Abstract
The heat shock protein 70 (Hsp70) family of chaperones play crucial roles in protein folding and have been linked to numerous diseases. We were interested in developing a generally applicable assay format for the Hsp70 family and have developed fluorescence polarization based assays for both the mammalian Hsp72 and its bacterial counterpart, DnaK. These assays are comparable in assay set-up, incubation conditions and buffer components. Both unfolded polypeptides and synthetic peptides can be utilized as tracers to detect binding although peptides meeting the minimum seven residue length for Hsp70 binders have weaken binding when modified with fluorescein presumably due to steric effects. Although we did not identify a suitable general substrate for all Hsp70 proteins, fluorescein tagged peptide substrates that gave high affinity binding were identified for both DnaK and hsp72. We would predict that these assays will be suitable for identifying both selective chemical probes of Hsp70 family members and “pan” Hsp70 inhibitors.
INTRODUCTION
Heat shock protein 70 (Hsp70) chaperones are highly conserved both structurally and functionally among different species [1]. Family members including mammalian Hsp70 and its bacterial ortholog DnaK, play important housekeeping roles in the cell assisting in protein folding and trafficking [2]. During stress conditions, Hsp70 proteins help prevention of insoluble aggregates by transiently binding to exposed hydrophobic regions of denatured or misfolded proteins. The mammalian Hsp70 family consist of multiple members [3] with the best characterized including a constitutive form (hsc70/Hsp73) and a stress-inducible form (hsp70/Hsp72)(i). A role for Hsp70 proteins in many therapeutic areas including autoimmunity, transplantation, viral infections, neurodegenerative diseases and cancer has been well documented [2, 4-6]. Induction of Hsp70 proteins in response to environmental stresses protects cells from apoptosis and such a mechanism is frequently observed in cancer cells [7]. The inducible Hsp72 is often constitutively expressed in cancer cells [8].
Hsp70 proteins consist of two domains, an ATP-binding domain and a polypeptide-binding domain [3]. Substrates bind with high affinity to the ADP-bound form of Hsp70 proteins and conversion of ADP to ATP results in rapid substrate release [9, 10]. Co-chaperones mediate the hydrolysis of ATP on Hsp70 converting it back to the high affinity ADP-bound state. Hsp70 proteins bind to short exposed hydrophobic stretches on substrate proteins. Hsp70 activity has been observed in vitro by incubating purified Hsp70 with unfolded substrates and measuring binding. Due to their function in binding a wide range of protein sequences, defining the peptide specificities of Hsp70 chaperones has been challenging and synthetic peptide and/or phage display libraries have been used extensively to map the binding requirements for various Hsp70 family members including DnaK [11-13], Hsc70 [14, 15] and Hsp70 [16, 17]. Linear peptides with a minimum peptide length of seven amino acids are required for binding [14, 17, 18]. The interaction of Hsp70 with their substrate peptides or unfolded polypeptides is a relatively weak interaction and for certain peptides Kd values in the submicromolar range have been reported [17, 19, 20].
Although bacterial DnaK and mammalian Hsp70 proteins show a high degree of homology (45-50% amino acid similarity) and share the same overall structure [21], there are reported differences in their peptide binding selectivity’s towards different peptides sequences [17, 19-21]. DnaK has a preference for a peptide binding motif containing a hydrophobic core sequence of 4-5 residues flanked by basic residues [12]. Hsp72 has a preference for peptides enriched in aromatic and hydrophobic amino acids with less binding to charged resides [16]. Other reports suggest a preference for charged peptides containing Lys or Arg [22] and no hydrophobic residues. These differences may reflect close or overlapping binding pockets for substrate [22].
Despite the interest in modulating Hsp70 activity there is a distinct lack of chemical probes or inhibitors. Hsc70 has been demonstrated to bind to certain immunosuppressive drugs such as 15-deoxyspergualin (DSG) [23] with moderate affinity (Kd = 0.5 µM) although the binding site for DSG seems to be distinct from the peptide binding site [24]. Peptides derived from HLA Class I heavy chain have also been shown to have immunosuppressive properties and to bind to Hsp70 proteins [25, 26]. Compounds targeting the ATP domain of Hsp70 such as 3’-sulfogalactolipids [27] have also been identified. Interestingly, a peptide encoding the minimum region of a Hsp70 substrate bound Hsp72 preferentially compared to Hsc70 [8]. For DnaK, some compounds binding to the substrate site display antibacterial activity [28]. Taken together these studies suggest differences in binding selectivity’s of Hsp70 family members and that a number of potential binding sites/pockets exist on Hsp70 proteins including; a hydrophobic substrate site, a charged substrate site, the ATPase site, a “DSG-site” and the co-chaperone binding site that could all potentially be modulated with chemical probes or inhibitors.
A number of approaches have been taken to measure Hsp70 activity and develop assays that could be utilized to identify chemical probes/inhibitors. We have previously developed polyacrylamide gel-based [29] and chromatographic-based [20] assays to assess the binding of peptide substrates by Hsp70 family members. Typically these assays are qualitative in nature and not robust enough for high throughput screening. HTS-amenable assays based on modulating the ATPase activity of DnaK [30] and fluorescence polarization (FP) assays for peptide binding to Hsp70 [31] have recently been reported.
In order to identify inhibitors with varying selectivity for Hsp70 family members we have taken a chemical genomics approach as has been taken for other target classes such as kinases [32] and begun to develop a panel of Hsp70 assays using an assay format based on fluorescence polarization and testing a range of potential substrates.
(i) Hsp70 refers to all members of the Hsp70 family; Hsc70 is the constitutive from of mammalian Hsp70; Hsp72 is the mammalian inducible form of Hsp70.
MATERIALS AND METHODS
Materials
Recombinant human heat shock protein 72 was either expressed in E.coli and purified as described previously [29] or purchased from Sigma (H7283). DnaK was obtained from Stressgen (SPP-630). Labeled and unlabeled synthetic peptides were synthesized using conventional Fmoc chemistry (Applied Biosystems Corp. and Anaspec Inc.). Prior to deprotection the resin for each peptide was divided into two portions. One portion of the resin was deprotected and cleaved as normal. The remainder of the resin was reacted with carboxyfluorescein succinimidyl ester (Invitogen) using a 20-fold molar excess. Unreacted NHS-fluorescein was washed away and the labeled peptide deprotected and cleaved as normal. Masses of peptides were confirmed by mass spectrum and HPLC. A model unfolded polypeptide, reduced and carboxymethylated lactabumin (RCMLA; Sigma) was labeled in solution with NHS-fluorescein (Invitrogen) as described previously [29]. MALDI-TOF MS analysis of the fluorescein labeled RCMLA indicated that there were on average three fluorescein groups per RCMLA molecule (data not shown).
Fluorescence Polarization Measurements
All assay and fluorescence polarization (FP) measurements were performed at room temperature and with a 10 min incubation. The assay buffer used consisted of 25 mM HEPES pH 7.2, 150 mM KCl. Binding was assessed by incubating increasing concentrations of a Hsp70 protein with a fixed concentration of fluorescently-labeled substrate and FP values measured. FP measurements were performed either on a Beacon Fluorescence Polarization system (Invitrogen) or PHERAstar microreader (BMG Labtech) using the FP filter set (emission 450 and excitation 520 nm). FP values are expressed in millipolarization (mP) units. All statistical analyses were performed with GraphPad Prism 5.0 software. Binding data was analyzed using a non-linear regression analysis method (single site binding model) in Prizm 5.0.
RESULTS AND DISCUSSION
Substrates for Hsp70 Family Members DnaK and Hsp72
We are interested in developing assays for a range of Hsp70 protein family members. As the best characterized, we began with mammalian Hsp72 and bacterial DnaK. Fluorescence polarization (FP) was chosen as the assay format as it has been generally applicable to measuring protein-protein or protein-ligand interactions [33] including other heat shock proteins [31, 34]. The advantages of FP include that it is a homogeneous format, utilizes small substrates and the assays are generally insensitive to fluorescence interference.
Unfolded Polypeptide Substrate
To provide a valuable readout, the FP substrate should bind specifically and with high affinity to the target protein. Substrate binding was first assessed using the permanently unfolded model substrate, reduced and carboxymethylated lactalbumin (RCMLA). We have previously shown in a gel shift assay format that both DnaK and Hsp72 can bind to RCMLA [29]. RCMLA was fluorescently labeled with fluorescein (Flu-RCMLA) and binding to Hsp72 and DnaK assessed as described in materials and methods. The data for Hsp72 binding Flu-RCMLA is shown in Fig. (1).
Fig. (1). Titration of Hsp72 with the model unfolded polypeptide RCMLA.
Various concentrations of Hsp72 were incubated with (1.67 µg/ml) fluorescein-labeled RCMLA (flu-RCMLA) for 10 min at room temperature and FP values measured.
The data in Fig. (1) yielded an apparent Kd value of 1.53 µM. A comparable value (apparent Kd value of 2.0 µM) was obtained for DnaK (data not shown). Although Flu-RCMLA could potentially fulfill the requirement for a universal Hsp70 substrate, the relatively low affinity, limited assay window of ~ 20 mP and the challenge in consistently preparing the substrate precluded its use. For these reasons we decided to look to small peptide substrates to facilitate Hsp70 assays.
Synthetic Peptide Substrates
Due to their function in binding a wide range of protein sequences, defining the peptide specificities of Hsp70 chaperones has been challenging. A number of groups have used synthetic peptides to map such motifs [12-14, 16-20, 22, 35]. For DnaK the bacterial form of Hsp70, a 21-mer peptide comprising the targeting sequence of the precursor of mitochondrial aspartate aminotransferase (ALLQSRLLLSAPRRAAATARA; peptide name ALLQ, Table 1) had been previously identified [11] as binding tightly. It was also shown [11] that this peptide could be fluorescently tagged on the N-terminus and used to follow real time binding by monitoring changes in fluorescence upon peptide binding.
Table 1.
Peptide Sequences Used as Hsp70 Substrates Protein/Tracer Optimization
Because of the reported relatively high affinity of this peptide for DnaK (63 nM) [11] we first used this sequence (Table 1) as the basis for developing a fluorescence polarization assay for measuring peptide binding to DnaK and potentially to other Hsp70 proteins. The ALLQ peptide was labeled with NHS-fluorescein on the N-terminus on resin (Flu-ALLQ). Because labeling is done prior to deprotection only the N-terminal amine is labeled. This is especially advantageous for labeling of lysine/arginine containing peptides.
A second peptide containing predominantly lysine and arginine residues (NIVRKKK; peptide name KKK) previously shown to bind Hsp70 proteins [17] was also labeled on-resin using NHS-fluorescein. Peptide substrates used in these studies are shown in Table 1.
The optimal trace concentration was determined for each fluorescent-peptide. Each peptide was titrated and the fluorescence intensity measured. The ratio of fluorescence intensity to background was plotted versus tracer concentration. For optimal sensitivity, the fluorescently labeled peptide tracer concentration was set to produce a total fluorescence signal-to-background ratio greater than 50. For example, for Flu-ALLQ this required a concentration of 50 nM (data not shown) and 10 nM for Flu-KKK (Fig. 2).
Fig. (2). Tracer Optimization.
The signal-to-background was calculated as the fluorescence intensity in the presence of labeled Flu-KKK divided by the fluorescence intensity of buffer.
Fluorescence Polarization Assays for DnaK and Hsp72 DnaK FP Assay
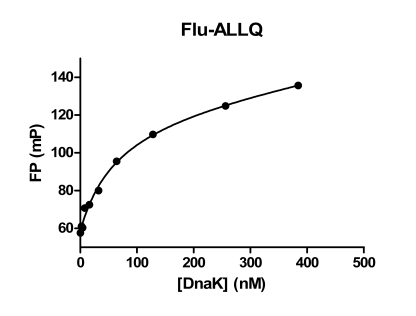
Binding of DnaK to a fixed concentration of Flu-ALLQ (50 nM) was determined. For this experiment it was determined that an incubation time of 10 min at room temperature was sufficient to reach equilibrium. A binding isotherm constructed by plotting DnaK concentration against millipolarization (mP) values shows that DnaK binds in a dose-dependent manner. As the concentration of DnaK is increased mP values increase until saturation is reached (Fig. 3). The assay window reached approximately 70 mP.
Fig. (3). Binding Isotherms for Flu-ALLQ to DnaK.
Increasing concentrations of DnaK were incubated with Flu-ALLQ at room temperature for 10 min and the FP values measured. Data were plotted in GraphPad Prizm 5.0.
The data in Fig. (3) was analyzed using a non-linear regression analysis method (single site binding model) in Prizm 5.0 and yielded an apparent Kd value of 64.4 nM, very close to that reported previously [11]. A Scatchard plot transformation of the same data (not shown) yielded a value of 53.7 nM for “target concentration” (x intercept) indicating stoichiometric 1:1 binding of peptide substrate ligand and DnaK in this FP assay.
It has been shown that Hsp70 members associate with polypeptide substrates or peptides in the presence of ADP whereas the ATP bound form of Hsp70 proteins have poor affinity for substrates [10]. We tested the effect of ADP and ATP on the FP assay. The addition of ADP to the assay did not affect the binding of labeled substrates (data not shown) suggesting that the Hsp72 proteins as tested exist in a predominantly ADP-bound form as a result of their purification using ATP affinity chromatography [29]. For DnaK binding to Flu-ALLQ, no peptide binding was measurable in the presence of 3 mM ATP/1 mM MgCl2 (data not shown) suggesting that this FP assay can detect conformational changes in DnaK.
Competition Binding Assay for DnaK
Using the information obtained in the binding isotherm experiments, we next developed a competition binding assay in which tracer and protein concentrations are fixed and increasing concentrations of compounds are added and assessed for inhibition. DnaK and Flu-ALLQ concentrations were fixed at 200 nM and 50 nM respectively to give ~90% binding. Compounds can then be added over a range of concentrations and mP values measured after incubation.
To further validate the competitive DnaK FP assay, the IC50 for a known DnaK binding peptide was to be determined. In the absence of any known DnaK binding small molecules, another peptide previously identified as a DnaK binder was assessed in this assay. The peptide with sequence NRLLLTG (Table 1; termed LLL) was previously identified as a DnaK binder from phage display screening [35] and has been well characterized as a DnaK binder [37-39]. The Kd for LLL mediated stimulation of DnaK ATPase was previously reported as 10 µM [35]. LLL was analyzed in our FP competition assay. Results are shown in Fig. (4).
Fig. (4). Competitive displacement assay for LLL binding to DnaK.
Assays were performed in triplicate with 200 nM DnaK and 50 nM Flu-ALLQ. Various concentrations of the LLL peptide were added and FP values measured after 10 min. The average values and standard deviation are plotted and the curve was fitted in GraphPad Prizm 5.0.
The LLL peptide inhibited Flu-ALLQ binding to DnaK, with an IC50 value of 2.4 µM comparable to that reported previously [35], and an R-squared value of 0.97 for curve fit. A preliminary analysis of the applicability of the assay for high throughput screening (HTS) was assessed using the maximum assay window from the data in Fig. (4). A Z’ value [36] of 0.64 was calculated from this data suggesting that this assay can be applied to HTS.
Hsp72 FP Assay
Next we assessed the ability of another Hsp70 protein, Hsp72 to also bind to the peptide substrate Flu-ALLQ. A binding isotherm of the data (Fig. 5) demonstrates that Hsp72 also binds Flu-ALLQ in a dose-dependent manner.
Fig. (5). Differential binding of DnaK and Hsp72 to Flu-ALLQ.
Various concentrations of DnaK and Hsp72 protein were each incubated with the Flu-ALLQ peptide (50 nM) and FP values measured.
Non-linear regression analysis yielded an apparent Kd value of 621.6 nM indicating that Hsp72 binds this peptide with ~10-fold weaker affinity than DnaK. Interestingly, using a smaller fragment of ALLQ, comparable values for binding to DnaK and Hsc70 were observed: 0.43 µM to Hsc70 [31] and 0.4 µM to DnaK [40]. Whether this reflects differences in Hsp72 and Hsc70 binding or differences in the lengths of the peptides in mediating binding requires further investigation.
For Hsp72, a seven residue peptide (NIVRKKK, termed KKK) was previously identified as a Hsp70 binder [14, 17] and shown to compete for RCMLA binding to Hsp72 [29]. KKK was labeled with fluorescein and also used to measure direct binding to Hsp72 in an FP assay (Fig. 6).
Fig. (6). Binding isotherms for Hsp72 binding to unfolded polypeptide and synthetic peptide substrates.
Various concentrations of Hsp72 protein were incubated with the Flu-KKK (20 nM) and Flu-RCMLA (1.67 µg/ml) and FP values measured.
Binding isotherms for Hsp72 binding to Flu-KKK and with Flu-RCMLA as a control (Fig. 6) yield apparent Kd values of 13.7 µM and 1.97 µM respectively. An IC50 value for KKK binding to Hsc70 of 2.8 µM has been reported previously [14]. These differences in binding for the KKK peptide may reflect different selectivity’s of Hsp72 and Hsc70 or may suggest that fluorescein labeling of this minimal binding sequence of seven residues may have steric effects on binding. The low affinity of the “charged” Hsp70 binder makes it a poor tracer to monitor binding and challenging to use in developing an assay to identify chemical probes.
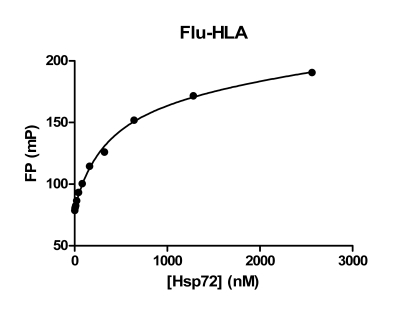
Another set of peptides have been identified as Hsp70 binders. Peptides derived from the MHC class I antigens have been shown to bind to Hsc70 and also have immunosuppressive properties [25,26]. We tested one of these peptides from HLA-B2702 (residues 75-84; RENLRIALRY; Table 1) in an FP assay. This 10-mer peptide was labeled on the N-terminus with fluorescein (termed Flu-HLA) and used as a tracer to measure Hsp72 binding (Fig. 7).
Fig. (7). Binding isotherm for a MHC class I-derived peptide sequence to Hsp72.
Various concentrations of Hsp72 protein were incubated with the Flu-HLA peptide (20 nM) and FP values measured.
Non-linear regression analysis of the data in Fig. (7) yielded an apparent Kd value of 0.37 µM for Flu-HLA binding to Hsp72. Using a biotinylated version of this peptide as tracer in an ELISA type competition binding assay, Woo and coworkers measured an IC50 value of 7 µM for binding of this peptide to Hsc70 [26]. These differences in binding may reflect differences in assay format or that the HLA peptide has higher affinity for Hsp72 versus Hsc70.
CONCLUSIONS
Our initial goal in these studies was to test the feasibility in developing a robust assay format that would be generally applicable to all members of the Hsp70 chaperone family. We have shown that fluorescence polarization proves to be an effective format for assessing both unfolded polypeptide and peptide substrate binding to a range of Hsp70 proteins including the mammalian inducible form Hsp72 and bacterial DnaK. A summary of the binding data is shown in Table 2.
Table 2.
Summary of Substrate Binding to Hsp70 Proteins
| Hsp70 Protein | Hsp72 | Dnak |
|---|---|---|
| Kd (µM) | ||
| Substrate | ||
| Flu-RCMLA | 1.53 | 2.0 |
| Flu-ALLQ | 0.62 | 0.064 |
| Flu-KKK | 13.7 | ND |
| Flu-HLA | 0.37 | ND |
ND = not determined.
One question we wanted to address was whether we would be able to use a single general substrate for all Hsp70 proteins or would individual substrates be required for each chaperone. We found that a 21-mer peptide Flu-ALLQ bound with high affinity to DnaK and as a tracer, inhibition by another DnaK binding peptide, LLL could be measured. However, binding of this peptide to Hsp72 was ~10-fold weaker suggesting it may not be useful as a general substrate even though a smaller fragment of this peptide has shown comparable binding to DnaK and Hsp70 [31]. Here we show that a 10-mer peptide derived from HLA binds well to Hsp72 compared to other peptides tested and should serve as a tracer for an Hsp72 binding assay. In conclusion, we have identified fluorescein tagged synthetic peptide substrates based on the targeting sequence of the precursor of mitochondrial aspartate aminotransferase and on a HLA Class I heavy chain sequence that gave high affinity binding for DnaK and Hsp72 respectively. We anticipate that these FP assays for Hsp70 proteins will be implemented in an HTS format for the discovery of both selective and pan-Hsp70 chemical probes. It will be interesting to test if screens of Hsp72 and DnaK yield similar or diverse hits.
ACKNOWLEDGEMENTS
This work was supported in part by funding from the Golden LEAF Foundation and the BIOIMPACT Initiative of the State of North Carolina through the Biomanufacturing Research Institute & Technology Enterprise (BRITE) Center for Excellence at North Carolina Central University. The authors would also like to thank Dr. Satish Jindal (Schering Plough Corp.).
REFERENCES
- 1.Gupta R, Golding G. Evolution of Hsp70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–82. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 2.Jindal S. Heat shock proteins: Applications in health and disease. Trends Biotechnol. 1996;14:17–20. doi: 10.1016/0167-7799(96)80909-7. [DOI] [PubMed] [Google Scholar]
- 3.Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–10. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 4.Calderwood S, Khaleque M, Sawyer D, Ciocca D. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–72. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky J, Chiosis G. Hsp70 molecular chaperones: Emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6:1215–25. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 6.Rohde M, Daugaard M, Jensen M, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–82. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didelot C, Lanneau D, Brunet M, et al. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr Med Chem. 2007;14:2839–47. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt E, Maingret L, Puig P, et al. Heat shock protein 70 neutralization exerts potent antitumor effects in animal models of colon cancer and melanoma. Cancer Res. 2006;66:4191–7. doi: 10.1158/0008-5472.CAN-05-3778. [DOI] [PubMed] [Google Scholar]
- 9.Buchberger A, Theyssen H, Schröder H, et al. Nucleotide-induced conformational changes in the ATPase and Substrate Binding Domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995;270:16903–10. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- 10.Wei J, Gaut J, Hendershot L. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–82. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 11.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–73. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 12.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–7. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega C, Kurt N, Chen Z, Rüdiger S, Cavagnero S. Binding specificity of an-helical protein sequence to a full-length Hsp70 chaperone and its minimal substrate-binding domain. Biochemistry. 2006;45:13835–46. doi: 10.1021/bi061432a. [DOI] [PubMed] [Google Scholar]
- 14.Takenaka I, Leung S, McAndrew S, Brown J, Hightower L. Hsc70-binding peptides selected from a phage display peptide library that resemble organellar targeting sequences. J Biol Chem. 1995;270:19839–44. doi: 10.1074/jbc.270.34.19839. [DOI] [PubMed] [Google Scholar]
- 15.Fourie A, Hupp T, Lane D, et al. HSP70 binding sites in the tumor suppressor protein p53. J Biol Chem. 1997;272:19471–9. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- 16.Hightower L, Sadis S, Takenaka I. Interactions of vertebrate hsc70 and hsp70 with unfolded proteins and peptides. The Biology of Heat Shock Proteins Mol Chaperones. 1994;26:179–207. [Google Scholar]
- 17.Fourie A, Sambrook J, Gething M. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994;269:30470–8. [PubMed] [Google Scholar]
- 18.Flynn G, Pohl J, Flocco M, Rothman J. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–30. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 19.Gragerov A, Gottesman M. Different peptide binding specificities of Hsp 70 family members. J Mol Biol. 1994;241:133–5. doi: 10.1006/jmbi.1994.1482. [DOI] [PubMed] [Google Scholar]
- 20.Williams KP, Evans DM, Rosenberg S, Jindal S. Use of a peptide library to characterize differential peptide binding specificities of bacterial and mammalian Hsp70. In: Marshak DR, editor. Techniques in Protein Chemistry VII. New York: Academic Press; 1996. pp. 57–64. [Google Scholar]
- 21.Erbse A, Mayer MP, Bukau B. Mechanism of substrate recognition by Hsp70 chaperones. Biochem Soc Trans. 2004;32:617–21. doi: 10.1042/BST0320617. [DOI] [PubMed] [Google Scholar]
- 22.Crouy-Chane A, Kohiyama M, Richarme G. Specificity of DnaK for arginine/lysine and effect of DnaJ on the amino acid specificity of DnaK. J Biol Chem. 1996;271:15486–90. doi: 10.1074/jbc.271.26.15486. [DOI] [PubMed] [Google Scholar]
- 23.Nadler S, Tepper M, Schacter B, Mazzucco C. Interaction of the immunosuppressant deoxyspergualin with a member of the Hsp70 family of heat shock proteins. Science. 1992;258:484–6. doi: 10.1126/science.1411548. [DOI] [PubMed] [Google Scholar]
- 24.Wisén S, Gestwicki J. Identification of small molecules that modify the protein folding activity of heat shock protein 70. Anal Biochem. 2008;374:371–7. doi: 10.1016/j.ab.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Krensky A, Clayberger C. HLA-derived peptides as novel immunotherapeutics. Clin Immunol Immunopath. 1995;75:112–6. doi: 10.1006/clin.1995.1059. [DOI] [PubMed] [Google Scholar]
- 26.Woo J, Iyer S, Cornejo M, et al. Immunosuppression by D-isomers of HLA class I heavy chain (amino acid 75 To 84)-derived peptides is independent of binding to Hsc70. Transplantation. 1997;64:1460–7. doi: 10.1097/00007890-199711270-00015. [DOI] [PubMed] [Google Scholar]
- 27.Mamelak D, Mylvaganam M, Whetstone H, et al. Hsp 70 contains a specific sulfogalactolipid binding site. Differential aglycone influence on sulfogalactosyl ceramide binding by recombinant prokaryotic and eukaryotic Hsp 70 family members. Biochemistry. 2001;40:3572–82. doi: 10.1021/bi001643u. [DOI] [PubMed] [Google Scholar]
- 28.Liebscher M, Jahreis G, Lucke C, Grabley S, Raina S, Schiene-Fischer C. Fatty Acyl benzamido antibacterials based on inhibition of DnaK-catalyzed protein folding. J Biol Chem. 2007;282:4437–46. doi: 10.1074/jbc.M607667200. [DOI] [PubMed] [Google Scholar]
- 29.Jindal S, Murray P, Rosenberg S, Young R, Williams KP. Human stress protein hsp 70: Overexpression in E.coli, purification and characterization. Biotechnology. 1995;13:1105–9. doi: 10.1038/nbt1095-1105. [DOI] [PubMed] [Google Scholar]
- 30.Chang L, Bertelsen E, Wisén S, Larsen E, Zuiderweg E, Gestwicki J. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–6. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Kang Y, Taldone T, Clement C, Fewell S, Aguirre J, Brodsky J, Chiosis G. Design of a fluorescence polarization assay platform for the study of human Hsp70. Bioorg Med Chem Lett. 2008;18:3749–51. doi: 10.1016/j.bmcl.2008.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janzen W, Bernasconi P, Cheatham L. Optimizing the chemical genomics process. In: Darvas F, Guttman A, Dorman G, et al., editors. Chemical Genomics. New York: Marcel Dekker; 2004. pp. 59–100. [Google Scholar]
- 33.Jameson D, Croney J. Fluorescence polarization: Past, present and future. Comb Chem High Through Screen. 2003;6:167–76. doi: 10.2174/138620703106298347. [DOI] [PubMed] [Google Scholar]
- 34.Du Y, Moulick K, Rodina A, et al. High-throughput screening fluorescence Polarization assay for tumor-specific Hsp90. J Biomol Screen. 2007;12:915–24. doi: 10.1177/1087057107306067. [DOI] [PubMed] [Google Scholar]
- 35.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman M. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–54. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Thomas D, Chung Y, Oldenburg KR. A Simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–74. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 37.Stevens S, Cai S, Pellecchia M, Zuiderweg E. The solution structure of the bacterial HSP70 chaperone protein domain DnaK (393-507) in complex with the peptide NRLLLTG. Protein Sci. 2003;12:2588–96. doi: 10.1110/ps.03269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X, Zhao X, Burkholder W, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–14. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swain J, Schulz E, Gierasch L. Direct Comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J Biol Chem. 2006;281:1605–11. doi: 10.1074/jbc.M509356200. [DOI] [PubMed] [Google Scholar]
- 40.Han W, Christen P. cis-Effect of DnaJ on DnaK in ternary complexes with chimeric DnaK/DnaJ-binding peptides. FEBS Lett. 2004;563:146–50. doi: 10.1016/S0014-5793(04)00290-X. [DOI] [PubMed] [Google Scholar]