Abstract
• Background and Aims The Cycas balansae complex is arguably a controversial group with regard to species delineation. Some taxonomists recognize a single polymorphic species while others distinguish five narrowly defined ones. The unresolved taxonomy has the potential to bring about significant problems for species conservation. Thus, an investigation to examine the genetic diversity and differentiation in the C. balansae complex was performed to determine the relationship of populations and to test whether the morphologically defined segregations represent genetically distinct units.
• Methods Inter-simple sequence repeat (ISSR) markers were employed to assess the genetic diversity in the C. balansae complex with a sample of 158 individuals from all extant populations in China.
• Key Results ISSR markers revealed low genetic diversity in all populations studied (HE and HO averaged 0·0639 and 0·0798 at the population level, respectively). Phenetic analysis showed that the C. balansae complex grouped into five clusters closely corresponding to the narrowly defined C. balansae, C. parvula, C. shiwandashanica, C. tanqingii and C. simplicipinna.
• Conclusions ISSR data suggest that the C. balansae complex has evolved into five genetically distinct units. These might be derived from a relatively widespread common ancestor through multiple vicariant events including geographical isolation resulting from the collision of the Indian plate with the Eurasian plate and from Pleistocene glaciations. In conservation, attention should be paid to each genetic unit.
Keywords: Conservation, Cycas balansae complex, ISSR, genetic differentiation, genetic units
INTRODUCTION
Knowledge about taxonomy and genetic variation patterns within and between populations are basic requirements in conservation biology (Franke and Soule, 1981). Species circumscription may have a strong impact on the estimate of genetic diversity within a species (Hedrén, 2004), the extent and structure of which is a prerequisite for evaluating their survival ability in a changing environment, and for establishment of conservation practices. With the goal to preserve species evolutionary processes, taxonomic uncertainty could result in negative impacts on biodiversity conservation, such as genetic contamination of local populations during in situ management activities (Moritz, 1994). Furthermore, ex situ conservation also requires an advanced understanding of the phylogenetic relationship and distinctiveness of the taxa in order to avoid interspecific hybridization, which is a major threat to the integrity of species as distinct breeding groups (Furman et al., 1997).
Cycads (Cycadaceae, Stangeriaceae, Zamiaceae) are the most primitive living seed plants with over 250 million years of history (Gao and Thomas, 1989; Norstog and Nicholls, 1997; Hill, 2003). Although extremely dominant during the Mesozoic, they are now largely relictual in highly fragmented habitats in the tropics and subtropics. Most cycad species are endangered and face the threat of extinction in the wild (Osborne, 1995). All species of cycads have been listed in the Convention on International Trade in Endangered Species of Wild Fauna and Flora. In China, all cycads have also been given First Grade conservation status. Cycads have recognizable morphological characteristics intermediate between less advanced plants such as ferns and more derived plants including the angiosperms (Brenner, 2003). Efforts must be undertaken to avoid further decline and extinction of cycads, which constitute a key node in plant evolution.
Members of the Cycas balansae complex are distributed in south-west China through Yunnan and Guangxi provinces into adjacent northern Indo-China, including Vietnam, Laos and northern Thailand. Their distribution is characterized by their geographically restricted and historically fragmented pattern. Like the Macrozamia pauli–guilielmi complex (Zamiaceae) (Sharma et al., 1998) and the Macrozamia plurinervia complex (Zamiaceae) (Sharma et al., 2004), the plants in the C. balansae complex lack distinct morphological characteristics and their taxonomic and phylogenic relationships are problematic and controversial. Chen and Stevenson (1999) regarded the complex as one species, C. balansae, characterized by acaulescent (versus caulescent) caudex and wide (versus narrow) leaflets. However, most researchers recognize five separate species, i.e. C. parvula, C. tanqingii, C. balansae, C. shiwandashanica and C. simplicipinnata, separated by characters such as the number and size of ovules (Wang and Liang, 1996; Huang, 2001). Of these species, C. parvula and C. tanqingii are restricted to south-west China, whereas the others, C. balansae, C. shiwandashanica and C. simplicipinnata, are mainly distributed in northern Indo-China, but with a few populations extending to south-west China. The choice of taxonomic treatment can profoundly affect not only the number of rare and endemic species within cycads in this region but also the establishment of reasonable and effective conservation strategies. In view of the controversy on the C. balansae complex, morphological information alone is inadequate to evaluate their conservation status. Traditional taxonomy based on morphological characters may unite cryptic species that are morphologically similar but genetically divergent, which could lead to gene contamination in the course of breeding programmes and reintroduction (Hammond, 2001). Taxon-specific molecular markers that measure the degree of differentiation between taxa can serve as a way to overcome the limitations of traditional morphology-based taxonomy (Hahn and Grifo, 1996). Inter-simple sequence repeats (ISSRs) are markers composed of a microsatellite sequence plus a short arbitrary, adjacent sequence primed by a single oligonucleotide. ISSR primers amplify the region between two closely spaced and oppositely oriented simple sequence repeats. ISSR technologies can reproducibility produce large numbers of polymorphic fragments at low cost (Fang et al., 1997; Moreno et al., 1998). The literature reveals that ISSR markers have a great potential for inferring genetic relationships among closely related species (Zietkiewicz et al., 1994; Salimath et al., 1995; Fang et al., 1998; Martin and Sanchez-Yelamo, 2000; Benharrat, 2002). ISSR markers have also been used to estimate genetic diversity within and between populations of C. guizhouensis (Xiao et al., 2004). Here ISSRs are used to analyse genetic diversity and differentiation in the C. balansae complex with the goal to understand genetic relationships between populations and their evolutionary history, and whether the morphologically defined segregated species represent genetically distinct units, and to provide information for species conservation.
MATERIALS AND METHODS
Sample collection
Leaf samples were obtained from 158 individuals from eight natural populations representing the five species proposed by Wang (2000) in the C. balansae complex (Table 1).The species were not compared with an out-group species since the purpose of this investigation was to establish inter-genetic relationships within the complex and not strictly phylogenetic relationships. Fresh leaflets were dried with silica gel and stored at 4 °C until the DNA extraction. Vouchers were collected from each population and deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Science (KUN).
Table 1.
Populations studied and sample size
| Species | Pop. ID | Locality* | Latitude (N) | Longitude (E) | Altitude (m) | Sample size | Voucher |
|---|---|---|---|---|---|---|---|
| C. shiwandashanica | FC | GX Fangchenggang | 21°45′ | 108°05′ | ? | 20 | Gong 03136 |
| C. parvula | YJ1 | YN Yuanjiang | 23°42′ | 101°47′ | 1454 | 18 | Gong 03105 |
| YJ2 | YN Yuanjiang | 23°40′ | 101°50′ | 1445 | 20 | Gong 03106 | |
| C. balansae | JP | YN Jinping | 22°41′ | 103°00′ | 389 | 20 | Gong 03110 |
| MG | YN Maguan | 22°51′ | 104°07′ | 610 | 20 | Gong 03111 | |
| PB | YN Pingbian | 23°03′ | 103°43′ | 745 | 20 | Gong 03112 | |
| C. tanqingii | LC | YN Lvchun | 22°37′ | 102°19′ | 640 | 20 | Gong 03118 |
| C. simplicipinna | ML | YN Mengla | 21°56′ | 101°15′ | 570 | 20 | Gong 03124 |
GX is the code for Guangxi Province and YN for Yunnan Province in China.
DNA extraction, PCR amplification and electrophoresis
Genomic DNA was extracted from leaves of single individuals following the CTAB protocol (Doyle, 1991). Extracted DNA was quantified using a spectrophotometer comparing band intensities with known standards of lambda DNA on 1 % agarose gel. The working solution of DNA (approx. 10 ng μL) was in sterile double-distilled water.
In a preliminary study, 100 ISSR primers, designed by the University of British Columbia (UBC), Vancouver, Canada, were tested in PCR-amplification on a subset of three random individuals. Twelve fragments [USB # 807 (AG) 8T, # 808 (AG)8C, # 810 (GA)8T, # 811 (GA)8C, # 815 (CT)8G, # 835 (AG)8YC, # 836 (AG)8YA, # 840 (GA)8YT, # 841 (GA)8YC, # 842 (GA)8YG, # 843 (CT)8RA and # 857 (AC)8YG] were selected for further analysis on all individuals.
DNA amplification was performed in a 20-μL reaction volume containing about 20 ng of template DNA, 2·0 μL 10× PCR buffer, 2·0–2·5 mm MgCl2, 0·1 mm dNTPs, 2 % formamide, 200 nm primer, 1·0 unit of Taq polymerase and double-distilled water. Amplification was performed in a PTC-100 thermal cycler (MJ Research, Inc.) programmed for an initial step of 1 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 45 s at 52 °C (or 50 °C) and 1·5 min at 72 °C, and a final 7-min extension at 72 °C. A negative control reaction in which the template DNA was replaced by water was also included to verify for absence of contamination.
Amplification products were electrophoretically separated in 1·5 % agarose gels buffered with 0·5× TBE. A 100-bp DNA ladder (New England Biolabs) was used as size marker. DNA fragments were identified by image analysis software for gel documentation (LabWorks Software Version 3.0; UVP, Upland, CA 91786, USA) following staining with ethidium bromide.
Data analysis
ISSR bands were scored as present or absent. Only those bands that showed consistent amplification were considered. Smeared and weak bands were excluded. The resulting presence/absence data matrix was analysed using POPGENE v. 1.31 (Yeh et al., 1999) to estimate four genetic diversity parameters: percentage of polymorphic loci (P), effective number of alleles (Ae), Shannon's Information index (HO) and Nei's gene diversity (HE) (Nei, 1973). To examine population genetic structure the proportion of genetic divergence was estimated within and among populations following Nei (1973) and GST as the coefficient of gene differentiation among the populations. The genetic identity (I) and the genetic distance (D) between populations were also computed using the model presented in Nei (1972). Gene flow estimates (Nm) were calculated as Nm = (1 − GST)/4GST (Slatkin and Barton, 1989). An unweighted pair group method using an arithmetic average (UPGMA) dendrogram was performed on the data matrix of mean character difference between pairs of samples with software Mvsp3.13b (downloaded from the website at http://www.kovcomp.com) and matrix of ISSRs.
An analysis of molecular variance, AMOVA Version 1.5 (Excoffier, 1993), was also used to analyse the proportion of the genetic differentiation among populations.
RESULTS
The 12 primers chosen for final analysis yielded a total of 103 clear ISSR markers. Of those, only 41 markers were present in all individuals and the percentage of polymorphic markers was 60·19 %. Table 2 describes the polymorphism in this complex revealed by ISSR in detail. Assuming Hardy–Weinberg equilibrium, Nei's genetic diversity (HE) and Shannon indices (HO) were estimated to be 0·2110 and 0·3166, respectively, in the total complex, while both indices averaged 0·0639 and 0·0798, respectively, at the population level.
Table 2.
Genetic diversity in C. balansae complex detected by ISSR analysis
| Species | Pop. ID | Ae (s.d.) | HO (s.d.) | HE (s.d.) | P (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| C. shiwandashanica | FC | 1·1179 (0·2831) | 0·0982 (0·2232) | 0·0669 (0·1548) | 17·48 | ||||
| C. parvula | YJ1 | 1·0852 (0·2137) | 1·0690 (0·2092) | 0·0833 (0·1920) | 0·0643 (0·1751) | 0·0538 (0·1278) | 0·0419 (0·1183) | 24·65 | 13·59 |
| YJ2 | 1·0880 (0·2242) | 0·0843 (0·1942) | 0·0546 (0·1301) | 17·48 | |||||
| C. balansae | JP | 1·2278 (0·3581) | 1·1126 (0·2650) | 0·1921 (0·2783) | 0·1013 (0·2154) | 0·1301 (0·1937) | 0·0669 (0·1472) | 34·95 | 20·39 |
| MG | 1·1730 (0·3263) | 0·1491 (0·2529) | 0·0996 (0·1755) | 29·13 | |||||
| PB | 1·1124 (0·2614) | 0·1031 (0·2141) | 0·0675 (0·1458) | 21·36 | |||||
| C. tanqingii | LC | 1·1058 (0·2681) | 0·0890 (0·2137) | 0·0606 (0·1475) | 15·53 | ||||
| C. simplicipinna | ML | 1·0768 (0·2222) | 0·0690 (0·1852) | 0·0458 (0·1260) | 13·59 | ||||
| Total complex | 1·3615 (0·3814) | 0·3166 (0·2837) | 0·2110 (0·1995) | 60·19 | |||||
Ae, the effective number of alleles per locus; HO, Shannon's information index; HE, Nei's genetic index; P, percentage of polymorphic loci.
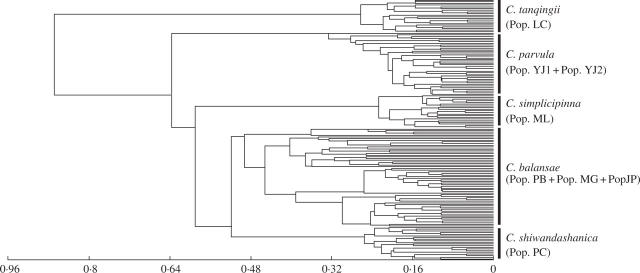
Generally, there is significant differentiation among the populations in this complex. The level of gene flow between populations (Nm) was estimated to be only 0·3303 individuals per generation, indicating that there is a low migration rate between populations. The coefficient of genetic differentiation between populations (GST) was 0·6022, of which only 41 % (0·2486) contributed to the differentiation between populations within the defined species. This finding is corroborated by AMOVA analysis showing that 67·83 % of the total variation was partitioned between populations, of which, 50·60 % was between the defined species (Table 3). Table 4 shows an estimate of Nei's genetic identities (I) and genetic distance (D) for every pairwise comparison between both populations and the defined species. The inter-specific genetic distances were 0·2286 on average, ranging from 0·1282 to 0·3610, which were higher than the intra-specific genetic distances with a mean value of 0·0691, and ranging from 0·0110 to 0·0791. A UPGMA dendrogram based on mean character difference visualized that this complex is grouped into five clusters corresponding to the five species, C. tanqingii, C. parvula, C. simplicipinna, C. shiwandashanica and C. balansae. It also showed that the genetic relationship among the three species, C. balansae, C. shiwandashanica and C. simplicipinnata, with main distribution in Indo-China, is closer than the other two species in south-west China, C. parvula and C. tanqingii.
Table 3.
Nei's (1972) original measures of genetic identity (above diagonal) and genetic distance (below diagonal)
|
C. parvula |
C. balansae |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pop. ID | C. shiwandashanica FC | YJ1 | YJ2 | JP | MG | PB | C. tanqingii LC | C. simplicipinna ML |
| FC | – | 0·8345 | 0·8363 | 0·8783 | 0·8665 | 0·8797 | 0·6970 | 0·8489 |
| YJ1 | 0·1809 | – | 0·9890 | 0·8371 | 0·8036 | 0·8309 | 0·7238 | 0·7689 |
| YJ2 | 0·1787 | 0·0110 | – | 0·8295 | 0·7970 | 0·8294 | 0·7148 | 0·7662 |
| JP | 0·1298 | 0·1779 | 0·1870 | – | 0·9239 | 0·9124 | 0·7252 | 0·8466 |
| MG | 0·1433 | 0·2186 | 0·2269 | 0·0791 | – | 0·9100 | 0·7068 | 0·8606 |
| PB | 0·1282 | 0·1853 | 0·1871 | 0·0917 | 0·0944 | – | 0·7367 | 0·8676 |
| LC | 0·3610 | 0·3233 | 0·3357 | 0·3214 | 0·3470 | 0·3055 | – | 0·7421 |
| ML | 0·1638 | 0·2629 | 0·2664 | 0·1665 | 0·1501 | 0·1420 | 0·2983 | – |
Table 4.
AMOVA for the Cycas balansae complex
| Source of variance | d.f. | Sum of squares | Mean squares | Variance component | % total variance | P value |
|---|---|---|---|---|---|---|
| Among species | 4 | 904·3267 | 226·082 | 6·109 | 50·60 | <0·0010 |
| Among populations within species | 3 | 134·3020 | 44·767 | 2·081 | 17·23 | <0·0010 |
| Within populations | 150 | 582·5611 | 3·884 | 3·884 | 32·17 | <0·001 |
DISCUSSION
Genetic diversity
Walters and Decker-Walters (1991) hypothesized that low within-population genetic variation with relatively high between-population genetic differentiation is a biological and evolutionary characteristic of cycads, a pattern that has been verified by most researchers (Barrett and Kohn, 1991; Ellstrand and Elam, 1993; Xiao et al., 2004). In the present study, every population in the C. balansae complex harbours rather low genetic diversity. This has been ascribed to genetic drift and inbreeding effects due to their small and isolated populations, as well as limited dispersal ability of their large, heavy gravity-dispersed seeds, and beetle-dispersed pollen (Barrett and Kohn, 1991; Ellstrand and Elam, 1993; Xiao et al., 2004). However, the genetic differentiation between populations of different species within the complex presents a striking contrast: Gst in C. parvula is 0·0978, whereas it is 0·4003 in C. balansae (no Gst values were obtained from the remaining species as they were only represented by a single population each). The difference in distances between populations of either species is postulated to be the main reason for the contrast. Gene flow distance between local populations is 2–7 km in cycads (Yang and Meerow, 1996; Xiao et al., 2004). The distance between C. parvula populations is just 7·3 km, while those of C. balansae populations are >30 km, which implies that much gene exchange occurs between the former populations, but is unlikely between the latter populations.
Genetic relationship
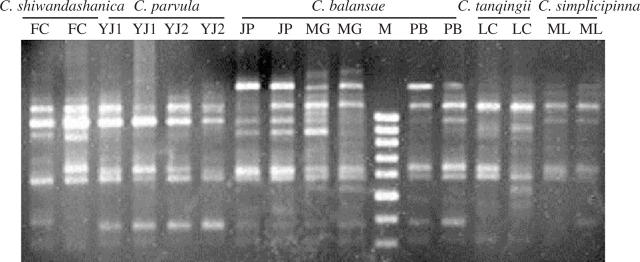
ISSR analysis is helpful to understand the taxonomy and phylogenetic relationships within the C. balansae complex. ISSR banding profiles showed that species-specific markers were detected in each defined species as indicated in Fig. 1. Tables 3 and 4 indicate that the majority of genetic differentiation (59 %) exists among the defined species in this complex. A UPGMA dendrogram, based on mean character difference, showed that the populations clustered into five distinct branches (Fig. 2), which correspond well with the defined species, C. tanqingii, C. parvula, C. simplicipinna, C. shiwandashanica and C. balansae. These indicate that the C. balansae complex has formed five genetically distinct units. The analyses of sclerotesta morphology (D. Y. Wang and J. P. Liao, unpubl. res) and isozyme data (Yang and Meerow, 1999) also indicate that five genetically divergent entities may be separated. Gottlieb (1977) and Crawford (1983, 1990) found that mean genetic identities of conspecific populations were usually above 0·90, whereas the genetic identities of congeneric taxa were usually below 0·7. In the present study, genetic identities of populations in each defined species were higher than 0·90, ranging from 0·9100 to 0·9890, but those between the recognized species were much lower than the conspecific threshold of 0·90 (average 0·8040) (Table 4). Evidence available from the ISSR data show that C. balansae complex is genetically divergent and has evolved into five genetically distinct units, corresponding well to the defined species. Accordingly, a taxonomy is accepted that separates out the five narrowly defined species, C. tanqingii, C. parvula, C. simplicipinna, C. shiwandashanica and C. balansae, at species level. The monophyly of these entities needs to be verified by character-based phylogenetic analyses.
Fig. 1.
ISSR banding profiles obtained on 1.5 % agarose gels with primer USB 843 for 16 individuals representative of eight populations of the five species in C. balansae complex. M = molecular size marker, 100-bp ladder.
Fig. 2.
UPGMA dendrogram based on mean character differences estimated from ISSR data from 158 plants in eight populations in the C. balansae complex.
The phylogeny relationships between the five species in the C. balansae complex have been reconstructed with a set of quantitative morphological traits (Wang and Liang, 1996; Hill and Stevenson, 1998, 1999; Huang, 2001). The UPGMA dendrogram presented here is largely congruent with the phylogeny proposed by Hill and Stevenson, in which, however, C. parvula was mistaken as C. diannanensis, a widely accepted species with tomentose ovules (versus glabrous ovules in C. parvula). Cycas tanqingii is hypothesized to have diverged from the others first, followed by C. parvula, The species mainly distributed in Indo-China, including C. simplicipinna, C. shiwandashanica and C. balansae, were more closely related to each other.
All investigated individuals in the C. balansae complex shared a considerable number of markers (41) indicating that these species form a genetic assemblage. Due to slow reproduction rates (Tang, 1990) and absence of a long-distance dispersal mechanisms for seeds (Bank et al., 2001), it seems reasonable to assume that the species in this complex have been derived from a common ancestor through multiple vicariant events, which arose from geographical isolation resulting from the collision of the Indian plate with the Eurasian plate and from glaciations in the Pleistocene. Morphological cladistics and biogeography indicate that a primary split occurred during cycad evolution between populations in south-west China and those in Indo-China (Hill, 1999) with C. tanqingii and C. parvula in south-west China separating away from the others species mainly in Indo-China. Cycas tanqingii has a characteristic of only two ovules (versus more than two ovules in the remaining species); and C. parvula of small ovules (versus big ovules). Cycas simplicipinna, which is distributed mainly in northern Indo-China and Thailand, has leaflets that become silvery-grey when dry (as opposed to blackish-brown in the other species). Cycas balansae (in north Vietnam) and C. shiwandashanica (in north-east Vietnam), which have a sister relationship, are separated by the Honghe river. Cycas balansae has papery leaflets, whereas those of C. shiwandashanica are leathery.
Conservation implications
Populations and individuals of the C. balansae complex appear to have declined dramatically during recent decades. The two principal threats to cycads are (a) severe habitat loss and (b) selective removal from the wild for trade or utilization due to their popularity among horticulturalists. Accordingly, it is essential to take action to preserve the species and their habitats, which includes setting up nature reserves and protection stations, education for cycad protection and strengthening the co-operation between governments in this region on wild cycad conservation management. The pronounced spatial genetic structure suggests that it is necessary to preserve as many populations as possible and that it is not appropriate to preserve only part of populations, since the low genetic diversity in populations implies a reduced ability to survive changing environments and increased susceptibility to diseases in the long term (Ellstrand and Elam, 1993). If methods to enhance genetic diversity, such as transplant programmes and artificial pollination between populations, are required to promote gene exchange and species survival, genetic contamination must be avoided between genetic units.
Acknowledgments
We thank Drs Xue-Jun Ge and Gang Hao for their help in laboratory work and data analysis, and Dr Mikael Hedrén and two anonymous reviewers for their valuable comments and language correction. This work was supported by the National Natural Science Foundation of China (30070081) and Yunnan Province Natural Science Foundation (2005C0033Q).
LITERATURE CITED
- Barrett SCH, Kohn JK. 1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE, eds. Genetics and conservation of rare plants. New York: Oxford University Press.
- Benharrat H, Veronesi C, Theodet C, Thalouarn P. 2002. Orobanche species and population discrimination using intersimple sequence repeat (ISSR). Weed Research 42: 470–475. [Google Scholar]
- Bnak H, Wink M, Vorster P, Treutlein J, Brand L, Bnak M, Hurter J. 2001. Allozyme and DNA sequence comparisons of nine species of Encephalartos (Zamiaceae). Biochemical Systematics and Ecology 29: 241–266. [DOI] [PubMed] [Google Scholar]
- Brenner ED, Stevenson DW, Twigg RW. 2003. Cycads: evolutionary innovations and the role of plant-derived neurotoxins. Trends in Plant Science 8: 446–452. [DOI] [PubMed] [Google Scholar]
- Chen JR, Stevenson DW. 1999. Cycadaceae. In: Wu CY, Raven P, eds. Flora of China. Beijing: Science Press/Saint Louis, MO: Missouri Botanical Garden Press.
- Crawford DJ. 1983. Phylogenetic and systematic inferences from electrophoretic studies. In: Tanksley SD, Orton TJ, eds. Isozymes in plant genetics and breeding, Part A. Amsterdam: Elsevier, 257–287.
- Crawford DJ. 1990. Plant molecular systematics: macromolecular approaches. New York, NY: Wiley.
- Doyle J. 1991. DNA protocols for plants—CTAB total DNA isolation. In: Hewitt GM, Johnston A, eds, Molecular techniques in taxonomy. Berlin: Springer.
- Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implication for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- Excoffier L. 1993. Analysis of molecular variance (AMOVA), Version 1.5. Geneva: Genetics and Biometry Laboratory, University of Geneva.
- Fang DQ, Krueger RR, Roose ML. 1998. Phylogenetic relationship among selected Citrus germplasm accessions revealed by inter-simple sequence repeat (ISSR) markers. Journal of the American Society of Horticultural Science 123: 612–617. [Google Scholar]
- Fang DQ, Roose ML, Krueger RR, Federici CT. 1997. Fingerprinting trifoliate orange germ plasm accessions with isozymes, RFLPs, and inter-simple sequences repeat marker. Theoretical and Applied Genetics 95: 211–219. [Google Scholar]
- Franke OH, Soule ME. 1981. Conservation and evolution. Cambridge: Cambridge University Press.
- Furman BJ, Grattapagia D, Dvorak WS, O'Malley DM. 1997. Analysis of genetic relationship of Central American and Mexican pines using RAPD markers that distinguish species. Molecular Ecology 6: 321–331. [Google Scholar]
- Gao ZF, Thomas BA. 1989. A review of fossil cycad megasporophylls, with new evidence of Crossozamia pommel and its associated leaves from the lower Permian of Taiyuan, China. Review of Paleobotany and Palynology 60: 205–223. [Google Scholar]
- Gottlieb LD. 1977. Electrophoretic evidence and plant systemtics. Annals of the Missouri Botanical Garden 64: 161–180. [Google Scholar]
- Hahn WJ, Grifo FT. 1996. Molecular markers in plant conservation genetics. In: Sobral BWS, ed. The impact of plant molecular genetics. Boston, MA: Birkhauser.
- Hammond RL, Macasero W, Flores B. 2001. Phylogenetic reanalysis of the Saudi gazelle and its implications for conservation. Conservation Biology 15: 1123–1133. [Google Scholar]
- Hedrén M. 2004. Species delimitation and the partitioning of genetic diversity—an example from the Carex flava complex (Cyperaceae). Biodiversity and Conservation 13: 293–316. [Google Scholar]
- Hill KD. 1999. Cycas—an evolutionary perspective. In: Chen CJ, ed. Biology and conservation of cycads. Proceedings of Fourth International Conference on Cycad Biology. Beijing: International Academic Publishers, 98–115.
- Hill KD. 2003. The families and genera of cycads: a molecular phylogenetic analysis of Cycadophyta based on nuclear and plastid DNA sequence. International Journal of Plant Science 164: 933–948. [Google Scholar]
- Hill KD, Stevenson DW. 1998, 1999. The cycad pages. http://plantnet.rbgsyd.gov.au/PlantNet/cycad/.
- Huang YY. 2001. Systematics and evolution of cycads in China. Beijing: Meteorology Press.
- Martin JP, Sanchez-Yelamo. 2000. Genetic relationship among species of the genus Diplotaxis (Brassicaceae) using inter-simple sequence repeat markers. Theoretical and Applied Genetics 101: 1234–1241. [Google Scholar]
- Moreno S, Martin JP, Ortiz JM. 1998. Inter-simple sequence repeats PCR for characterization of closely related grapevine germplasm. Euphytica 101: 117–125. [Google Scholar]
- Moritz C. 1994. Applications of mitochondrial DNA analysis in conservation: critical review. Molecular Ecology 3: 401–411. [Google Scholar]
- Nei M. 1972. Genetic distance between populations. American Naturalist 106: 283–292. [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norstog KJ, Nicholls TJ. 1997. The biology of the cycads. New York, NY: Cornell University Press.
- Osborne R. 1995. The world cycad census and a proposed revision of the threatened species status for cycad taxa. Biological Conservation 71: 1–12. [Google Scholar]
- Salimath SS, de Oliveira AC, Godwin ID, Bennetzen JL. 1995. Assessment of genome origins and genetic diversity in the genus Eleusine with DNA marker. Genome 38: 757–763. [DOI] [PubMed] [Google Scholar]
- Sharma IK, Jones DL, Forster PI, Young AG. 1998. The extent and structure of genetic variation in the Macrozamia pauli-guilielmi complex (Zamiaceae). Biochemical Systematics and Ecology 26: 45–54. [Google Scholar]
- Sharma IK, Jones DL, Forster PI. 2004. Genetic differentiation and phonetic relatedness among seven species of the Macrozamia plurinervia complex (Zamiaceae). Biochemical Systematics and Ecology 32: 313–327. [Google Scholar]
- Slatkin M, Barton NH. 1989. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368. [DOI] [PubMed] [Google Scholar]
- Tang W. 1990. Maturity in cycads. Encephalartos 24: 24–26. [Google Scholar]
- Walters TW, Decker-Walters DS. 1991. Patterns of allozyme diversity in the West Indies cycad Zamia pumila (Zamiaceae). American Journal of Botany 78: 436–445. [Google Scholar]
- Wang DY. 2000. Studies on morphology, anatomy, taxonomy and evolution of Cycadaceae. PhD Dissertation, Nanjing Forestry University, Nanjing.
- Wang FX, Liang HB, eds. 1996. Cycads in China. Guangzhou: Guangdong Science and Technology Press.
- Xiao LQ, Ge XJ, Gong X, Hao G, Zheng SX. 2004. ISSR variation in the endemic and endangered plant Cycas guizhouensis (Cycadaceae). Annals of Botany 94: 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Meerow AW. 1996. The Cycas pectinata (Cycadaceae) complex structure and gene flow. International Journal of Plant Sciences 157: 468–483. [Google Scholar]
- Yang SL, Meerow AW. 1999. Genetic variation in Chinese cycad population. In: Chen CJ. ed, Biology and conservation of cycads. Proceedings of the Fourth International Conference on Cycad Biology. Beijing: International Academic Publishers, 175–186.
- Yeh FC, Yang RC, Boyle T. 1999. POPGENE. Microsoft Windows-based freeware for population genetic analysis. Release 1.31. Edmonton: University of Alberta.
- Ziekiewicz E, Rafalski A, Labuda D. 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20: 176–183. [DOI] [PubMed] [Google Scholar]