Abstract
• Background and Aims Despite considerable investment in elaborate floral displays, Tacca chantrieri populations are predominantly selfing. It is hypothesized that this species might possess considerable spatial or temporal variation in outcrossing rates among populations. To test this hypothesis, genetic variability and genetic differentiation within and among T. chantrieri populations were investigated to find out if they are in agreement with expectations based on a predominantly inbred mating system.
• Methods Genetic diversity was quantified using inter-simple sequence repeats (ISSR) in 303 individuals from 13 populations taken from known locations of T. chantrieri in China, and from one population in Thailand.
• Key Results Of the 113 primers screened, 24 produced highly reproducible ISSR bands. Using these primers, 160 discernible DNA fragments were generated, of which 145 (90·62 %) were polymorphic. This indicated considerable genetic variation at the species level. However, there were relatively low levels of polymorphism at population levels, with percentages of polymorphic bands (PPB) ranging from 8·75 % to 55 %. A high level of genetic differentiation among populations was detected based on different measures (Nei's genetic diversity analysis: GST = 0·5835; AMOVA analysis: FST = 0·6989). Furthermore, based on levels of genetic differentiation, the 14 populations clustered into two distinct groups separated by the Tanaka Line.
• Conclusions High levels of differentiation among populations and low levels of diversity within populations at large spatial scales are consistent with earlier small-scale studies of mating patterns detected by allozymes which showed that T. chantrieri populations are predominantly selfing. However, it appears that T. chantrieri has a mixed-mating system in which self-fertilization predominates, but there is occasional outcrossing. Significant genetic differences between the two distinct regions might be attributed to vicariance along the Tanaka Line. Finally, possible mechanisms of geographic patterns based on genetic differentiation of T. chantrieri are discussed.
Keywords: Tacca chantrieri, floral display, population genetic structure, gene flow, ISSR markers, mating system, geographic differentiation, Tanaka Line
INTRODUCTION
The spatial distribution of genetic diversity in plant populations, which is characterized by the genetic variability and genetic differentiation within and among populations, is primarily determined by the life history, including its reproductive traits, of the plant (Schoen, 1982a, b; Schoen and Clegg, 1985; Hamrick and Godt, 1996), but population history is also a determinant factor of the genetic variation within species (Schaal et al., 1998).
Population history, represented as fluctuations in the number and size of populations, and the evolutionary and biogeographic histories of species, may have played critical roles in determining its current genetic composition (Schaal et al., 1998). Contemporary biogeographic patterns of genetic variation are determined by historical patterns of gene flow and vicariance among populations (Hewitt, 1996; Soltis et al., 1997; Avise, 2000). This history should be reflected in the genetic structure and phylogeography of extant populations, which should provide information enabling evolutionary processes to be inferred and for biogeographical scenarios that underlie patterns of genetic differentiation to be tested. As such, if the genetic structure of a population of a species is understood, its evolutionary history can be elucidated (Bauert et al., 1998).
Plant reproductive traits also determine the population genetic structure via the influence of the plant's mating system (Hamrick and Godt, 1990; Schoen et al., 1996). The close relationship between the mating system and the level of genetic variation and genetic structure has been documented in many studies using different methods (Brown et al., 1989; Hamrick and Godt, 1990). Inbreeding species are expected to have less genetic diversity and heterozygosity within populations, as well as more genetic differentiation among populations, than outcrossing species (Charlesworth and Charlesworth, 1995; Hamrick and Godt, 1996). Therefore, genetic diversity within and among populations can reflect, in a certain extent, the relative rates of inbreeding versus outcrossing in a species.
As an important reproductive trait, floral design and display affect plant mating systems by attracting animal pollinators and thereby promoting pollen dispersal and cross-pollination (Harder and Barrett, 1996; Emms et al., 1997). It is believed that plants with greater investment in floral structures attractive to pollinators will benefit from increased fitness via cross-pollination. In this case, species with a high investment in extravagant floral displays are expected to be largely outcrossing (Charlesworth and Charlesworth, 1987).
Tacca chantrieri is a widespread species that occurs in humid tropical regions of south-east Asia. Its distribution has contracted due to overexploitation, habitat destruction, and forest fragmentation. Tacca chantrieri has extravagant floral displays and a high investment in reproductive structures that lead to the expectation that it is largely outcrossing (Drenth, 1972; Saw, 1993). Furthermore, dark floral colours, the presence of long filiform appendages or bracts, and the absence of nectar are commonly associated with fly pollination by deceit (sapromyophilous pollination syndrome) (Proctor et al., 1996). Surprisingly, in field pollination experiments and a mating system study, it was found that, despite considerable investment in extravagant floral display, most seeds produced by plants in four populations in south-west China resulted from autonomous self-pollination (Zhang et al., 2005). This mismatch between floral morphology and mating system might cause T. chantrieri to exhibit spatial and temporal variation in population genetic structure. To test this hypothesis, larger surveys of its population genetic structure are required.
Here, inter-simple sequence repeat (ISSR) markers are employed to determine the levels and distribution of genetic differentiation within and among populations of T. chantrieri. These markers have recently become widely used in population genetic studies because they require less investment in time, money and labour than other methods, are highly variable (Wolfe and Liston, 1998; Harris, 1999) and exhibit Mendelian inheritance (Gupta et al., 1994; Tsumura et al., 1996). Inter-simple sequence repeats also generate higher percentages of polymorphic loci than other methods (Esselman et al., 1999). The most recent review that evaluated among- and within-population diversity in wild angiosperms and gymnosperms using different nuclear DNA markers concluded that estimates of genetic variation derived by the dominantly inherited markers RAPD, AFLP and ISSR are very similar and may be directly comparable to codominant marker types; however, ISSRs tend to produce somewhat higher estimates of within-population variation (Nybom, 2004).
The main objective of this study was to quantify genetic variation in T. chantrieri from a range of populations and to compare it with expectations for selfing species. The following specific questions are addressed: (1) What are the levels of genetic variation within and among populations of T. chantrieri? (2) Are patterns of genetic diversity consistent with expectations based upon the highly selfing mating system estimates from previous work? (3) Are there any geographical patterns of genetic differentiation in this species? Following the presentation of the results, the implications of observed genetic patterns on the possible areas of origin and diversification of T. chantrieri are discussed.
MATERIALS AND METHODS
Plant species
Tacca chantrieri André (also called the ‘bat flower’ or ‘bat plant’) is one of the most widespread species in the genus Tacca, which is distributed on the Thailand–Indo-Chinese Peninsula and on the Malay Peninsula to southern China. Its distribution in China extends from southern Yunnan to Guangxi and Hainan, and it inhabits moist and shady understorey habitats in tropical forests (Drenth, 1972; Wu et al., 2003) (Fig. 1). Plants are 50–100 cm tall, with tubers or creeping rhizomes and alternate elliptic entire leaves. It has a curious inflorescence which is bat-like, both in shape and colour, with wide-spreading wing-like bracts of rich maroon-black accompanied by long trailing filaments or ‘whiskers’. If pollinated, the small black flowers develop into large black berries. In the Yunnan Province, south-west China, where the present studies were conducted, the species flowers from April to July, and by September–October the berries are ripe.
Fig. 1.
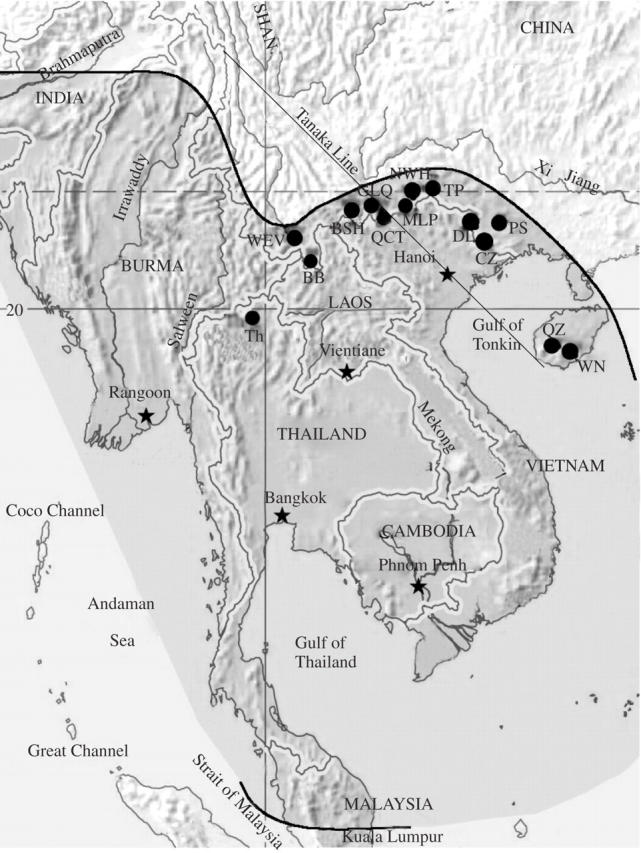
A map of the known distribution of Tacca chantrieri (shaded area between two thick black lines) and locations of sampled populations with the abbreviations used in this study (see Table 1 for explanations of population abbreviations). This map was based on data from Drenth (1972).
Study sites and sampling
From 2003 to 2004, leaves of T. chantrieri were collected from 14 populations in south China and Thailand. The populations were chosen because they represented the overall distribution of the species in China (Fig. 1), as well as one population from Thailand. The fourteen populations are geographically separated with each other by at least 60 km, and the size of the populations ranged from 600 to 3500 m2, with the exception of the CZ population which is much smaller (about 600 m2 with some 50 plants) (Table 1). In each of the 14 populations, a random sample of 15–35 plants was taken. Considering the sampling strategy, a Pearson's correlation test was used to analyse the effect of sample size on genetic diversity estimates. The total sample included 303 individuals. Distances between adjacent samples were at least 10 m to increase the possibility of detecting genetic variation within each population. Leaves were collected in the field and dried directly with silica gel.
Table 1.
Details of all the populations of Tacca chantrieri sampled
| Abbreviation | Sample size | Locality | Position | Altitude (m) | Size (individuals) |
|---|---|---|---|---|---|
| QCT | 22 | Qincaitang, Hekou County, Yunnan Province | 22°40′N, 104°01′E | 680–780 | ∼120 |
| GLQ | 21 | Gulinqing, Maguan County, Yunnan Province | 22°45′N, 103°58′E | 700–1200 | ∼200 |
| BSH | 20 | Bashahe, Lvchun County, Yunnan Province | 22°53′N, 101°56′E | 680–960 | ∼600 |
| BB | 20 | Wangtianshu, Mengla County, Yunnan Province | 21°37′N, 101°35′E | 680 | ∼180 |
| WEV | 20 | Yexianggu, Jinghong County, Yunnan Province | 22°10′N, 100°51′E | 760 | ∼150 |
| MLP | 21 | Balihe, Malipo County, Yunnan Province | 22°58′N, 104°51′E | 800–1000 | ∼100 |
| NWH | 15 | Nanwenhe, Malipo County, Yunnan Province | 22°59′N, 104°44′E | 720–920 | ∼50 |
| TP | 20 | Tianpeng, Funing County, Yunnan Province | 23°12′N, 105°32′E | 900–1000 | ∼100 |
| DL | 35 | Delong, Napo County, Guangxi Province | 23°17′N, 105°50′E | 520–680 | ∼180 |
| CZ | 16 | Ecological park, Chongzuo County, Guangxi Province | 22°24′N, 107°30′E | 100 | ∼50 |
| PS | 25 | Pingshan, Longan County, Guangxi Province | 22°53′N, 107°39′E | 200 | ∼300 |
| WN | 23 | Xinlong tropical flower garden, Wanning County, Hainan Province | 18°41′N, 110°12′E | 80 | ∼100 |
| QZ | 25 | Baihualing, Qiongzhong County, Hainan Province | 19°00′N, 109°49′E | 350 | ∼1000 |
| Th | 20 | Chiangmai, Thailand | 19°15′N, 98°55′E | 500 | ∼100 |
DNA extraction and ISSR–polymerase chain reaction (PCR) amplification
DNA was extracted using a modified CTAB method (Doyle, 1991). Total DNA was dissolved in 1× TE for subsequent use. ISSR–PCR amplifications were performed in a GeneAmp PCR System 9700 DNA Thermal Cycler (PerkinElmer, USA) with two cycling profiles: (1) 5 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 45 s at 48 °C, 1·5 min at 72 °C, and ending with 7 min at 72 °C; and (2) 5 min at 95 °C, followed by 35 cycles of 45 s at 94 °C, 30 s at 50 °C, 1·5 min at 72 °C, and ending with 7 min at 72 °C. One hundred and thirteen primers (UBC primer set no. 9, Biotechnology Laboratory, University of British Columbia) were screened initially to identify well-amplified polymorphic bands among the populations. Of the 113 primers tested, 24 produced strong, clear and reproducible bands. These were selected for further study of the 303 T. chantrieri individuals. PCR was carried out in a total volume of 20 μL, which included 100 ng of template DNA, 2 μL 10× PCR buffer (Mg2+ Plus), 0–1 μL MgCl2 (25 mM), 0·8 μL dNTPs Mixture (2·5 mM), 1·5 U of Taq polymerase (TaKaRa Biotechnology Dalian Co., Ltd, Dalian, China), 0·75 mM primers (Shanghai Sangon Biological Engineering Technology and Service Co. Ltd, Shanghai, China) and double-distilled water. Amplification products were separated via electrophoresis on 1·5 % (w/v) agarose gels with 0·5× TBE buffer at 120 V for 3–4 h along with a GeneRuler100 ladder (Fermentas UAB, Inc.), stained with ethidium bromide (0·1 mg μL−1). They were then photographed with an Epson digital still Af camera. Negative controls, which lacked template DNA, were included in each PCR set to test for possible contamination.
Genetic diversity analysis
Because ISSR markers are dominant, it was assumed that each band represented the phenotype at a single biallelic locus (Williams et al., 1990). Amplified fragments were scored for the presence (1) or absence (0) of homologous bands. The resulting presence/absence data matrix of the ISSR phenotypes was analysed using POPGENE version 1.31 (Yeh et al., 1997) to estimate the following genetic diversity parameters at the species level: the percentage of polymorphic bands (PPB), expected heterozygosity (HE) and genetic diversity measures (HT, total population gene diversity; GST, coefficient of gene differentiation). An analysis of molecular variance (AMOVA) was used with ARLEQUIN version 2000 (Schneider et al., 2000) to estimate the partitioning of ISSR phenotypic variation within populations, among populations and between two regions (south Yunnan–Thailand and south-east Yunnan–Guangxi–Hainan). The AMOVA variance components were used as estimates of genetic diversity within and between populations. The significance of this F-statistic analogue was tested with 1000 random permutations. A dendrogram was generated from pairwise Nei's genetic distances among the populations with the neighbour-joining algorithm using MEGA v. 2.1 (Kumar et al., 2001).
Gene flow was estimated indirectly using the formula: Nm= 0·5(1−GST)/GST (McDermott and McDonald, 1993). Test for a correlation between Nei's genetic distance and geographical distances (in kilometres) between populations, a Mantel test was performed using tools for population genetic analysis (Miller, 1997) (computing 999 permutations). To detect the geographical pattern of genetic differentiation, significance correlation between the genetic divergence of populations with geographic distance was also tested.
RESULTS
Genetic diversity released by ISSR markers
The 24 selected primers generated 160 bands that ranged in size from 2 kb to 320 bp in T. chantrieri, which corresponded to an average of 6·67 bands per primer. Of these bands, 90·62 % (145 in total) were polymorphic among the 303 individuals. The PPB for a single population ranged from 8·75 % (Thailand) to 55 % (TP), with an average of 32·7 ± 12·8 %, and the average effective number of alleles per locus was 1·19. Assuming Hardy–Weinberg equilibrium, the average gene diversity was estimated as 0·165 within populations (HE), and 0·264 at the species level (HT) (Table 2). Although the sample size varied among populations (15–35 individuals per population), there was no relationship between the sample size and genetic diversity (Pearson's correlation test; n = 14, r = 0·455, P = 0·102). Among the 14 populations, populations TP and DL exhibited the greatest level of variability (PPB, 55 % and 50 %; HE, 0·181 and 0·165, respectively). Populations Th and WEV exhibited the lowest level of variability (PPB, 8·75 % and 15 %; HE, 0·034 and 0·041, respectively).
Table 2.
Pooled values and mean genetic variabilities within populations of T. chantrieri detected by ISSR analysis
| Population | n | HE | PPB (%) | GST | Nm | FST | |
|---|---|---|---|---|---|---|---|
| West region | BSH | 20 | 0·098 (0·164) | 27·5 | |||
| QCT | 22 | 0·111 (0·174) | 35 | ||||
| BB | 20 | 0·079 (0·159) | 25 | ||||
| Th | 20 | 0·034 (0·116) | 8·75 | ||||
| WEV | 20 | 0·041 (0·117) | 15 | ||||
| GLQ | 21 | 0·085 (0·164) | 27·5 | ||||
| Mean | 0·0747 (0·031) | 23·125 | 0·6678 | ||||
| Pooled | 123 | 0·168 | 71·25 | 0·563 | 0·388 | ||
| East region | |||||||
| MLP | 21 | 0·156 (0·204) | 41·25 | ||||
| NWH | 15 | 0·09 (0·164) | 27·5 | ||||
| TP | 20 | 0·181 (0·20) | 55 | ||||
| DL | 35 | 0·165 (0·191) | 50 | ||||
| CZ | 16 | 0·104 (0·181) | 28·12 | ||||
| PS | 25 | 0·151 (0·201) | 40·62 | ||||
| WN | 23 | 0·16 (0·201) | 45 | ||||
| QZ | 25 | 0·095 (0·159) | 31·88 | ||||
| Mean | 0·1378 (0·036) | 39·92 | 0·5601 | ||||
| Pooled | 180 | 0·234 | 77·5 | 0·4168 | 0·6997 | ||
| Average species level | 21·64 | 0·165 (0·068) 0·264 | 32·72 (12·82) 90·62 | 0·5835 | 0·3568 | 0·6829 | |
n, Sample size; HE, expected heterozygosity; PPB, percentage of polymorphic loci; GST, genetic differentiation between populations estimated by using POPGENE 1·31; Nm, estimated gene flow; FST, genetic differentiation between populations estimated by using Arlequin. Standard deviations are shown in parentheses.
Population genetic structure
Most HT in T. chantrieri was distributed among populations. The mean GST for all populations was estimated as 0·5835, which indicated that 58·35 % of the genetic variability was distributed among populations. The AMOVA analysis is consistent with the results of Nei's genetic structure in that there is a high degree of population differentiation. Populations of T. chantrieri were grouped into two geographic regions: the south-east Yunnan–Guangxi–Hainan region (NWH, PS, CZ, DL, TP, MLP, WN and QZ); and the south Yunnan–Thailand region (BSH, BB, WEV, Th, QCT and GLQ). Highly significant (P < 0·001) genetic differences were detected between regions, among populations (within regions) and among individuals (within both populations and regions) (Table 3). Of the total molecular variance, 33·44 % was attributable to regional divergence, 36·46 % to population differences within regions and 30·10 % to individual differences within populations. When the total variance was partitioned without considering the regional distribution of populations, 69·89 % was attributable to populations (FST) and 30·1 % to individual differences within populations. The number of migrants (Nm) was estimated as 0·3568 individuals per generation between populations, and the strong genetic differentiation in T. chantrieri suggests that the two regions examined are isolated and that gene flow between the two regions is limited.
Table 3.
Analysis of molecular variance (AMOVA) within/among populations and between geographic regions in Tacca chantrieri
| Source of variation | d.f. | SSD | Variance component | Variation (%) | F statistics | P* |
|---|---|---|---|---|---|---|
| Between regions† | 1 | 1289·84 | 7·56 | 33·44 | FCT = 0·3344 | <0·001 |
| Among populations within regions | 12 | 2204·49 | 8·24 | 36·46 | FSC = 0·5477 | <0·001 |
| Within populations | 288 | 1960·54 | 6·81 | 30·1 | FST = 0·6989 | =0·00196 |
| Total | 301 | 5454·86 | 22·61 | |||
| Analysis of the populations within west region | ||||||
| Among populations | 5 | 854·06 | 9·052 | 66·78 | FSC = 0·6082 | <0·001 |
| Within populations | 116 | 526·93 | 4·504 | 33·22 | FST = 0·6678 | <0·001 |
| Analysis of the populations within east region | ||||||
| Among populations | 7 | 1359·41 | 10·673 | 56·01 | FSC = 0·4337 | <0·001 |
| Within populations | 172 | 1441·9 | 8·383 | 43·99 | FST = 0·5601 | <0·001 |
d.f., Degrees of freedom; SSD, sums of squares; FCT, total deviation from Hardy–Weinberg expectations; FSC among-population deviations from Hardy-Weinberg expectations; FST, deviation from Hardy–Weinberg expectations due to population subdivision.
* P values are the probabilities of having a more extreme variance component than the observed values by chance alone. Probabilities calculated by 1000 random permutations of individuals across populations.
† Geographic regions for T. chantrieri are those in the west (BSH, BB, WEV, QCT, GLQ, Th) and those in east (MLP, NWH, TP, DL, PS, CZ, WN, QZ).
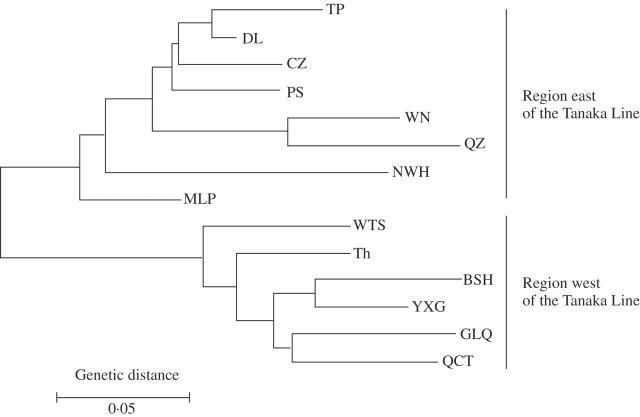
The neighbour-joining dendrogram based on the genetic distance between populations revealed a similar pattern: the genetic distances among the populations showed a spatial pattern that corresponded to their geographic locations (Fig. 2). Moreover, all 14 populations were clustered into two geographical groups: a clear geographical pattern of genetic diversity was identified along the Tanaka Line (Fig. 1), and a phytogeographical Line was identified between Sino-Japanese and Sino-Himalayan genera of East Asian flora (Tanaka, 1954; Li and Li, 1997). On the west side of the Line (south Yunnan–Thailand region), 66·78 % of the variance was attributable to populations (FST), and 33·22 % to individual differences within populations. On the east side (south-east Yunnan–Guangxi–Hainan region), 56·01 % of the variance was attributable to populations (FST) (Table 3).
Fig. 2.
Neighbour-joining tree of T. chantrieri based on pairwise Nei's genetic distance between populations (for explanation of codes see Table 1).
The result of a Mantel test with 1000 permutations revealed that the genetic divergence of populations (Nei's genetic distance) was significantly correlated with geographic distance in T. chantrieri (r = 0·5862, P = 0·001) (Fig. 3).
Fig. 3.
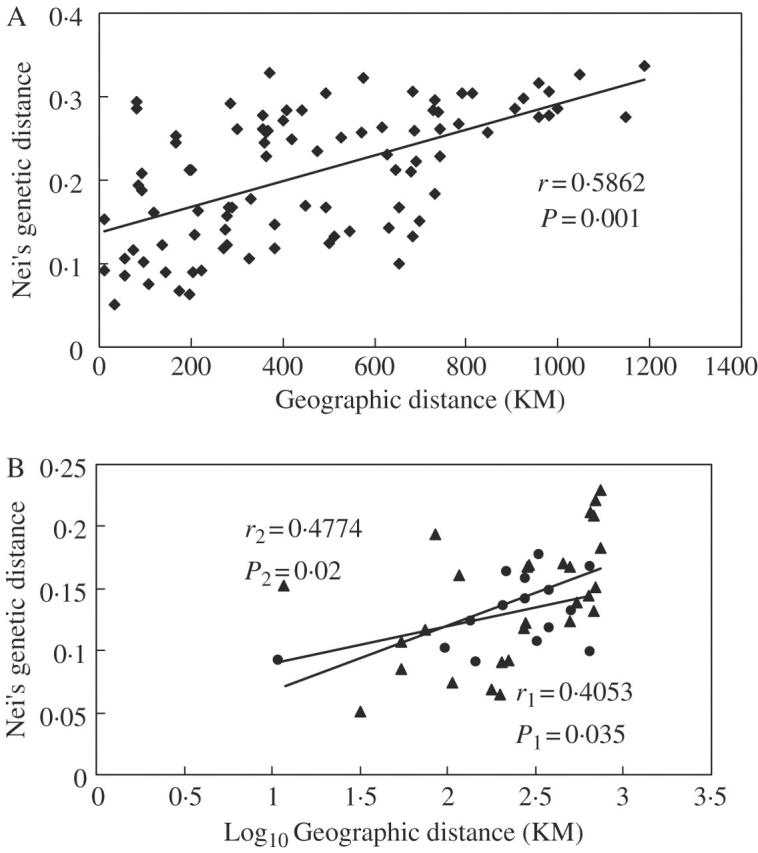
Correlation between geographical distance and Nei's genetic distance revealed by the Mantel test (TFPGA; Miller, 1997): (A) all populations; (B) considering separation by the Tanaka Line (circles, populations located on the west side of the Tanaka Line; triangles, populations located on the east side of the Tanaka Line).
DISCUSSION
Genetic variation in T. chantrieri and its correlation with the mating system
The ISSR survey of 14 populations of T. chantrieri revealed a high level of genetic variation at the species level, with 90·62 % of bands displaying polymorphism. However, there was considerable variation in PPB, with values ranging from 8·75 % to 55 %, and an average of 32·72 ± 12·82 %. This implied that a large proportion of genetic variation was partitioned among populations. In general, selfing species usually possess lower genetic diversity within populations and higher genetic differentiation among populations relative to outcrossing species (Hamrick and Godt, 1996). Therefore, the present data on the population genetic structure in T. chantrieri at large spatial scales are consistent with the highly selfing mating system documented at smaller spatial scales (Zhang et al., 2005). Among these 14 populations, the mating systems of WEV and BB have been quantified previously by allozyme markers. The WEV population had the highest selfing rate (Sm = 0·941), and exhibited the lowest genetic diversity (PPB = 15 %, HE = 0·041). The BB population also had a quite high selfing rate (Sm = 0·859) and contained very low genetic variation (PPB = 25 %, HE = 0·079) (Zhang et al., 2005). Among the four populations of T. chantrieri examined previously, estimates of the population level maternal selfing rate (Sm) averaged 0·86 (range 0·76–0·94). Such a figure is high, and is similar to that of other obligately selfing species. Consistent with this, the average genetic diversity in the south Yunnan–Thailand region was very low (PPB = 23·125 %, HE = 0·075). Similarities between the conclusions based on ISSR markers in this investigation and previous studies of reproductive biology and the mating system of T. chantrieri illustrate the potential of ISSR markers for population genetic studies.
Another feature of the population genetic variation of T. chantrieri is that populations maintain quite a large amount of ISSR variation, but it is not correlated with population size. The highest genetic variation was in population TP (PPB = 55 %, HE = 0·181), which had quite a small population size (only about 100 reproductive plants). Population QZ possessed the greatest size (>1000 reproductive individuals), but maintained only a medium level of genetic variation (PPB = 31·88 %, HE = 0·095). This result indicated that current population size cannot be a criterion for population genetic variation in this species.
Genetic structure patterns among populations of T. chantrieri and their possible causes
The present analyses of the data obtained from ISSR markers using different approaches (Nei's genetic diversity analysis and AMOVA) demonstrated similar patterns of genetic structure for populations of T. chantrieri. The AMOVA indicated that 69·89 % of the total genetic variation was partitioned among populations. In comparison with genetic variation and structure based on RAPD analyses of other wild plant populations (Nybom and Bartish, 2000; Nybom, 2004), the amount and pattern of genetic variation in T. chantrieri is more comparable to selfing or mixed mating taxa than to outcrossing species. The GST among populations was 0·5835, which was similar to the average for selfing plant species (0·51) in the analysis by Hamrick and Godt (1990).
A high level of population differentiation may be explained by several factors, such as the species breeding system, genetic drift, demographic fluctuations, or the genetic isolation of populations (Hogbin and Peakall, 1999). When populations are small and geographically and genetically isolated from one another, genetic drift influences the genetic structure and increases differentiation among populations (Barrett and Kohn, 1991; Ellstrand and Elam, 1993). Estimates of the effective gene flow per generation (Nm) of T. chantrieri were lower (0·3568) than one successful migrant per generation. This indicated limited gene flow among populations, which may be insufficient to counteract the effect of genetic drift. While inferences of the migration rate from estimates of Nm are not definitive for populations that do not exhibit metapopulation dynamics or large demographic shifts (Whitlock and McCauley, 1999), the method is still a reasonable guide to levels of gene flow among populations. The low estimates of migration among T. chantrieri populations correspond well with the geographic isolation of the populations, in which genetic differentiation among populations appears to be correlated with geographic distance between populations. For example, TP and DL are geographically close, and the genetic distance between them is also relatively small.
Geographical patterns of genetic differentiation of T. chantrieri and their implications
The neighbour-joining dendrogram based on the genetic distance between populations revealed a similar pattern to that of the genetic distances among populations: both showed a spatial pattern that corresponded to their geographic location (Fig. 2). From the neighbour-joining tree, significant genetic differences were found between the south Yunnan–Thailand and south-east Yunnan–Guangxi–Hainan regions, and coincidently separated by a presumed biogeographic boundary, The Tanaka Line (Tanaka, 1954; Li and Li, 1997). It is suggested that current genetic diversity distribution pattern of T. chantrieri populations might be due to a possible evolutionary event under vicariance from a single common ancestor through fragmentation of its original geographic range, and this vicariance could be explained by the different history of the geological structure on each side of the Tanaka Line.
The Tanaka Line is considered to be a boundary between the Sino-Japanese plate/biogeographic region in the east and the Sino-Himalayan plate/biogeographic region in the west. The approximate position of the Tanaka Line can be shown as a straight Line starting at the intersection of 28 °N, 98 °E southward to approximately 18 °45′ or 19 °N, 108 °E (Fig. 1). In general, the floral components of the Sino-Japanese region are relictual, and the elements of the Sino-Himalayan are evolved. The south-east Yunnan–Guangxi–Hainan area is part of an important floristic region in China called the Dian–Qian–Gui biogeographic region, and is located on the east side of the Tanaka Line. This region is noted for species abundance, endemism and historically high rates of speciation. The results of the present study indicated that expected heterozygosity of T. chantrieri was higher to the east (HE = 0·234) than to the west (HE = 0·168) of the Tanaka Line (Table 2). However, genetic differentiation among populations was greater to the west (GST = 0·563, FST = 0·6678) than to the east of the Line (GST = 0·417, FST = 0·5601) (Table 2). All these observations are consistent with an evolutionary origin for T. chantrieri in the Dian–Qian–Gui region, with a relatively recent range expansion to the west, resulting in reduced diversity and a higher population differentiation in the western region.
The genetic structure of plant populations is also influenced by the long-term evolutionary and ecological history of the species, which would include shifts in distribution, habitat fragmentation and population isolation (Schaal et al., 1998). Wu et al. (2003) proposed that Tacca originated from the southern marginal area of the Palaearctic continent when Pangaea expanded to the Pacific Ocean for the first time. Later, this genus became differentiated in a succession of nearby environments. Moreover, they also hypothesized that the northern part of the Indo-Chinese peninsular, stretching from Yunnan to Tibet, might be the site of ancient differentiation of this genus. The pattern of genetic structure of T. chantrieri is consistent with this: the genetic diversity was quite high south-east of Yunnan (TP and MLP populations), Guangxi (DL and PS populations) and Hainan (WN population) areas. This geographic area exactly corresponds to one of the centres of endemism in China (south-east of the Yunnan, Guizhou and Guangxi regions) (Li, 1996) and Hainan, which became separated from the south of China five million years ago (Zhu and Roos, 2004). The present results might support Wu's hypothesis to a certain extent: the original geographic region of this species lies from Vietnam to the northern edge of the subtropics in south-east Yunnan, Guizhou, Guangxi and Hainan (the Dian–Qian–Gui region) (Wu et al., 2003). At the very least, the populations on the east side of the Tanaka Line originated earlier than those on the west side.
CONCLUSIONS
For ISSR markers, T. chantrieri exhibits low levels of diversity within populations, and significant genetic variation among populations. This genetic structure is unexpected for a species with an extravagant floral display, but corresponds with the mating system of this species as previously quantified. Geographical patterns of genetic differentiation of T. chantrieri provide strong evidence for both its evolutionary and ecological history, and its vicariance among populations. Additional studies measuring genetic variation in other populations in southern distribution areas of this species, and in other species sympatric to it, would be especially helpful in determining if the population genetic structure detected in T. chantrieri is unique among other Tacca species.
Acknowledgments
We thank Dr Zhi-Yong Zhang, Mr Jie Cai for assistance in some laboratory work, Dr Wan-Jin Liao and Dr Xue-Jun Ge for assistance in ISSR analysis, as well as Mr Jiang-Yun Gao, Mr Zhi-Lin Bai and Mr Fan Chen for their help during fieldwork and collection of samples. Special thanks to Dr Spencer Barrett and Dr David Erickson for their comments and suggestions to help improve early drafts of the manuscript, and Dr David Westcott for improving the English. The research was supported by a grant from the Ministry of Science and Technology (2004 DKA30430) to D.-Z. Li, the Yunnan Provincial Natural Science Foundation grant 2003C0024Q to L. Zhang, and the National Natural Science Fundation of China (30225007) to Q.-J. Li.
LITERATURE CITED
- Avise JC. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press.
- Barrett SC, Kohn JR. 1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In Falk DA, Holsinger KE, eds. Genetics and conservation of rare plants. New York, NY: Oxford University Press, 3–30.
- Bauert MR, Kälin M, Baltisberger M, Edwards PJ. 1998. No genetic variation detected within isolated relict populations of Saxifraga cernua in the Alps using RAPD markers. Molecular Ecology 7: 1519–1527. [Google Scholar]
- Brown ADH, Burdon JJ, Jarosz AM. 1989. Isozyme analysis of plant mating systems. In: Soltis DE, Soltis PS, eds. Isozymes in plant biology. Portland, OH: Dioscorides Press, 73–86.
- Charlesworth D, Charlesworth B. 1987. The effect of investment in attractive structures on allocation to male and female functions in plants. Evolution 41: 948–968. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. 1995. Quantitative genetics in plants: the effect of the breeding system on genetic-variability. Evolution 49: 911–920. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. 1991. DNA protocols for plants: CTAB total DNA isolation. In: Hewitt GM, Johnston A, eds. Molecular techniques in taxonomy. Berlin: Springer-Verlag, 283–293.
- Drenth E. 1972. A revision of the family Taccaceae. Blumea 20: 367–406. [Google Scholar]
- Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- Emms SK, Stratton DA, Snow AA. 1997. The effect of inflorescence size on male fitness: experimental tests in the andromonoecious lily, Zigadenus paniculatus. Evolution 51: 1481–1489. [DOI] [PubMed] [Google Scholar]
- Esselman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD. 1999. Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Molecular Ecology 8: 443–451. [Google Scholar]
- Gupta M, Chyi Y-S, Romero-Severson J, Owen JL. 1994. Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theoretical and Applied Genetics 89: 998–1006. [DOI] [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. 1990. Allozyme diversity in plant species. In Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding, and germplasm resources. Sunderland: Sinauer, 43–63.
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London, Series B 351: 1291–1298. [Google Scholar]
- Harder LD, Barrett SCH. 1996. Pollen dispersal and mating patterns in animal pollinated plants. In: Lloyd DG, Barrett SCH, eds. Floral biology: studies on floral evolution in animal-pollinated plants. New York, NY: Chapman and Hall, 140–190.
- Harris J. 1999. RAPDs in systematics—a useful methodology? In: Hollingsworth PM, Bateman RM, Gornall RJ, eds. Molecular systematics and plant evolution. London: Taylor and Francis, 221–228.
- Hewitt GM. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Botanical Journal of the Linnean Society 58: 247–276. [Google Scholar]
- Hogbin PM, Peakall R. 1999. Evaluation of the contribution of genetic research to the management of the endangered plant Zieria prostrata. Conservation Biology 13: 514–522. [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Arizona State University, Tempe, AZ, USA. [DOI] [PubMed]
- Li XW. 1996. Floralristic statistics and analyses of seed plants from China. Acta Botanica Yunnanica 18: 363–384. [Google Scholar]
- Li XW, Li J. 1997. The Tanaka-Kaiyong Line—an important floristic Line for the study of the flora of East Asia. Annals of Missouri Botanical Garden 84: 888–892. [Google Scholar]
- McDermott JM, McDonald BA. 1993. Gene flow in plant pathosystems. Annual Review of Phytopathology 31: 353–373. [Google Scholar]
- Miller MP. 1997. Tools for population genetic analysis. Version 1.3. Department of Biological Sciences, Northern Arizona University, Flagstaff.
- Nybom H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143–1155. [DOI] [PubMed] [Google Scholar]
- Nybom H, Bartish IV. 2000. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3/2: 93–114. [Google Scholar]
- Proctor M, Yeo P, Lack A. 1996. The natural history of pollination. Portland, MA: Timber Press.
- Saw LG. 1993. Tacca: flowering and fruiting behaviour. Nature Malaysiana 18: 3–6. [Google Scholar]
- Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA. 1998. Phylogeographic studies in plants: problems and prospects. Molecular Ecology 7: 465–474. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. 2000. Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Schoen DJ. 1982a. The breeding system of Gilia achileifolia: variation in floral characteristics and outcrossing rate. Evolution 36: 352–360. [DOI] [PubMed] [Google Scholar]
- Schoen DJ. 1982b.. Genetic variation and the breeding system of Gilia achileifolia. Evolution 36: 361–370. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Clegg MT. 1985. The influence of flower color on outcrossing rate and male reproductive success in Ipomoea purpurea. Evolution 39: 1242–1249. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Morgan MT, Bataillon T. 1996. How does self-pollination evolve? Inferences from floral ecology and molecular genetic variation. Philosophical Transactions of the Royal Society of London, Series B 351: 1281–1290. [Google Scholar]
- Soltis DE, Gitzendanner MA, Strenge DD, Soltis PS. 1997. Chloroplast DNA intraspecific phylogeography of plants from the Pacific Northwest of North America. Plant Systematics and Evolution 206: 353–373. [Google Scholar]
- Tanaka T. 1954. Species problems in Citrus. Tokyo: Japanese Society for the Promotion of Science.
- Tsumura Y, Ohba K, Strauss SH. 1996. Diversity and inheritance of inter-simple sequence repeat polymorphisms in Douglas-fir (Pseudotsuga menziesii) and sugi (Cryptomeria japonica). Theoretical and Applied Genetics 92: 40–45. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, McCauley D. 1999. Indirect measures of gene flow and migration: FST ≠ 1/(4Nm +1). Heredity 82: 117–125. [DOI] [PubMed] [Google Scholar]
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research 18: 6531–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Liston A. 1998. Contributions of PCR-based methods to plant systematics and evolutionary biology. In Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants. II. DNA sequencing. Norwell: Kluwer Academic Publishers, 43–86.
- Wu Z-Y, Lu A-M, Tang Y-C, Chen Z-D, Li D-Z. 2003. The families and genera of angiosperms in China: a comprehensive analysis. Beijing: Science Press, 217–218 [in Chinese].
- Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX. 1997. POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton, Alberta, Canada.
- Zhang L, Barrett SCH, Gao J-Y, Chen J, Cole WW, Liu Y, Bai Z-L, Li Q-J. 2005. Predicting mating patterns from pollination syndromes: the case of ‘sapromyiophily’ in Tacca chantrieri (Taccaceae). American Journal of Botany 92: 517–524. [DOI] [PubMed] [Google Scholar]
- Zhu H, Roos MC. 2004. The tropical flora of southern China and its affinity to Indo-Malesian flora. Telopea 10: 639–648. [Google Scholar]