to have the highest level of transcripts. A relatively high level of CcDH2 transcripts was also detected in all the grain-development stages of arabica and the last three stages of the robusta grain examined (Fig. 4). In contrast to CcDH1, no CcDH2 transcripts were detected by RT–PCR in the other tissues studied.
Fig. 4.
RT–PCR expression analysis of the coffee dehydrins and CcLEA1 transcripts in different organs of Coffea arabica and C. canephora. 100 bp, 100-bp molecular weight marker ladder; SG, LG, Y, R, small green, large green, yellow and red for the grain and pericarp, respectively; R, S, L, F, root, stems, young leaves and flowers, respectively.
RT–PCR analysis of CcDH3 gene expression demonstrated that these transcripts were also detected in all the arabica grain samples, as well as in the last three stages of robusta grain development (Fig. 4). CcDH3 transcripts could also be clearly detected in some of the other tissues, such as the red pericarp, stem and flowers. Close examination of the original data showed that CcDH3 transcripts could actually be detected in all of the other arabica and robusta tissues examined. A significant difference was noted between the transcript levels for the first three pericarp stages of robusta and arabica. Whether this difference reflects an intrinsic difference between arabica and robusta cherries, or simply reflects some environmental difference between the samples remains to be determined. Also the expression of CcLEA1 was examined using RT–PCR. The data obtained confirms that this gene has a very unique expression pattern, with transcripts being detected only in the small green stage of arabica and the large green stage of robusta grain (Fig. 4). No expression was detected in any of the other arabica or robusta tissues sampled. These data are consistent with the distribution pattern of ESTs for this gene in the EST libraries, which indicated that this gene is expressed in robusta grain at 30 weeks after flowering (WAF) but not in any of the other grain libraries, or in the cherry, pericarp and leaf libraries.
Isolation and sequence analysis of the CcDH2 promoter
It is desirable to develop a repertoire of grain-specific promoters for future studies on transcriptional control in coffee, and for use in the construction of model recombinant genes to be expressed specifically in the grain. The data presented earlier demonstrate that the promoter of the CcDH2 gene is a relatively strong grain-specific promoter. Thus, it was decided to isolate a genomic fragment which incorporates this promoter by employing the Genome Walker technique. To ensure the strength of this promoter was associated with a single/low copy gene, first the copy number of the CcDH2 gene was estimated by Southern blotting. The data presented in Fig. 5 shows that each restriction enzyme digestion produced a single band, strongly suggesting that CcDH2 is encoded by a single gene in the C. canephora genome. Using a CcDH2-specific Genome Walker primer designed from the centre of the cDNA pcccs30w8a4 (DH2a primer 1), together with the AP1 primer of the Genome Walker kit, a genomic fragment from C. canephora that contained over 1.5 kb of DNA upstream of the pcccs30w8a4 cDNA sequence (pJMc1) was successfully PCR amplified and cloned. To amplify a single complete genomic fragment containing the DH2 promoter region and transcribed region, a new round of PCR was performed as described in Materials and methods using C. canephora DNA. This experiment resulted in the generation of the genomic fragment cloned in pVC1 which extended 1.43 kb upstream of the corresponding cDNA sequence (pcccs30w8a4). There were six base changes between the genomic sequences of pJMc1 and pVC1; one was in the promoter region, two were in the intron, and three were in the protein-coding sequence (Thr/Ala, Thr/Ser and Gly/Gly). Examination of an alignment of the four sequences available for CcDH2 (two genomic and two cDNA) indicates that these sequences represent three different alleles. These alleles are manifest by the SNPs associated with the amino acid changes noted above. The cDNA sequence in CcDH2a is identical to the coding sequence of the genomic clone pJMc1, indicating that these sequences probably represent the same allele.
Fig. 5.
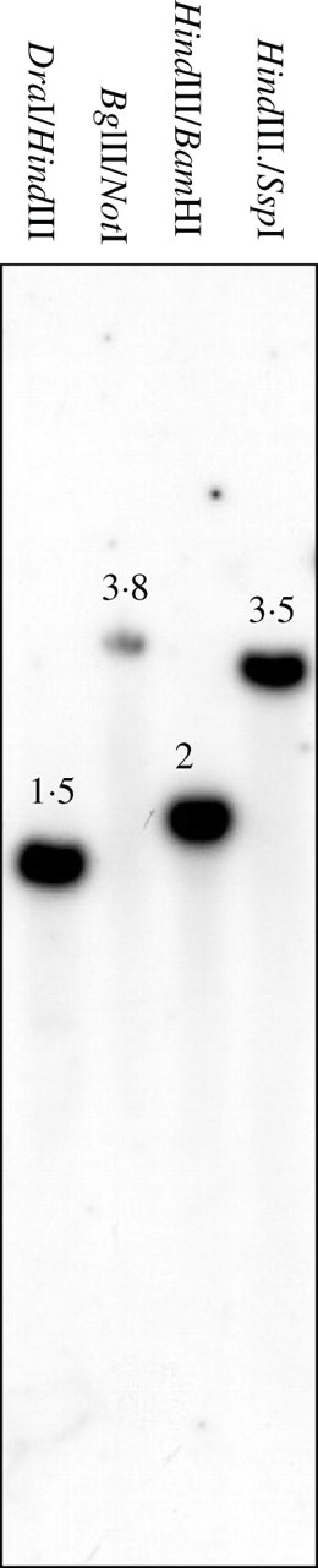
Southern blot analysis of the CcDH2 dehydrin.
Sequence analysis of the composite sequence obtained from the plasmids containing both genomic and cDNA sequences showed that the CcDH2 gene contains a single intron (231 bp) located within the ORF region (Fig. 6). Further analysis of the promoter region of this gene indicates the presence of a putative TATA sequence 30 bp upstream of the 5′ end of the cDNA sequence. Several potential regulatory elements previously shown to be involved in the regulation of gene expression during seed development and during osmotic stress can also be identified in the 5′ upstream region of the CcDH2 gene. For example, three regions with similarity to the arabidopsis ABA-responsive element RYACGTGGYR (Iwasaki et al., 1995) were found. Two other elements were found that shared significant similarity with the RY repeat (CATGCA(T/a) (A/g) of the ‘legumin’ box that is involved in regulating the expression of genes encoding the legumin type storage proteins (Shirsat et al., 1989; Baumlein et al., 1992). The presence of one dehydration-responsive element/C-Repeat cis-acting sequence motif (DRE/CRT; G/ACCGAC) has also been identified. A second DRE/CRT element is found in the first exon. DRE/CRT motifs have been shown to interact with DREBs/CBF transcription factors to control the response of linked genes to dehydration and other stresses in arabidopsis and rice (Dubouzet et al., 2003). Finally, several E-box motifs (CANNTG), which are well defined components in the promoters of seed-specific genes such as the 2S storage protein (Chatthai et al., 2004) and oleosins (Simkin et al., 2006), have been identified in the CcDH2 promoter region.
Fig. 6.

DNA sequence of the CcDH2a promoter and transcribed sequence from Coffea canephora. The nucleic acid sequence of pVC1 (accession number DQ338457) insert is presented, and its corresponding amino acid sequence. The first base in the cDNA (C) is marked by a circle. The putative TATA box is shown in bold and underlined. The RY repeat sequences are marked with grey, the ABA responsive elements are shown in boxes, and the DRE/CRT sequence is boxed with heavy lines.
DISCUSSION
The dehydrins are a group of specialized plant proteins that are involved in protecting cells from dehydration-related stresses (Close, 1996; Allagulova et al., 2003). Dehydrins also participate in two important and rather specific aspects of seeds. (1) They are widely perceived to participate, with other LEA proteins, in the dehydration process that occurs during the late stages of seed maturation by assisting the acclimatization of seed tissues to the lower water content found in mature seeds (Close, 1996; Nylander et al., 2001). (2) It is presumed that the dehydrins synthesized in seeds during maturation continue to stabilize the associated cellular structures during seed quiescence. In this latter context, it should also be noted that it has recently been proposed that dehydrins may also possess a radical-scavenging capability (Hara et al., 2003) and have metal-binding properties (Alsheikh et al., 2003), both characteristics that are likely to be useful during long periods of seed storage.
Despite the involvement of dehydrin proteins in plant resistance to osmotic stresses such as drought stress, and the probable importance of the dehydrins during grain development, little information is available on these genes in coffee. Thus it has been decided to take advantage of the new coffee EST collection to isolate cDNAs encoding highly expressed coffee dehydrins, and to establish the expression patterns of the dehydrins in different tissues, as well as during coffee grain and pericarp maturation. Five cDNAs from C. canephora, which represent three distinct dehydrin genes (CcDH1, CcDH2 and CcDH3) are presented here, as well as a cDNA representing an LEA gene (CcLEA1) with an extremely restricted expression pattern. The gene CcDH1 is represented by two cDNAs that differ by 21 bases and have over 95 % identity. The small sequence differences translate into six amino acid changes, three of which result from a 9 bp deletion in the ORF of CcDH1a. Because these two cDNAs show such a high level of identity, and the fact that the protein sequence changes fall outside the highly conserved regions in this protein sequence family (Fig. 1), it is considered that these two cDNAs represent different alleles of one gene.
Two distinct unigene sequences were found to encode an identical dehydrin protein (CcDH2a and CcDH2b). However, closer examination of full-length cDNA clones representing these two unigenes indicated that unigene #123405, which consisted of two ESTs, actually contained an intron within the predicted ORF sequence. This intron containing cDNA (CcCH2b; pcccs4630p1) also contained a poly (A) tail sequence indicating that the corresponding mRNA was polyadenylated but not spliced. Interestingly, the 3′ splice site of the intron in CcDH2b was slightly different from the equivalent genomic sequence of CcDH2a [Fig. 6; ttatgg/TCG versus genomic sequence ttatag/T(or A)CG]. The only other differences between the intron sequence of the cDNA and the intron sequences in three independently obtained genomic sequences were a few punctual base changes. None of these changes in the intron were conserved in each of the three genomic sequences. The fact that the two ESTs of unigene #123405 (CcDH2b) had the same splice site mutations and were found in two different grain specific libraries (30-week and 46-week libraries), strongly suggests that this intron-containing transcript is real and not a cloning artefact or sequencing error. Therefore, the available data strongly suggest that the absence of splicing in the two cDNAs of unigene #123405 were due to the single base pair change in the splice site (gg versus ag). As the 30-week and 46-week grain libraries were made from more than one variety, it is currently not known which varieties harbours this mutation. It is also not known if this mutation has any biological consequences. Interestingly, it has been been found recently that approx. 5 % of the cDNAs in a large set of full-length arabidopsis cDNAs harbour unspliced introns, which the authors have called ‘retained introns’ (Iida et al., 2004). This result is consistent with the observation that cDNA with poly (A) tails and containing unspliced introns can be found relatively easily in the new coffee EST libraries (J. McCarthy et al., unpubl. res.). Much further work is needed to determine to what extent such ‘retained introns’ are the result of problems such as slow primary transcript processing, or base changes causing the reduced/loss of splicing, or alternatively, whether more complex mechanisms/functions are involved.
Although their primary amino acid sequences are relatively different (CcDH1a shares 47.3 % identity with CcDH2a), both CcDH1 and CcDH2 can be classified as Y3SK2 dehydrins based on the distribution of the Y, S, and K motifs in the respective sequences. In addition to being relatively close in motif composition, CcDH1 and CcDH2 also exhibit some similarities in their patterns of expression. Both genes are strongly expressed during the later stages of grain development (Fig. 4). However, it appears that CcDH1 is also weakly expressed in several other tissues, albeit at different levels in arabica and robusta. In contrast, the expression of CcDH2 is limited to the grain in both species. It is noted that one CcDH2 EST was found in the leaf EST library, suggesting that this gene may, under some conditions, be expressed in the leaf. Further experiments are necessary to determine whether CcDH2 expression is inducible, for example, by dehydration stress or by exposure to cold temperatures. Given that the expression patterns for CcDH1 and CcDH2 are different, and the fact that the proteins vary slightly, even in the highly conserved Y, S and K motifs, could imply that these proteins may have related, but not identical roles. For example, it is possible that the CcDH1 and CcDH2 dehydrins are involved in protecting different functional targets. Alternatively, they could have similar functional targets, but their expression is controlled differently to enable these genes to be induced differentially in particular tissues by specific developmental and/or environmental cues. The coffee dehydrin CcDH3 has the structure SK3, and is also expressed during the late stages of grain development (Fig. 4). In addition, CcDH3 expression is detected in several other tissues, as was CcDH1 expression. This latter point suggests that the expression of CcDH1 and CcDH3 could be controlled by relatively similar signals, although differences in the absolute levels of transcripts indicates transcription induction and/or transcript stabilities are not identical for both genes. As for the majority of the dehydrins previously described from other plants, the precise functions of the coffee dehydrins described here are not yet known. Nonetheless, the significant expression of the CcDH1, CcDH2 and CcDH3 genes during the late stages of grain maturation clearly suggests that the three dehydrins play a significant role(s) in conditioning the coffee grain for maturation-induced dehydration, and probably are also important in protecting the mature grain tissues before germination. Future experiments will be aimed at determining whether one or more of these dehydrins are induced in coffee under conditions of water and low temperature stress.
The LEA gene described here (CcLEA1) has an unusual expression pattern, with transcripts being detected only during one relatively short period of grain development, and not in any other tissues. In robusta, CcLEA1 expression was only detected at the large green grain stage, and not in either the small green or yellow stages of grain development. This stage of grain development spans the period when the perisperm tissue undergoes a substantial size reduction and the endosperm expands significantly (unpubl. res.), thus suggesting that the CcLEA probably plays a specific role during the perisperm/endosperm transition. A Blast analysis of the protein database with the CcLEA1 protein uncovered three potentially related protein sequences from arabidopsis, Picia (white spruce), and maize. Although the highest level of identity was only 47.9 % for the Picia sequence, the protein alignment seen in Fig. 3 shows that the three related proteins did share several highly conserved blocks of homology, especially after the more variable N-terminal end of the protein. None of the homologous proteins have been assigned a function. However, the maize cDNA sequence, which is called ‘root cap protein 2’, was isolated with another highly similar cDNA called ‘root cap protein 1’ from maize root caps using a differential screen (Matsuyama et al., 1999). Transcription of both genes was restricted to the outermost cells of the maize root cap. These cells are believed to be first associated with the production of mucilage and then, later, are sloughed off (Moore and McClelen, 1983). It also appears that, as they sloughed off, the root cap cells undergo cell death (Matsuyama et al., 1999). Although expression of CcLEA1 was not detected in roots, it is quite possible that the root cap fraction of the total root samples tested here were very small and thus genes specifically expressed in root caps were below the level of detection. Further expression analysis using particular regions of the root should clarify whether CcLEA1 is expressed in the root cap region of coffee. It is interesting to speculate that this class of protein may play a similar role in both the root cap and the perisperm region as it develops into endosperm during grain maturation. Although no data concerning the putative arabidopsis LEA homologue (accession number NP200248) have been found in the literature, an examination of the arabidopsis MPSS expression database (http://mpss.udel.edu) shows that this arabidopsis gene is expressed in both the root and in germinating seeds. No expression was detected for this gene in any of the other tissues described in the arabidopsis MPSS database. The detection of transcripts for the arabidopsis homologue in root supports the idea that the arabidopsis protein is a homologue to the maize root cap protein and possibly CcLEA1. It will be interesting in the future to investigate whether the putative homologue expressed during the germination of arabidopsis seeds is associated with tissues undergoing cell death, and similarly, if the root cap proteins of maize are expressed during maize seed development and/or germination. Finally, one striking similarity between CcLEA1 and its putative homologues is the near absolute conservation of the cysteines, a feature which was first observed in the maize root cap proteins (Matsuyama et al., 1999). However, the functional significance of these conserved cysteines is currently unknown.
Because the CcDH2 gene appears to be grain specific, the promoter of this gene was isolated for further study. The promoter sequence obtained (Fig. 6) has several previously recognized regulatory sequence motifs that are expected to be involved in the temporal and spatial control of CcDH2 dehydrin expression. Considering that the detailed testing of coffee grain-specific promoters can take more than 3–4 years due to the slow development and flowering of coffee, the possibility of testing the capacity of this promoter sequence is being examined to direct grain-specific expression in model plants such as arabidopsis or tomato. It is now well established that dehydrin proteins somehow participate in protecting plants from water-related environmental stresses (Allagulova et al., 2003). Therefore the detailed information on the coffee dehydrins presented here, and the identification of other LEA cDNA in the coffee EST bank (Table 2), should open some new avenues for research on stress tolerance in coffee. For example, it is now possible to study the induction of these genes in different tissues in response to various stresses. It is also possible to examine whether there are variations in the expression of these genes in coffee varieties exhibiting dramatically different tolerances to drought and low temperatures. The information on the coffee dehydrin and LEA sequences presented here also opens up the possibility of investigating the potential relationship(s) between the poor storage capability of coffee grain and the expression levels of various dehydrin and LEA genes.
Acknowledgments
We thank Dr Vincent Petiard and the members of the coffee gene discovery group for their support and helpful discussions. In particular, we thank Maud Lepelley for her assistance with the DNA sequencing and bioinformatic analysis of the cDNA clones, and Dr Andy Simkin and Dr Isabelle Privat for comments on the manuscript. Finally, we thank Dr Ir Zaenudin Su and Dr Surip Mawardi of the Indonesian Coffee and Cacao Research Institute (ICCRI) for supplying samples of C. canephora ‘BP409’.
LITERATURE CITED
- Allagulova CR, Gimalov FR, Shakirova FM, Vakhitov VA. 2003. The plant dehydrins: structure and putative functions. Biochemistry –Moscow 68: 945–951. [DOI] [PubMed] [Google Scholar]
- Alsheikh MK, Heyen BJ, Randall SK. 2003. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. Journal of Biological Chemistry 278: 40882–40889. [DOI] [PubMed] [Google Scholar]
- Baumlein H, Nagy I, Villarroel R, Inze D, Wobus U. 1992. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. The Plant Journal 2: 233–239. [PubMed] [Google Scholar]
- Chatthai M, Forward BS, Yevtushenko D, Stefanov I, Osuska L, Osusky M, et al. 2004. 2S storage protein gene of Douglas-fir: characterization and activity of promoter in transgenic tobacco seeds. Plant Physiology and Biochemistry 42: 417–423. [DOI] [PubMed] [Google Scholar]
- Choi DW, Close TJ. 2000. A newly identified barley gene, Dhn12 encoding a YSK2 DHN, is located on chromosome 6H and has embryo-specific expression. Theoretical and Applied Genetics 100: 1274–1278. [Google Scholar]
- Close T. 1996. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiologia Plantarum 97: 795–803. [Google Scholar]
- Close TJ. 1997. Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiologia Plantarum 100: 291–296. [Google Scholar]
- Crouzillat D, Lerceteau E, Petiard V, Morera J, Rodriguez H, Walker D, et al. 1996. Theobroma cacao L.: a genetic linkage map and quantitative trait loci analysis. Theoretical and Applied Genetics 93: 205–214. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, et al. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal 33: 751–763. [DOI] [PubMed] [Google Scholar]
- Dure L. 1993. Structural motifs in LEA proteins of higher plants. In: Close TJ, Bray EA, eds. Response of plants to cellular dehydration during environmental stress. Rockville, MD: American Society of Plant Physiologists, 91–103.
- Dure L, Greenway S, Galau G. 1981. Biochemistry 20: 4162–4178. [DOI] [PubMed] [Google Scholar]
- Godoy JA, Lunar R, Torres-Schumann S, Moreno J, Rodrigo M, Pintor-Toro JA. 1994. Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Molecular Biology 26: 1921–1934. [DOI] [PubMed] [Google Scholar]
- Hara M, Terashima S, Fukaya T, Kuboi T. 2003. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 217: 290–298. [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, et al. 2004. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Research 32: 5096–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J, Bartels D. 1996. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47: 377–403. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Yamaguchi-Shinozaki K, Shinozaki K. 1995. Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose introduction by dehydration is mediated by abscisic acid and requires protein synthesis. Molecular and General Genetics 247: 391–398. [DOI] [PubMed] [Google Scholar]
- Koag MC, Fenton RD, Wilkens S, Close TJ. 2003. The binding of maize DHN1 to lipid vesicles: gain of structure and lipid specificity. Plant Physiology 131: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Mueller LA, McCarthy J, Crouzillat D, Petiard V, Tanksley SD. 2005. Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts. Theoretical and Applied Genetics 112: 114–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T, Yasumura N, Funakoshi M, Yamada Y, Hashimoto T. 1999. Maize genes specifically expressed in the outermost cells of the root cap. Plant Cell Physiology 40: 469–476. [DOI] [PubMed] [Google Scholar]
- Moore R, McClelen C. 1983. Ultrastructural aspects of cellular differentiation in the root cap of Zea mays. Canadian Journal of Botany 61: 1566–1572. [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. 2001. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Molecular Biology 45: 263–279. [DOI] [PubMed] [Google Scholar]
- Puhakainen T, Hess MW, Makela P, Svensson J, Heino P, Palva ET. 2004. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Molecular Biology 54:743–753. [DOI] [PubMed] [Google Scholar]
- Roberts J, DeSimone N, Lingle W, Dure L. 1993. Cellular concentrations and unifomity of cell-type accumulation of two LEA proteins in cotton embryos. The Plant Cell 5: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsat A, Wilford N, Croy R, Boulter D. 1989. Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Molecular and General Genetics 215: 326–331. [DOI] [PubMed] [Google Scholar]
- Simkin AJ, Qian T, Caillet V, Michoux F, Ben Amor M, Lin C, Tanksley S, McCarthy J. 2006. Oleosin gene family of Coffea canephora: quantitative expression analysis of five oleosin genes in developing and germinating coffee grain. J Plant Physiol (in press). [DOI] [PubMed]
- Skriver K, Mundy J. 1990. Gene expression in response to abscisic acid and osmotic stress. The Plant Cell 2: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulages JL, Kim K, Arrese EL, Walters C, Cushman JC. 2003. Conformation of a group 2 late embryogenesis abundant protein from soybean: evidence of poly (L-proline)-type II structure. Plant Physiology 131: 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M, Tunnacliffe A. 2004. POPP the question: what do LEA proteins do? Trends in Plant Science 9: 13–17. [DOI] [PubMed] [Google Scholar]
- Zhu B, Choi DW, Fenton R, Close TJ. 2000. Expression of the barley dehydrin multigene family and the development of freezing tolerance. Molecular and General Genetics 264: 145–153. [DOI] [PubMed] [Google Scholar]