Abstract
• Background Heat shock transcription factors (Hsfs) are modular transcription factors encoded by a large gene family in plants. They bind to the consensus sequence ‘nGAAnnTCCn’ found in the promoters of many defence genes, and are thought to function as a highly redundant and flexible gene network that controls the response of plants to different environmental stress conditions, including biotic and abiotic stresses. Hsf proteins encoded by different genes exhibit a high degree of complexity in their interactions. They can potentially bind and activate their own promoters, as well as the promoters of other members of their gene family, and they can form homo- or heterotrimers resulting in altered nuclear localization, as well as enhanced or suppressed transcription.
• Scope In this review, we summarize recent studies on Hsf function in Arabidopsis and tomato and present evidence obtained from microarray expression studies in Arabidopsis that the Hsf gene network is highly flexible and specialized, with specific members and/or member combinations controlling the response of plants to particular stress conditions. In addition, we describe recent studies that support the hypothesis that certain Hsfs function as molecular sensors that directly sense reactive oxygen species (ROS) and control the expression of oxidative stress response genes during oxidative stress.
Keywords: Heat shock transcription factors (Hsfs), reactive oxygen species (ROS), abiotic stress, gene network, sensor
INTRODUCTION
The heat shock (HS) response is a highly conserved response, characterized by rapid induction of heat shock proteins (HSPs) that primarily function as molecular chaperones to ensure the correct function of many cellular proteins under conditions of elevated temperature. The HS response was shown to be controlled by heat shock transcription factors (Hsfs) that act by binding to the highly conserved heat shock element (HSE; a palindromic motif of nGAAn) in the promoters of target genes. A key step in the activation process of Hsfs, in response to different stress conditions, involves the formation of homotrimers with high affinity for the HSE. In addition to mediating a relatively large part of the defence response of eukaryotes to heat stress, Hsfs are also thought to be involved in different pathological conditions, cellular responses to oxidative stress, heavy metals, amino acid analogues and metabolic inhibitors, and certain developmental and differentiation processes (Sorger and Pelham, 1988; Park and Craig, 1989; Jedlicka et al., 1997; Morimoto, 1998; Hahn et al., 2004).
An intimate relationship appears to exist between oxidative stress and the HS response (Liu and Thiele, 1996; McDuffe et al., 1997; Ahn and Thiele, 2003). When the HS response was first identified in Drosophila by Ritossa et al. in 1962, it was also shown to be induced during recovery from anoxia, which results in oxidative stress. Heat stress was shown to cause impairments in mitochondrial functions that result in the induction of oxidative damage (Davidson and Schiestl, 2001; Larkindale and Knight 2002; Vacca et al., 2004). In plants, the steady-state transcript and protein level of many reactive oxygen species (ROS)-scavenging enzymes was found to be elevated by heat stress (Rainwater et al., 1996; Sato et al., 2001; Rizhsky et al., 2002; Mittler et al., 2004; Vacca et al., 2004). In addition, acquired thermotolerance, i.e. the ability of plants to develop heat tolerance following a mild heat pre-treatment, was shown to be mediated in part by enhancing cellular mechanisms that prevented oxidative damage under heat stress (Bergmüller et al., 2003; Larkindale and Huang, 2004). HS leading to programmed cell death in plants was also shown to be associated with an enhanced production of ROS and the activation of the oxidative burst (Vacca et al., 2004). The intimate relationship between the HS and oxidative stress responses, the activation of Hsfs during these processes, and recent genetic and biochemical studies (described below) suggest that Hsfs might function as direct sensors of hydrogen peroxide in plants. In this review, we will summarize evidence supporting this hypothesis, as well as propose a model for the function of the Hsf network in plants.
EUKARYOTIC HSF GENES
Yeast and Drosophila contain only one Hsf gene, while vertebrates have four Hsfs. In contrast, plants show a much higher complexity, with Hsf genes comprising whole networks of approx. 18 (tomato) to 34 (soybean) Hsf genes (Nover et al., 1996, 2001; Kotak et al., 2004). The yeast Hsf protein was shown to be vital for survival. It constitutively binds the HSE and displays a basal level of transcriptional activation required under normal conditions. The yeast Hsf gene is therefore involved in many aspects of cell functioning such as protein degradation, carbohydrate metabolism and energy generation (Sorger and Pelham, 1988; Yamamoto et al., 2005). In contrast to yeast, Drosophila Hsf is required for early developmental stages, but is dispensable for general cell growth and viability (Jedlicka et al., 1997). In vertebrates, that have four Hsf proteins, Hsf1 is the primary Hsf activated during HS (Sarge et al., 1993). It is, however, not essential for survival under normal conditions. In addition to Hsf1, Hsf3 is also inducible during HS. Interestingly, the four vertebrate Hsfs show diverse regulatory responses to a wide spectrum of environmental and developmental signals, demonstrating a high degree of complexity exhibited by a relatively small gene family (Rabindran et al., 1991; Sarge et al., 1991; Schuetz et al., 1991; Nakai and Morimoto, 1993; Nakai et al., 1997; Morimoto, 1998; Tanabe et al., 1998).
The model plant Arabidopsis thaliana contains 21 Hsf genes, as well as several genes encoding Hsf-like proteins. More than 16 Hsf genes are found in tomato, and many other Hsf genes were identified in rice, maize and other species. Plant Hsf genes were assigned to three different classes (classes A, B and C) according to their unique structural characteristics (Nover et al., 2001). Class A HSF proteins comprise the largest group of Hsfs with 15 proteins in Arabidopsis. They contain an activation domain at the C-terminus and are thought to be involved in transcriptional activation. Class B and class C Hsfs lack a defined aromatic/hydrophobic/acidic (AHA)-type activation domain. The absence of an activation domain, as well as their inability to rescue the yeast Hsf1 mutation, has led to the assumption that class B Hsfs function as repressors (Boscheinen et al., 1997; Czarnecka-Verner et al., 2000, 2004). However, HsfB1 was recently demonstrated to function as a novel co-regulator of the tomato HsfA1 or HsfA2 enhancing their transcriptional activity (Bharti et al., 2004).
HSF STRUCTURE
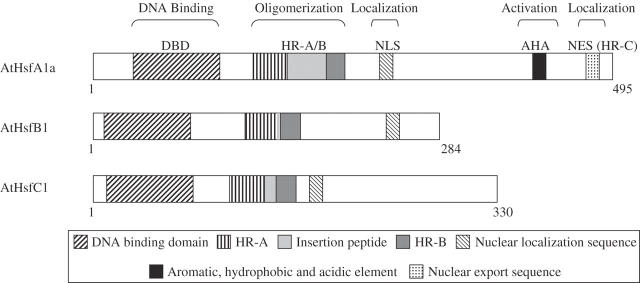
Hsfs have a modular structure, conserved among eukaryotes (Fig. 1). All Hsfs contain a DNA-binding domain (DBD), an oligomerization domain, a nuclear localization sequence (NLS) and in most cases a nuclear export sequence (NES) (Nover et al., 1996, 2001). The DBD is the most conserved domain among eukaryotic Hsfs. It contains a helix–turn–helix motif (H2–T–H3), that allows the specific recognition and binding of the palindromic HSEs (Damberger et al., 1994; Harrison et al., 1994; Vuister et al., 1994; Schulthsiss et al., 1996). The DBD of all plant Hsf genes contains an intron, located immediately downstream of the H2–T–H3 motif (Nover et al., 2001). Downstream of the DBD and separated by a flexible linker peptide which is variable in size, two adjacent hydrophobic heptad repeats (HR-A and HR-B) comprise the oligomerization domain. It is thought that the primary role of the HR-A/B is to provide hydrophobic surfaces for Hsf trimerization (Zuo et al., 1994).
Fig. 1.
Arabidopsis Hsf protein structure. A model representing the structure of Arabidopsis HsfA1a (A-Class Hsf), HsfB1 (B-Class Hsf) and HsfC1 (C-Class Hsf) is shown. The different domains are indicated on the top and different structural features present in each domain are outlined below. The models presented in the figure were generated according to Nover et al. (2001).
The three classes of plant Hsf genes were defined based on peculiarities in the flexible linker and the HR-A/B domains (Nover et al., 2001). Class A, that comprise the largest group, contain a C-terminal activation domain (CTAD), that is the least conserved in sequence and size (Nover et al., 2001). The function of the class A Hsfs as transcriptional activators depends on a short AHA motif found in the CTAD, enriched with aromatic, large hydrophobic and acidic amino acids (Nover and Scharf, 1997; Doring et al., 2000). AHA domains are crucial for the interaction of Hsfs with the transcriptional machinery, as was shown by pull-down experiments and functional analysis of class A Hsfs (Yuan and Gurley, 2000; Kotak et al., 2004). Another hydrophobic repeat (HR-C) is found at the C-terminal domain (Nover et al., 2001). This region is conserved among animal Hsfs but it is poorly conserved in yeast and plant Hsfs (Wu, 1995). The NES is a leucine-rich export signal found at the C-terminus of the CTAD. The subcellular distribution of Hsfs is dictated by the balance between nuclear import and nuclear export as determined by the relative strengths of the NLS and NES, and possibly by intermolecular interactions between different Hsf monomers (Gorlich and Kutay, 1999; Heerklotz et al., 2001). In support of this model, disruption of the C-terminal domain of Arabidopsis Hsfs restricts Hsf proteins to the nucleus (Kotak et al., 2004). In contrast to plant Hsfs, the C-terminal HR-C domain of animals is highly conserved (Wu et al., 1995). Under basal conditions, the human Hsf1, as well as the Drosophila Hsf, are maintained as inactive monomers via intramolecular interactions between HR-C and HR-A/B, suppressing trimer formation (Rabindran et al., 1993; Zuo et al., 1994). This type of intramolecular suppression is thought not to exist in plants Hsfs.
THE PLANT HSF NETWORK
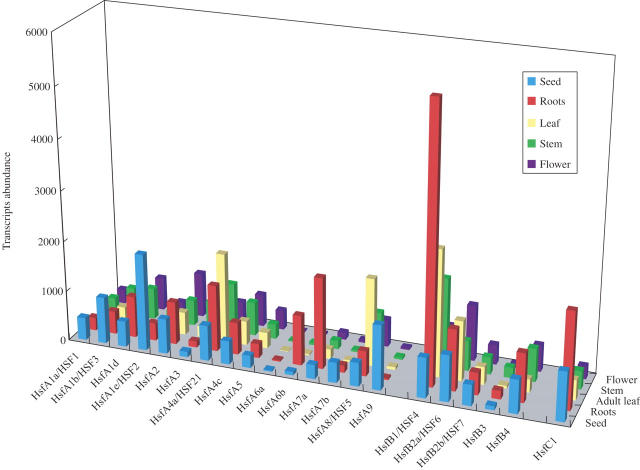
The number of plant Hsfs continues to grow. A new signature domain of the CTAD containing the AHA and NES motifs of plants allowed the identification of >60 new class A Hsfs from expressed sequence tag (EST) databases, including 19 new Hsfs in soybean (34 in total) and at least 23 Hsfs in rice (Kotak et al., 2004). The complexity of the plant Hsf gene family is thought to allow a highly flexible and efficient response to rapid changes in environmental conditions that accompany the stationary lifestyle of plants (Nover et al., 2001; Kotak et al., 2004). Figure 2, generated from Genevestigator microarray data sets (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch/), reveals a surprisingly diverse basal level of expression of the Arabidopsis Hsf gene network in different plant tissues. Previously, transcripts encoding Hsfs A3, A6a, A6b, A7b, B2a and B3 could not be detected using RNA from control or heat-stressed tissue-cultured cells or leaves (Nover et al., 2001). However, as shown in Fig. 2, apart from HsfA6a that was not expressed in all tissues, all other Hsf genes demonstrated a low to moderate level of expression in most tissues, or in a tissue-specific manner.
Fig. 2.
Steady-state transcript level of Arabidopsis HSFs in different tissues. The basal steady-state transcript level of all Arabidopsis Hsfs was obtained from the Genevestigator microarray database using the ‘Meta-analyzer’ tool (Zimmermann et al., 2004). The values represent the signal intensity of each probe set as given by Genevestigator (https://www.genevestigator.ethz.ch/).
The best-studied plant Hsf system is that of tomato (Lycopersicon peruvianum). The tomato HsfA1, a constitutively expressed Hsf, is the master regulator of the heat response and is essential for thermotolerance (Mishra et al., 2002; Baniwal et al., 2004). Plants with suppressed LpHsfA1 do not survive a moderate heat stress (1 h at 45 °C), suggesting that the function of LpHsfA1 could not be compensated for by other LpHsfs. In contrast, enhanced expression of LpHsfA1 in transgenic plants enhances thermotolerance, even to severe heat stress conditions (Mishra et al., 2002; Baniwal et al., 2004). The tomato HsfA2 is considered the ‘work horse’ of the HS response in tomatoes. It accumulates in cells after heat stress induction and becomes the dominant Hsf of cells. The synthesis of LpHsfA2 is controlled by the activation of LpHsfA1 (Mishra et al., 2002; Baniwal et al., 2004). LpHsfA2 is found in cells in three different forms: (1) a soluble cytoplasmic form in heat-acclimated cells; (2) a nuclear form found in HS-treated cells; and (3) a stored form in cytoplasmic HS granules (HSGs), which represents a major site for HSP accumulation (Nover et al., 1983, 1989). Hsp17.4-CII was found to interact with the C-terminal domain of LpHsfA2 and to act as a co-repressor forming aggregates that can be solubilized in the presence of class CI sHsps, or by heteo-oligomerization with LpHsfA1 (Port et al., 2004). The tomato HS-induced HsfA2 is dependent on LpHsfA1 expression for nuclear localization and for its nuclear retention by formation of an LpHsfA1–LpHsfA2 hetero-oligomer (Scharf et al., 1998; Heerklotz et al., 2001; Port et al., 2004). In contrast, Arabidopsis HsfA1a is not required for the HS-dependent expression of AtHsfA2 (Busch et al., 2005), nor for its nuclear localization (Kotak et al., 2004).
In Arabidopsis, in contrast to tomato, no AtHsf master regulator could be identified. The Arabidopsis loss-of-function mutants AthsfA1a and AthsfA1b alone had no obvious effects on the HS response, only the AthsfA1a/1b double mutant was impaired in HS gene expression, showing lower transcript levels of HSPs at early stages of the HS response (Lohmann et al., 2004; Busch et al., 2005). The lack of a strong negative effect of the double mutant might suggest that in Arabidopsis other Hsf proteins can compensate for AtHsfA1a and AtHsfA1b; however, there was no increase in the expression of any of the other 13 class A Hsf genes in the AtHsfA1a/1b double mutant. These results show that AtHsfA1a and AtHsfA1b are necessary for early onset of HS gene expression at the transcriptional level, but they are not the sole regulators of the HS response in Arabidopsis (Lohmann et al., 2004). Only a small fraction (4 %) of the HS-regulated genes were associated with AtHsfA1a/1b function (Busch et al., 2005).
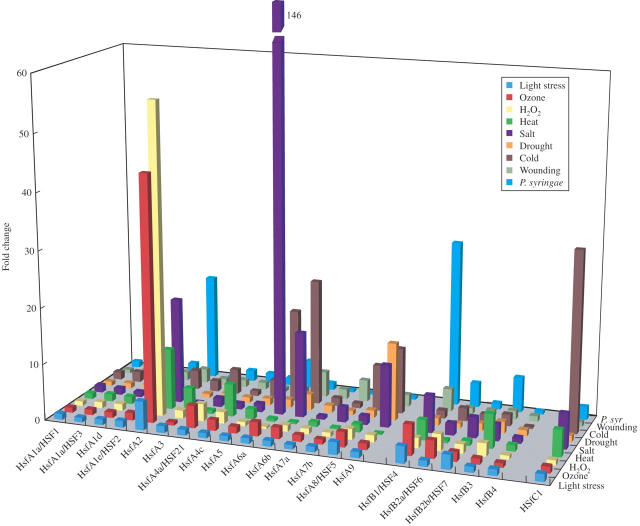
Figure 3 summarizes changes in the steady-state transcript level of different members of the Arabidopsis Hsf gene family in leaves of plants subjected to different abiotic stresses. The data presented in Fig. 3 reveal a high diversity in the response of different AtHsf genes to different abiotic stresses. Based on the data presented in Fig. 3, it is suggested that there is a high degree of specialization in the response of specific Hsfs to particular stress conditions. Thus, for example, AtHsfA9 appears to be specific to salt, drought and cold stress, while AtHsfA6a and AtHsfA6b appear to be cold and salt specific. With the exception of AtHsfA2 and AtHsfB1, the pattern of Hsf expression during heat stress was different from the pattern of Hsf expression during other stresses. Because HSEs are found in the promoters of many defence genes (e.g. Rizhsky et al., 2004a), it is possible that different Hsfs, expressed during different stresses, activate or control different defence pathways. When the HSP response of plants subjected to drought, heat or drought combined with heat was compared, for example, it was found that all HSPs expressed during heat were also expressed during drought combined with heat and/or drought. However, leaves subjected to drought or drought combined with heat expressed specific HSPs not found in heat stress-treated leaves (Rizhsky et al., 2004b). The combinatorial function of Hsfs could therefore be responsible for stress-specific expression of HSPs or other defence genes, and specific stress conditions could therefore cause activation of a particular set(s) of different Hsfs (Fig. 3; Rizhsky et al., 2004b). Because most of the experiments presented in Fig. 3 include only a limited number of time points, the complete expression pattern of the Hsf network during different stress responses might not be captured by the figure. The data presented in Fig. 3 suggest that in addition to being potentially redundant, the Hsf gene network is highly flexible and specialized. It controls the response of plants to diverse stress conditions, as well as potentially their combination (Rizhsky et al., 2004b; Mittler, 2006).
Fig. 3.
Arabidopsis Hsf transcript accumulation in response to different environmental stress conditions. The values are presented as fold change, relative to control treatment or t = 0 min of the experiment. All data, except for the light stress experiment, were obtained from the Genevestigator microarray database using the ‘digital-northern’ tool (Zimmermann et al., 2004). The relative induction levels of Hsfs during moderate light stress were obtained from a microarray experiment previously described by Davletova et al. (2005a). To demonstrate maximal fold increase for each Hsf transcript in each stress treatment, in each experiment that contained several time points, the time point in which a particular Hsf showed the highest expression level was chosen for representation. In general, the following time points were used: light stress (30 min), salt (12 h), drought (24 h), cold (24 h) and wounding (24 h). The following exceptions were made for particular Hsfs showing the highest level of induction at other time points: Light stress: HsfB1, 3 h; HsfA8, 6 h. Salt stress: HsfA1a, HsfA4a, HsfA6a, HsfB2a, 6 h; A1e, A8, A9, B1, B2b and B3, 24 h. Cold stress: A4a and B2a, 6 h; A1d and A5, 12 h. Wounding: B1 and 6a, 6 h.
The promoters of all AtHsf genes contain clusters of HSEs (Nover et al., 2001), suggesting that the expression of this transcription factor gene family could be self-regulated. Potentially, each AtHsf could bind to the HSEs of every AtHsf, including its own, and activate or repress its expression, although there is still no evidence to support such self-regulation. In addition, approx. 33 % of 22 810 Arabidopsis genes analysed were found to contain the consensus motif (nGAAnnTCCn) of HSE within 1000 bp of their putative promoter sequence (Busch et al., 2005). This may suggest that there are other requirements for Hsf-mediated gene activation/suppression, since it is hard to expect that every gene's promoter containing HSE is regulated by Hsfs.
Functional interdependence studies between Hsfs, co-immunoprecipitation and yeast one-hydrid assays suggest that all class A LpHsfs can interact with each other, potentially forming hetero-oligomers (Scharf et al., 1998; Bharti et al., 2000). For example, the LpHsfA3 was isolated from a heat stress cDNA library by a two-hybrid screen using LpHsfA1 as a bait (Bharti et al., 2000). Furthermore, different Hsfs can associate with each other potentially functioning as co-activators or co-repressors. According to an as yet unpublished observation reported by Baniwal et al. (2004), the tomato and Arabidopsis HsfA4a specifically interact with AtHsfA5 and function as co-repressors. In addition, the activity of AtHsfA4a was strongly repressed when co-expressed with AtHsfB1 (Czarnecka-Verner et al., 2000). AtHsfB1 was later shown to repress the transcriptional activity of class A Hsfs through an active mechanism that involves its C-terminal regulatory region (Czarnecka-Verner et al., 2004). In contrast, the tomato LpHsfB1 acts as a co-activator of class A LpHsfs. The co-activation of A-type LpHsfs by LpHsfB1 depends on a histone-fold-like motif in its C-terminal domain, which is required for the recruitment of the plant CREB-binding protein orthologue HAC1. The stimulation effect of LpHsfB1 was not restricted only to interaction with LpHsfA1, but could also be observed with LpHsfA2 (Bharti et al., 2004). AtHsfB1 on the other hand, lacking the crucial lysine residue in the histone-fold motif, does not function as a co-activator and potentially even interferes with the activation of class A Hsfs via competition for HSE binding (Bharti et al., 2004).
The complexity of the Hsf gene network of plants is evident at at least five different levels: (1) a large number of Hsf genes are present in the plant genome; (2) each Hsf gene can potentially bind to its own promoter, as well as to the promoters of all other Hsf genes; (3) monomers encoded by different Hsf genes can interact leading to activation or suppression of transcription; (4) monomers encoded by different Hsf genes can interact affecting nuclear targeting and retention; and (5) spatial and temporal expression patterns of Hsfs could affect different responses in different tissues. These features make the Hsf gene network a highly redundant and specialized network that functions in a stress- or developmental-specific manner.
REDOX REGULATION OF HSFS IN EUKARYOTES
Considerable evidence supporting a possible role for plant Hsfs as direct sensors of ROS can be found in studies of mammalian, Drosophila and yeast Hsfs. In general, the transcriptional activation of Hsfs is achieved in two stages. First, inactive Hsf monomers form a homo-trimer upon receiving a stress signal. The DNA-binding trimer can then be modified further by phosphorylation, or subjected to conformational changes that activate transcription (Fig. 4; Larson et al., 1988; Lee et al., 2000). Human Hsf1 and Drosophila Hsf were shown to sense hydrogen peroxide directly and assemble into a homotrimer in a reversible and redox-regulated manner (Zhong et al., 1998; Ahn and Thiele, 2003). It was demonstrated that two cysteine residues, located within and near the DBD, are required for intramolecular disulfide bond formation in response to heat or H2O2 stress. The conformational change induced by the disulfide formation was shown in turn to be essential for the formation of HsHsf1 homotrimers, nuclear translocalization and gene activation (Hahn and Thiele, 2004). H2O2 was also found to induce transactivation, nuclear translocation and DNA binding activity of Drosophila Hsf1 (Jacquier-Sarlin and Polla, 1996). Furthermore, the DNA binding of the Drosophila Hsf1 protein was shown to be reversibly regulated by H2O2 as well as by high temperature (Fig. 4; Zhong et al., 1998).
Fig. 4.
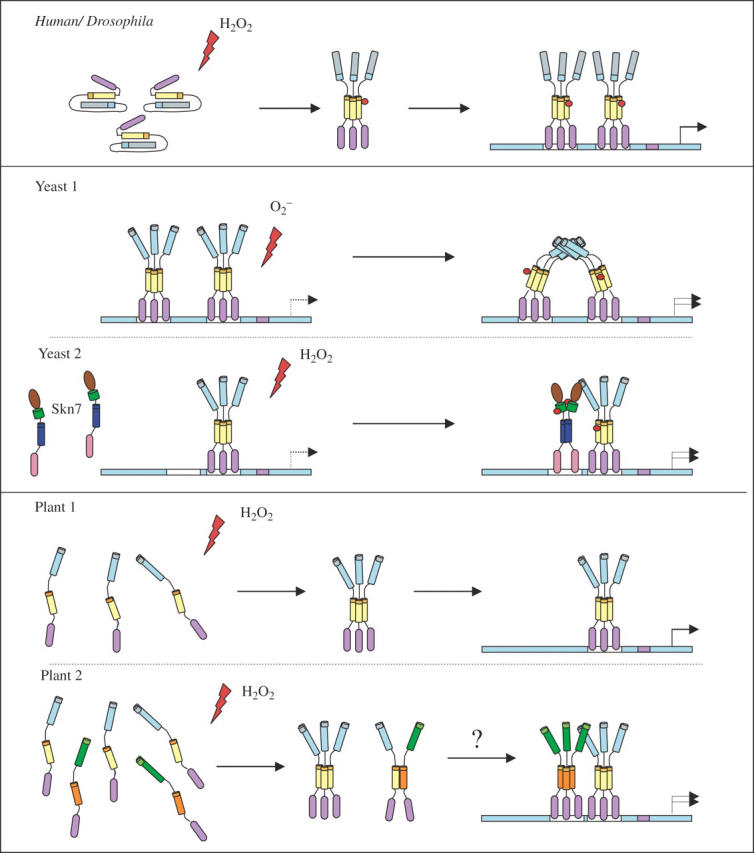
A putative model for Hsf function in different organisms. The human and Drosophila Hsfs (upper panel) are inactive under non-stress conditions due to intramolecular interactions between HR-A/B and the HR-C domains. In response to hydrogen peroxide, the protein forms a disulfide bond between two cysteine residues inside and near the DNA-binding domain. The active Hsf is then phosphorylated and forms a transcriptionaly active homotrimer that is transported to the nucleus and activates ROS-responsive gene expression. Two different models are presented for oxidative stress-mediated activation of Hsf in yeast (middle panels). In the upper section, both Hsfs are bound to the HSEs of the target gene promoter as a homotrimer; superoxide anions directly interact with the protein inducing a conformational change that forms a cooperative interaction between them that increases their transcriptional activity. The second yeast model (lower section) shows the increase of Hsf activity by the co-activation of Skn7. Upon perception of H2O2 stress, Skn7, which is localized in the nucleus, is phosphorylated at the receiver domain by a histidine kinase sensor inducing the formation of an Skn7 homodimer. The active homodimer can bind to HSE adjacent to Hsf and increase its transcriptional activation. Two hypothetical models for plants are presented in the lower panels. A simplistic model is shown in the upper section in which Hsf (AtHsfA4a or AtHsfA2a) forms a homotrimer in response to interaction with ROS and is transported to the nucleus to activate oxidative stress gene expression. In the lower section, oxidative stress induces the homo-trimerization of a particular Hsf, which in response interacts with another Hsf to mediate its transport to the nucleus. The two active Hsfs can cooperatively induce gene expression. For example, AtHsfA8a could act as the co-activator. AtHsfA8a is expressed during oxidative stresses (Fig. 3), and in KO-Apx1 plants (see Fig. 5). It is localized to the cytoplasm and it could be dependent on other Hsfs for nuclear localization. Alternatively, the homotrimer of AtHsfA4a can be transported to the nucleus where it can cooperate with a class B Hsf (AtHsfB2b) or another class A Hsf.
In yeast, superoxide anions (O2−) directly induced a conformational change in the Hsf DNA-binding trimer from its low-activity mode to a high-activity mode. The O2− signal was shown to be perceived by the region that links the DBD and the trimerization domains of the yeast Hsf, promoting the physical interaction of two DNA-bound homotrimers (Lee et al., 2000). The yeast Hsf is required under oxidative stress for the activation of Cu, Zn superoxide dismutase (SOD), presenting a direct circle of regulation in which O2− activates the yeast Hsf that induces CuZnSOD which scavenges O2− and suppresses the activation of Hsf (Liu and Thiele, 1996). In yeast, Skn7, a response regulator that contains a two-component system receiver domain, was shown to be required for the activation of HS gene expression specifically in response to hydrogen peroxide. Skn7 was found to bind to HSEs and physically associate with Hsf1, functioning as an essential co-activator during oxidative stress conditions (Raitt et al., 2000). Interestingly, a familial form of amyotropic lateral sclerosis (ALS) neurodegenerative disease was shown to be associated with autosomal dominant mutations of Cu/ZnSOD1, which accumulate in a non-native conformation and aggregate. An HsHsf1 dominant positive mutant was shown to protect neurons from mutant Cu/ZnSOD-1 toxicity by stimulating Hsp70 expression (Bruening et al., 1995; Batulan et al., 2003). Thus, it was suggested that the sensitivity of motor neurons to stress could result from maladjusted mechanisms of stress activation of Hsf1 (Voellmy, 2004).
HSFS AND OXIDATIVE STRESS IN PLANTS
The promoter of the central H2O2-scavenging enzyme cytosolic ascorbate peroxidase 1 (Apx1), as well as the promoters of many defence genes and transcription factors involved in H2O2 signalling and defence, contain an Hsf-binding motif (Mittler and Zilinskas, 1992; Rizhsky et al., 2004a; Davletova et al., 2005a). Promoter analyses, as well as overexpression studies of AtHsfA1b in Arabidopsis, suggest that the Hsf binding site at the Apx1 promoter is functional (Storozhenko et al., 1998; Panchuk et al., 2002). Furthermore, several other Arabidopsis Apx genes showed enhanced transcript accumulation in response to a short-term heat shock in an AtHsfA1b-dependent mechanism (Panchuk et al., 2002). The activity of Arabidopsis Apx2 is specifically induced by high light stress and high temperature. Apx2 was also shown to be AtHsfA1b dependent (Panchuck et al., 2002). This result coincides with the observation of Lohmann et al. (2004, data were not provided) of a strong negative effect on the induction of Apx2 in the AthsfA1a/1b double mutant.
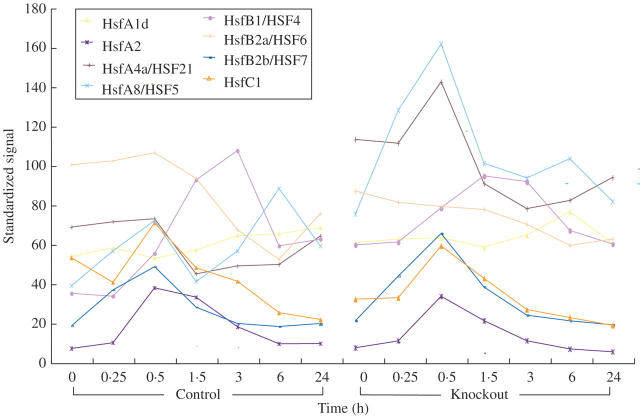
ROS such as H2O2, O2− and 1O2 are thought to function as early signals for high light stress in plants (Pnueli et al., 2003; Rizhsky et al., 2003; Apel and Hirt, 2004; Mittler et al., 2004; Davletova et al., 2005a). Accordingly, Arabidopsis mutants deficient in Apx1 accumulate higher levels of H2O2 compared with wild-type plants during light stress (Pnueli et al., 2003; Davletova et al., 2005a). Figure 5 compares the change in steady-state transcript level of all Arabidopsis Hsfs in wild-type and in knockout Apx1 (KO-Apx1) plants during light stress. The most dramatic difference was observed for AtHsfA4a and AtHsfA8 transcripts showing a constant higher level of expression in the Apx1 mutant (Davletova et al., 2005a), while the expression pattern of other AtHsfs was only slightly affected or remained essentially unchanged in the absence of the Apx1 gene. The Hsfs whose transcripts transiently peaked during light stress in wild type as well as in the KO-Apx1 mutant are AtHsfs A2, A4a, A8, B1, B2b and C1 (Fig. 5). Of these, only AtHsfs A2, A4a, A8 and B1 showed elevated levels, ≥2-fold, during oxidative stress conditions, i.e. H2O2 or ozone or both (Fig. 3). HsfA2 increases approx. 50-fold under oxidative stresses, which suggest that its function might be important under these conditions; however, it did not increase in KO-Apx1 above its expression level in the wild type, suggesting that different AtHsfs might also respond differently to different types of oxidative stress, or to different levels of ROS. It could also be that the high level of expression of AtHsfA2 during oxidative stress conditions requires a transcriptional activator that is absent during light stress.
Fig. 5.
Time course analysis of Arabidopsis Hsf accumulation in response to light stress in wild-type and knockout plants lacking the ROS defence enzyme cytosolic ascorbate peroxidase (KO-Apx1). The expression of the eight light stress-responsive Hsfs is presented, in the wild type (left) and KO-Apx1 (right) during time course exposure to moderate light stress. The steady-state transcripts level was obtained from microarray experiments previously described by Daveltova et al. (2005a).
Thirty-two percent of the transcripts elevated in KO-Apx1 plants in response to a moderate light stress were also elevated in wild-type plants in response to H2O2 application (Davletova et al., 2005b). HsfA4a and Zat12, as well as other genes that might be associated with H2O2 signalling, including NADPH oxidase (RbohD), MAPK3 and several WRKY transcription factors, were elevated under both conditions (Davletova et al., 2005b). The zinc finger protein Zat12 is required for the expression of Apx1 during oxidative stress (Rizhsky et al., 2004a; Davletova et al., 2005b). Interestingly, a dominant negative construct for AtHsfA4a, when expressed in Arabidopsis, suppressed the expression of Zat12 and Apx1 during light stress (Davletova et al., 2005a). This finding suggests that AtHsfA4a functions upstream of Zat12 and Apx1 (both containing an HSE element in their promoters; Rizhsky et al., 2004a). Its rapid response to hydrogen peroxide stress, its control of Apx1 and Zat12 expression and its constitutive expression in cells in the absence of stress makes AtHsfA4a a prominent candidate to function as an Hsf H2O2 sensor in Arabidopsis. Furthermore, AtHsfA4a as well as AtHsfs A1a, A1b, A1d, A1e and A2 are evenly distributed in the cytoplasm and nucleus (Kotak et al., 2004); this characteristic is highly important for the function of a sensor to detect changes in the cytosol and affect expression of the appropriate gene in the nuclei. In rice, HsfA4a is encoded by the Spl7 gene. A point mutation in Spl7 (tryptophan to arginine substitution in the DBD) caused a ‘disease lesion mimics’ phenotype, which suggests a role for HsfA4a as an anti-apoptotic factor (Yamanouchi et al., 2002). Overproduction of H2O2 and O2− is known to be associated with programmed cell death, causing lesion formation (Mittler et al., 1996; Torres et al., 2002; Vacca et al., 2004), supporting the hypothesis that HsfA4a is a redox-sensitive hydrogen peroxide sensor in plants. Nevertheless, based on the microarray results presented in Fig. 5, HsfA8 might also function as a potential H2O2 sensor. Further work is of course required to determine whether Hsfs such as HsfA4a or HsfA8 function as ROS or redox sensors in plants.
Acknowledgments
Work at the laboratory of R.M. is supported by funding from The National Science Foundation (NSF- 0431327; NSF-0420033) and The Nevada Agricultural Experimental Station (Publication number 03055517).
LITERATURE CITED
- Ahn SG, Thiele DJ. 2003. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes and Development 17: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, et al.2004. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. Journal of Biosciences 29: 471–487. [DOI] [PubMed] [Google Scholar]
- Batulan Z, Shinder GA, Minotti S, He BP, Doroudchi MM, Nalbantoglu J, Strong MJ, Durham HD. 2003. High threshold for induction of the stress response in motor neurons is associated with failure to activate Hsf1. Journal of Neuroscience 23: 5789–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmüller E, Porfirova E, Dörmann P. 2003. Characterization of an Arabidopsis mutant deficient in γ-tocopherol methyltransferase. Plant Molecular Biology 52: 1181–1190. [DOI] [PubMed] [Google Scholar]
- Bharti K, Schmidt E, Lyck R, Heerklotz D, Bublak D, Scharf KD. 2000. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant Journal 22: 355–365. [DOI] [PubMed] [Google Scholar]
- Bharti K, Von Koskull-Doring P, Bharti S, Kumar P, Tintschl-Korbitzer A, Treuter E, Nover L. 2004. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf KD. 1997. Heat stress transcription factors from tomato can functionally replace Hsf1 in the yeast Saccharomyces cerevisiae. Molecular and General Genetics 255: 322–331. [DOI] [PubMed] [Google Scholar]
- Bruening W, Winnett E, Pelletier J. 1995. Wilms' tumor: a paradigm for insights into development and cancer. Cancer Investigation 13: 431–443. [DOI] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schoffl F. 2005. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant Journal 41: 1–14. [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Yuan CX, Scharf KD, Englich G, Gurley WB. 2000. Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Molecular Biology 43: 459–471. [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Pan S, Salem T, Gurley WB. 2004. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Molecular Biology 56: 57–75. [DOI] [PubMed] [Google Scholar]
- Damberger FF, Pelton JG, Harrison CJ, Nelson HC, Wemmer DE. 1994. Solution structure of the DNA-binding domain of the heat shock transcription factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Protein Science 3: 1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JF, Schiestl RH. 2001. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Molecular and Cellular Biology 21: 8483–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. 2005a. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. 2005b. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology 139: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L. 2000. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12: 265–278. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. 1999. Transport between the cell nucleus and the cytoplasm. Annual Review of Cell and Developmental Biology 15: 607–660. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ. 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. Journal of Biological Chemistry 279: 5169–5176. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Molecular and Cellular Biology 24: 5249–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HC. 1994. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263: 224–227. [DOI] [PubMed] [Google Scholar]
- Heerklotz D, Doring P, Bonzelius F, Winkelhaus S, Nover L. 2001. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Molecular and Cellular Biology 21: 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier-Sarlin MR, Polla BS. 1996. Dual regulation of heat-shock transcription factor (Hsf) activation and DNA-binding activity by H2O2: role of thioredoxin. Biochemical Journal 318: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C. 1997. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO Journal 16: 2452–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Doring P. 2004. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant Journal 39: 98–112. [DOI] [PubMed] [Google Scholar]
- Larkindale J, Huang B. 2004. Thermotolerance and antioxidant systems in Agrostis stolonifera: involvement of salicylic acid, abscisic acid, calcium, hydrogen peroxide, and ethylene. Journal of Plant Physiology 161: 405–413. [DOI] [PubMed] [Google Scholar]
- Larkindale J, Knight MR. 2002. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiology 128: 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JS, Schuetz TJ, Kingston RE. 1988. Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature 335: 372–375. [DOI] [PubMed] [Google Scholar]
- Lee S, Carlson T, Christian N, Lea K, Kedzie J, Reilly JP, Bonner JJ. 2000. The yeast heat shock transcription factor changes conformation in response to superoxide and temperature. Molecular Biology of the Cell 11: 1753–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Thiele DJ. 1996. Oxidative stress induced heat shock factor phosphorylation and Hsf-dependent activation of yeast metallothionein gene transcription. Genes and Development 10: 592–603. [DOI] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schoffl F. 2004. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Molecular Genetics and Genomics 271: 11–21. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. 2002. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes and Development 16: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. 2006. Abiotic stress, the field environment and stress combination. Trends in Plant Science 11: 15–19. [DOI] [PubMed]
- Mittler R, Zilinskas BA. 1992. Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. Journal of Biological Chemistry 267: 21802–21807. [PubMed] [Google Scholar]
- Mittler R, Shulaev V, Seskar M, Lam E. 1996. Inhibition of programmed cell death in tobacco plants during pathogen-induced hypersensitive response at low oxygen pressure. Plant Cell 8: 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9: 490–498. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes and Development 12: 3788–3796. [DOI] [PubMed] [Google Scholar]
- Nakai A, Morimoto RI. 1993. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Molecular and Cellular Biology 13: 1983–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Ngata K. 1997. Hsf4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Molecular and Cellular Biology 17: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD. 1997. Heat stress proteins and transcription factors. Cellular and Molecular Life Sciences 53: 80–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. 1983. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Molecular and Cellular Biology 3: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. 1989. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Molecular and Cellular Biology 9: 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. 1996. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. 2001. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schoffl F. 2002. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiology 129: 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Craig EA. 1989. Positive and negative regulation of basal expression of a yeast HSP70 gene. Molecular and Cellular Biology 9: 2025–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R. 2003. Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant Journal 34: 187–203. [DOI] [PubMed] [Google Scholar]
- Port M, Tripp J, Zielinski D, Weber C, Heerklotz D, Winkelhaus S, Bublak D, Scharf KD. 2004. Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiology 135: 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Giorgi G, Clos J, Wu C. 1991. Molecular cloning and expression of a human heat shock factor, Hsf1. Proceedings of the National Academy of Sciences of the USA 88: 6906–6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. 1993. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science 259: 230–234. [DOI] [PubMed] [Google Scholar]
- Rainwater DT, Gossett DR, Millhollon EP, Hanna HY, Banks SW, Lucas MC. 1996. The relationship between yield and the antioxidant defense system in tomatoes grown under heat stress. Free Radical Research 25: 421–435. [DOI] [PubMed] [Google Scholar]
- Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Molecular Biology of the Cell 11: 2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. 1962. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18: 571–573. [Google Scholar]
- Rizhsky L, Liang H, Mittler R. 2002. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiology 130: 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. 2003. The water–water cycle is essential for chloroplast protection in the absence of stress. Journal of Biological Chemistry 278: 38921–38925. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. 2004a. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. Journal of Biological Chemistry 279: 11736–11743. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. 2004b. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134: 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge D, Zimarino V, Holm K, Wu C, Morimoto RI. 1991. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes and Development 5: 1902–1911. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. 1993. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Molecular and Cellular Biology 13: 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Murakami T, Funatsuki H, Matsuba S, Saruyama H, Tanida M. 2001. Heat shock-mediated APX gene expression and protection against chilling injury in rice seedlings. Journal of Experimental Botany 52: 145–151. [PubMed] [Google Scholar]
- Scharf KD, Heider H, Hohfeld I, Lyck R, Schmidt E, Nover L. 1998. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Molecular and Cellular Biology 18: 2240–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. 1991. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proceedings of the National Academy of Sciences of the USA 88: 6911–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss J, Kunert O, Gase U, Scharf KD, Nover L, Ruterjans H. 1996. Solution structure of the DNA-binding domain of the tomato heat-stress transcription factor Hsf24. European Journal of Biochemistry 236: 911–921. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54: 855–864. [DOI] [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montagu M, Inze D, Kushnir S. 1998. The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiology 118: 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kawazoe Y, Takeda S, Morimoto RI, Nagata K, Nakai A. 1998. Disruption of the Hsf3 gene results in the severe reduction of heat shock gene expression and loss of thermotolerance. EMBO Journal 17: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proceedings of the National Academy of Sciences of the USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, De Gara L. 2004. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiology 134: 1100–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy R. 2004. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister GW, Kim SJ, Wu C, Bax A. 1994. NMR evidence for similarities between the DNA-binding regions of Drosophila melanogaster heat shock factor and the helix–turn–helix and HNF-3/forkhead families of transcription factors. Biochemistry 33: 10–16. [DOI] [PubMed] [Google Scholar]
- Wu C. 1995. Heat shock transcription factors: structure and regulation. Annual Review of Cell and Developmental Biology 11: 441–469. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Mizukami Y, Sakurai H. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. Journal of Biological Chemistry 280: 11911–11919. [DOI] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. 2002. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proceedings of the National Academy of Sciences of the USA 99: 7530–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CX, Gurley WB. 2000. Potential targets for Hsf1 within the preinitiation complex. Cell Stress Chaperones 5: 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C. 1998. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Molecular Cell 2: 101–108. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Baler R, Dahl G, Voellmy R. 1994. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Molecular and Cellular Biology 14: 7557–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]