Abstract
Desaturation of coenzyme-A esters of saturated fatty acids is a common feature of sex pheromone biosynthetic pathways in the Lepidoptera. The enzymes that catalyze this step share several biochemical properties with the ubiquitous acyl-CoA Δ9-desaturases of animals and fungi, suggesting a common ancestral origin. Unlike metabolic acyl-CoA Δ9-desaturases, pheromone desaturases have evolved unusual regio- and stereoselective activities that contribute to the remarkable diversity of chemical structures used as pheromones in this large taxonomic group. In this report, we describe the isolation of a cDNA encoding a pheromone gland desaturase from the cabbage looper moth, Trichoplusia ni, a species in which all unsaturated pheromone products are produced via a Δ11Z-desaturation mechanism. The largest ORF of the ≈1,250-bp cDNA encodes a 349-aa apoprotein (PDesat-Tn Δ11Z) with a predicted molecular mass of 40,240 Da. Its hydrophobicity profile is similar overall to those of rat and yeast Δ9-desaturases, suggesting conserved transmembrane topology. A 182-aa core domain delimited by conserved histidine-rich motifs implicated in iron-binding and catalysis has 72 and 58% similarity (including conservative substitutions) to acyl-CoA Δ9Z-desaturases of rat and yeast, respectively. Northern blot analysis revealed an ≈1,250-nt PDesat-Tn Δ11Z mRNA that is consistent with the spatial and temporal distribution of Δ11-desaturase enzyme activity. Genetic transformation of a desaturase-deficient strain of the yeast Saccharomyces cerevisiae with an expression plasmid encoding PDesat-Tn Δ11Z resulted in complementation of the strain’s fatty acid auxotrophy and the production of Δ11Z-unsaturated fatty acids.
Keywords: acyl-CoA desaturase, Lepidoptera, pheromone biosynthesis
Acyl-CoA Δ9Z-desaturases occur ubiquitously in the animal and fungal kingdoms, where they play essential roles in fatty acid metabolism (for review, see ref. 1) and the regulation of cell membrane fluidity in response to temperature fluctuations (2, 3). These non-heme iron-containing enzymes typically use saturated 16- and 18-carbon fatty acyl thioesters of CoA (palmitoyl-CoA and stearoyl-CoA, respectively) as substrates (4) and catalyze the NADH- and oxygen-dependent removal of hydrogen atoms from carbon atoms 9 and 10 to form the Z double bond of palmitoleic and oleic acids, respectively (5).
Early biochemical investigations of acyl-CoA Δ9-desaturases of vertebrates showed that the enzyme is an integral membrane protein of the endoplasmic reticulum and that only a small portion of it, containing the iron-containing catalytic center, is exposed to the cytoplasm (5–7). Investigations of the enzyme’s functional association with other proteins and membrane lipids showed that the active desaturase is a complex of three proteins, of which two are components of the NADH-dependent electron transport system of the endoplasmic reticulum, NADH-cytochrome b5 reductase (a flavoprotein) and cytochrome b5 (a hemoprotein) (8–12).
Characterization of a stearoyl-CoA Δ9-desaturase cDNA isolated from rat liver revealed an encoded 358-aa apoprotein with >60% hydrophobic residues (13), consistent with the finding of earlier biochemical studies. Subsequent characterizations of additional vertebrate acyl-CoA desaturase cDNAs from mouse (14, 15), carp (3), and humans (16) showed extensive sequence similarity (identity plus conservative substitutions exceeding 90%, excluding the initial 40–60 amino-terminal amino acids) among the encoded proteins. Two cDNAs that encode proteins having significant sequence similarity to vertebrate acyl-CoA Δ9-desaturases also have been isolated recently from the tick Amblyomma americanum and from Drosophila melanogaster (17).
The palmitoyl-CoA Δ9-desaturase (OLE1) of the yeast Saccharomyces cerevisiae (18) also has been deduced from its encoding cDNA and has been shown to be a protein of 510 amino acids (251 hydrophobic) with a predicted molecular mass of ≈57 kDa (19). Optimal alignment of the S. cerevisiae OLE1 amino acid sequence with that of the rat stearoyl-CoA Δ9-desaturase revealed a 257-aa region having 36% identity and 60% similarity (19). The 113-aa carboxyl terminus of the yeast OLE1 protein, which has no structural correlate among characterized animal desaturases, has regions of high sequence identity to cytochrome b5 and has been shown to be essential for desaturase function (20). This cytochrome b5-like carboxyl-terminal extension is also present in the orthologous OLE1 desaturase of the oleaginous yeast, Cryptococcus curvatus (21). The functional equivalence of the S. cerevisiae and rat desaturases was shown by complementation of the desaturase-deficient ole1 strain’s unsaturated fatty acid (UFA) auxotrophy with a plasmid encoding the rat desaturase (19). Further molecular genetic investigations of the heterologously expressed rat desaturase identified eight histidine residues occurring in highly conserved sequence motifs that are essential for catalytic function (22).
The existence of a family of desaturases in the Lepidoptera possessing unusual catalytic properties initially was suggested by the elucidation of the chemical structures of the diverse UFA derivatives that are species-specific constituents of lepidopteran sex pheromones (reviewed in refs. 23–25). Subsequent studies of pheromone biosynthetic pathways revealed the basis of this molecular diversity arising from variations in a discrete number of conserved enzymatic steps involving synthesis of saturated fatty acids from acetate and acetyl-CoA, desaturation, limited chain-shortening by β-oxidation, and reduction of the terminal functional group (23–25). The number of unique regio- and stereospecific desaturation mechanisms discovered in these pathways, including Z9, E9, Z10, Z11, E11, Z14, and E14 mechanisms (23–27), indicates that the evolution of novel functional properties among pheromone desaturases has played a significant role in generating the remarkable diversity of chemical structures used as species-specific mate recognition signals in this large taxonomic group (25).
Biochemical studies of Δ11-desaturases from pheromone glands of Trichoplusia ni (28) and Spodoptera littoralis (29) showed that these enzymes have many similarities with the ubiquitous metabolic acyl-CoA Δ9-desaturases. In this report, we describe the isolation from the T. ni pheromone gland of a cDNA encoding a protein that is homologous to animal and fungal acyl-CoA Δ9-desaturases, the spatial and temporal occurrence of its corresponding mRNA at the level of Northern blot analysis, and its expression in an ole1-containing yeast strain resulting in complementation of the strain’s UFA auxotrophy and the formation of Z11-UFAs.
MATERIALS AND METHODS
RNA Isolations.
Pheromone glands were dissected from the abdomens of adult T. ni females reared in captivity, and total RNA was extracted as described (31, 32). RNA was isolated by the same procedure from dissected fat bodies of larvae (both sexes) and adult females and from abdominal muscle of adult females from which pheromone glands had been removed previously. Poly(A)+ RNA was selected from total RNA by using either oligo(dT) cellulose (Ambion, Austin, TX) or oligo(dT) paramagnetic beads (Dynal, Great Neck, NY).
Isolation of Pheromone Desaturase cDNAs.
First-strand oligo(dT)-primed cDNA was made from poly(A)+ RNA (Superscript kit, GIBCO/BRL) according to the manufacturer’s protocol. A pheromone gland cDNA library was made in λZAP (Stratagene) according to the manufacturer’s protocol. A second library used has been described (32). A hybridization probe was made by using a 550-bp segment of the T. ni Δ11-desaturase cDNA in a PCR-based procedure (33, 34). Degenerate primers were designed to encode histidine-rich sequence motifs conserved in acyl-CoA Δ9-desaturases of rat (13) and yeast (19) as follows (where [N] is all four bases): 5′-d9d1 = 5′-CCCCA[T/C]C[G/A][N]CT[G/C]TGG[T/A]C[N]CA-3′; and 3′-d9d2 = 5′-CCCTCTAGA[G/A]TG[G/A][G/A][T/A]A[G/A]TT[G/A]TG[G/A][T/A]A-3′. Primers were incubated with 100 ng of cDNA template and Taq polymerase (Perkin–Elmer), and PCR was performed for 55 cycles of 94°C for 2 min, 48°C for 1 min, and 68°C for 1 min. Parallel reactions were performed by using the cloned rat and yeast acyl-CoA desaturase cDNAs as templates (kindly provided by Philipp Strittmatter, Univ. of Connecticut Health Center, Farmington, CT and Charles Martin, Rutgers Univ., Piscataway, NJ, respectively). The PCR product was digested with XbaI to produce a sticky end at the 3′-d9d2 terminus and was ligated into a plasmid linearized by digestion with XbaI and SmaI. Escherichia coli strain XL1-Blue (Stratagene) was transformed with the ligation reaction, and insert-containing plasmids obtained from the resultant clones were sequenced by using an automated sequencer (Applied Biosystems).
The cloned PCR product was labeled with digoxigenin (Genius kit, Boehringer Mannheim) according to the manufacturer’s protocol and was hybridized under standard conditions to plaque lifts of the cDNA libraries. Probe-positive λZAP clones were isolated, and plasmids were obtained by the manufacturer’s automatic excision protocol for sequencing as above.
Northern Blot Analysis.
Northern blots of poly(A)+ RNA isolated from various tissues were hybridized with digoxigenin-labeled probes as described (35). The equivalence of RNA loadings was assessed by spectrophotometric measurement at A260/A280, methylene blue staining of RNA on the blot, and quantitation of β-actin sequences by reverse transcription–coupled PCR (Superscript kit, GIBCO/BRL). A 784-bp digoxigenin-labeled hybridization probe was made in a PCR (33 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 1.5 min) containing the PDesat-TnΔ11Z cDNA, the specific primers 5′-d11–1 = 5′-GAAGCTCGCACGATGAC-3′ and 3′-d11–3 = 5′-CTAATTCTGCTGTGCGGTAATCCCATGGAAAGAC-3′, 400 μM dATP, 400 μM dCTP, 400 μM dGTP, 260 μM dTTP, 140 μM alkali-labile digoxigenin-dUTP (PCR DIG labeling mixture, Boehringer), and Taq Polymerase (Perkin–Elmer). Reverse transcription–coupled PCRs containing the β-actin primers 5′-βact = 5′-ATGTG[C/T]AA[A/G]GC[N]GG[N]TT[T/C]GC-3′ and 3′-βact = 5′-GGNGC[A/G/T]AT[A/G/T)AT[C/T]TT[A/G/T]AT[C/T]TTCAT-3′, derived from the invariant amino acid sequences MCKAGFA and MKIKIIAP (36), respectively, were done as follows: 35 cycles of 94°C for 2 min, 60°C for 1 min, and 72°C for 1 min.
Plasmid Construction and Yeast Transformation.
A cDNA fragment containing the 349-aa ORF of the PDesat-TnΔ11Z cDNA and flanked by BamHI and SacI restriction sites at its 5′ and 3′ ends, respectively, was obtained by PCR by using specific primers complementary to the 20-nt terminal sequences of the ORF. This product was digested with BamHI and SacI and was subcloned into a plasmid derivative of YEp352/OLE4.8 in a procedure identical to that used to construct a functional yeast/rat desaturase gene (19). The final plasmid, designated YEpOLEX-CLR7, contains the 349-aa PDesat-TnΔ11Z-encoding ORF ligated via a four-codon linker in-frame with and downstream from the 5′ end of the OLE1 ORF encoding the first 27 amino acids of the yeast Δ9-desaturase. Sequences flanking the chimeric ORF consisted of the promoter and terminator elements of the OLE1 gene contained on the original YEp352/OLE4.8 plasmid. The latter and the S. cerevisiae strain L8–14C (MATα, ole1Δ∷LEU2, leu2–3, leu2–112, trp1–1, ura3–52, his4) (19) used in this investigation were generously provided to us by Charles Martin.
Before transformation, L8–14C cells were grown to ≈1 × 107 cells/ml at 30°C in yeast extract/peptone/dextrose medium (2% Bacto-peptone, 1% yeast extract, 2% glucose) containing 1% Tergitol (Type Nonidet P-40, Sigma–Aldrich; to solubilize UFAs), 0.5 mM oleic acid, and 0.5 mM palmitoleic acid. The strain was transformed with the YEpOLEX-CLR7 plasmid DNA by a standard method (37) according to the manufacturer’s protocol (alkali-cation yeast transformation kit, Bio 101) and was plated onto complete synthetic dextrose medium containing 1% Tergitol (Type Nonidet P-40, Sigma–Aldrich; to solubilize UFAs), 0.5 mM oleic acid, and 0.5 mM palmitoleic acid and lacking uracil.
Functional Assays.
To test for genetic complementation of the ole1 auxotrophy by the YEpOLEX-CLR7 plasmid, URA+ transformant colonies were selected and patched onto complete (yeast extract/peptone/dextrose) medium lacking supplemental UFAs and were incubated at 30°C for 24–48 hours. The transformants then were grown to ≈1 × 107 cells/ml at 30°C in yeast extract/peptone/dextrose medium and were washed and extracted with chloroform/methanol (3:1). After solvent evaporation, the residue was treated with 0.5 M KOH/methanol, and the resulting fatty acid methyl esters were analyzed by GC/MS (HP 5970). The positions of double bonds in unsaturated 16- and 18-carbon fatty acids were confirmed by analysis of the degradation products of their dimethyl disulfide adducts (38).
RESULTS AND DISCUSSION
Rationale for Homology Probing Strategy.
The discovery that pheromone desaturases have significant biochemical similarities to the ubiquitous metabolic acyl-CoA Δ9-desaturases (28, 29) led us to hypothesize that both classes of enzymes are related by descent from a common ancestral gene and that structural domains that are conserved among metabolic acyl-CoA Δ9-desaturases from phylogenetically distant species also will be present in the lepidopteran pheromone gland desaturases. To test this hypothesis, we used a PCR-based homology probing methodology that used degenerate primers encoding two highly conserved, histidine-rich sequence motifs that have been implicated in iron binding and catalytic function of acyl-CoA Δ9-desaturases (22).
The cDNA templates used in PCR reactions were synthesized from mRNA isolated from the pheromone glands of T. ni, which was chosen for this investigation for the following reasons: (i) The pheromone desaturase of this species is one of only two biochemically well characterized pheromone desaturases (28, 29); (ii) the unsaturated products present in the T. ni pheromone gland are all derivatives of a Z11 desaturation mechanism that is present only in the pheromone gland (28); (iii) the T. ni pheromone gland is a relatively large discrete eversible sac that can be neatly dissected away from other cuticular structures and fatty tissues; and (iv) the temporal induction of pheromone biosynthetic enzymes and protein expression patterns in the pheromone gland of this species have been characterized (30), enabling us to use a developmental stage in which pheromone desaturase mRNA levels are most likely to be at their highest levels.
Isolation of the PDesat-TnΔ11Z cDNA from T. ni Pheromone Gland.
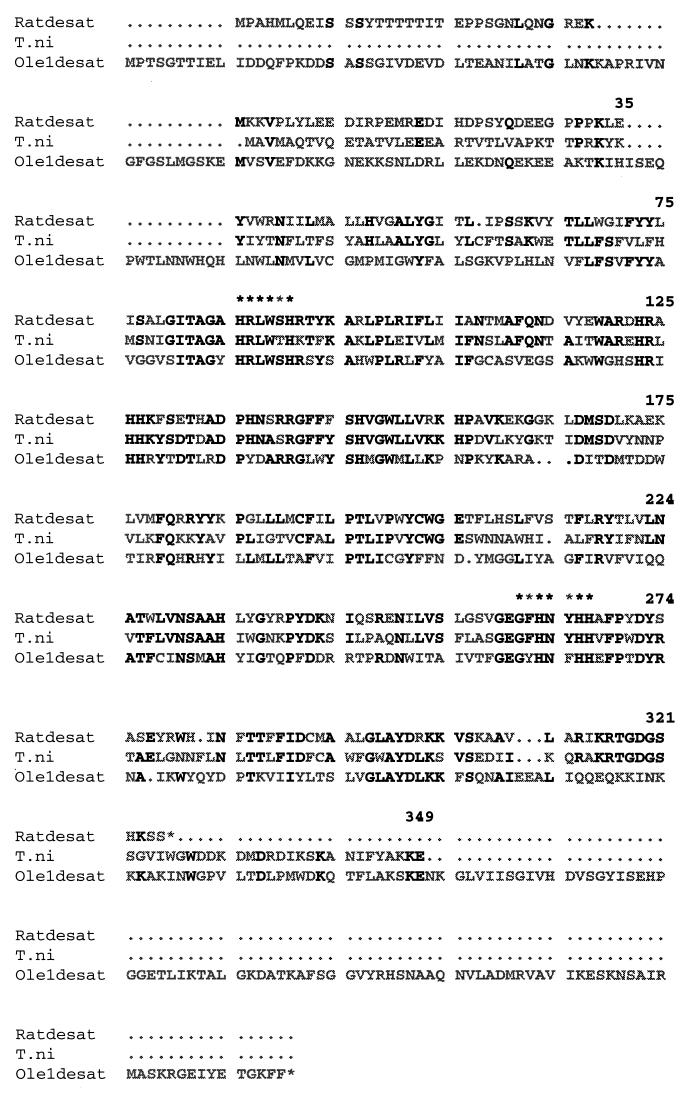
The experimental procedures that we used resulted in the cloning of a full length cDNA with an ORF of 1,047 nt encoding the 349-aa protein designated PDesat-TnΔ11Z (Fig. 1). The 560-bp product obtained in the initial PCRs containing the degenerate primers 5′-d9d1 and 3′-d9d2 and T. ni pheromone gland cDNA was labeled and hybridized to two independently prepared pheromone gland cDNA libraries. An extremely high percentage (>2%) of clones was labeled, consistent with a high abundance of the corresponding mRNAs in the pheromone glands from which the libraries were made. Several probe-positive clones were isolated and sequenced. All cDNAs longer than 1.2 kilobases contained an ORF of 1,047 nt encoding a 349-aa protein. Several sequence polymorphisms were observed among the clones, with the most common ones occurring at positions 22 (Val or Met), 58 (Cys or Ser), 70 (Ser, Thr or Ala), and 315 (Lys or Glu). The protein representing the consensus amino acid sequence shown in Fig. 1 has a predicted molecular mass of 40,240 Da and a pI of 9.12. It is possible that translation could be initiated at a second ATG occurring in-frame only three codons downstream from the ATG initiating the ORF, which would result in a slightly smaller protein. These two inferred PDesat-TnΔ11Z polypeptides are similar in size to one of about a dozen proteins, the expression of which is associated with the onset of maximal pheromone biosynthetic capability in the T. ni pheromone gland (30).
Figure 1.
The PDesat-TnΔ11Z consensus amino acid sequence (T. ni) compared with acyl-CoA Δ9-desaturases of rat (Ratdesat) and yeast (Ole1desat). Numbering begins with the first methionine of the T. ni sequence. Identities in the aligned sequences are shown in bold, and nonidentities are shown in gray. Starred amino acids are the motifs to which 5′-d9d1 and 3′-d9d2 target primers were made.
Homology of PDesat-Tn Δ11Z to Acyl-CoA Δ9-Desaturases.
Alignment of the PDesat-TnΔ11Z amino acid sequence with those of rat (13) and yeast (19) acyl CoA Δ9-desaturases (Fig. 1) shows that the deduced PDesat-TnΔ11Z protein is similar in size to the acyl-CoA Δ9-desaturase from rat and lacks the cytochrome b5-like carboxyl-terminal extension of the OLE1 desaturases of S. cerevisiae (20) and C. curvatus (21). The highest level of sequence conservation between PDesat-TnΔ11Z and any acyl-CoA Δ9-desaturase occurs in the region corresponding to the primary 560-bp PCR product delimited by amino acid positions 86 and 267 (for rat, 55% amino acid identity and 72% similarity; for yeast, 34% identity and 58% similarity). Of the 13 histidine residues occurring in the PDesat-TnΔ11Z core domain, 11 are conservative, including all of those that are required for acyl-CoA Δ9-desaturase catalytic function (22). Conservation outside the core domain is somewhat lower, particularly in the amino terminal domain, resulting in overall conservation values as determined by the blast 2.0 program (39) of 52% identity and 70% similarity vs. rat and 30% identity and 50% similarity vs. yeast. As with the rat and yeast acyl-CoA Δ9-desaturases, the PDesat-TnΔ11Z protein has a high percentage of hydrophobic residues (77%), and its Kyte–Doolittle hydrophobicity plot (40) is similar to those of the rat and yeast acyl-CoA Δ9-desaturases (data not shown), consistent with conservation of the proposed acyl-CoA Δ9-desaturase transmembrane topology (19).
Comparisons of the sequences of the PDesat-TnΔ11Z core domain to the corresponding regions of two presumptive arthropod acyl-CoA Δ9-desaturases (sequences not shown) reveal that the PDesat-TnΔ11Z is about as similar to the fly acyl-CoA Δ9-desaturase as it is to the tick acyl-CoA Δ9-desaturase (for D. melanogaster, 61% identity and 85% similarity; for A. americanum, 61% identity and 80% similarity). Comparisons of the corresponding region of the rat acyl-CoA Δ9-desaturase to the latter two sequences gave similar values (for D. melanogaster, 64% identity and 81% similarity; for A. americanum, 61% identity and 78% similarity). In the context of the similarity of all of these values, it is of interest to note that fossil evidence supports the divergence of lepidopteran and dipteran orders in the Permian Era between 240 and 280 million years ago (41) whereas recent molecular analyses support the divergence of insects and chelicerates (the latter including contemporary tick species) from a common ancestral form in the Precambrian Era >550 million years ago (42, 43).
PDesat-TnΔ11Z mRNA Occurrence Coincides with Δ11-Desaturase Activity.
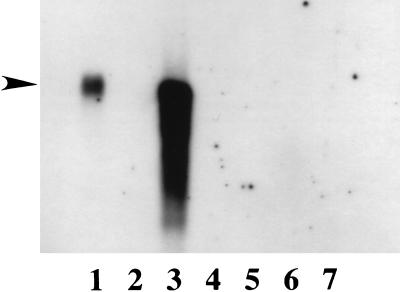
Northern blot experiments showed the presence of abundant levels of the PDesat-TnΔ11Z transcript in poly(A)+ RNA from pheromone glands of 48-hour posteclosion adult females (Fig. 2, lane 1), consistent with our finding of an extremely high percentage of clones containing PDesat-TnΔ11Z sequences in the pheromone gland cDNA libraries made from the same developmental stage. The mobility of the PDesat-TnΔ11Z transcript is indistinguishable from that of a transcript synthesized in vitro by using the linearized PDesat-TnΔ11Z cDNA template and SP6 RNA polymerase (Fig. 2, lane 3). The latter finding is consistent with the interpretations, based on the sequence analysis presented above, that the largest PDesat-TnΔ11Z cDNA clone isolated is either full length or nearly full length and that it encodes the entire PDesat-TnΔ11Z protein. The PDesat-TnΔ11Z transcript is not detected in loadings of similar amounts of poly(A)+ RNA isolated from fat bodies of adults (Fig. 2, lane 5) and larvae (Fig. 2, lane 7). We performed additional Northern blots and reverse transcription–coupled PCR experiments (data not shown) designed to increase the sensitivity of detection (by manipulating probe strength and duration of signal integration) and to quantify the levels of transcripts detected relative to those encoding β-actin, respectively. These experiments revealed PDesat-TnΔ11Z transcript in pheromone gland poly(A)+ RNA from newly eclosed (0–12 hour) adult females at levels approximately two orders of magnitude lower than those present on a mass equivalent basis in poly(A)+ RNA from pheromone glands of 48-hour posteclosion adult females but failed to detect PDesat-TnΔ11Z transcript in poly(A)+ RNA from adult thoracic muscle or fat bodies from larvae and adults. Reverse transcription–coupled PCR experiments performed on the RNA samples used for the Northern blots indicated that the maximal level of PDesat-TnΔ11Z transcript present in pheromone glands of 48-hour posteclosion females is at least three orders of magnitude greater than the level of β-actin transcripts present in the same developmental stage. Previous investigations have shown that the Δ11-desaturase activity of T. ni is limited to the pheromone gland of adult females whereas Δ9-desaturase activity is present in other fatty tissues throughout development in both sexes (26, 31). Furthermore, investigations of the induction of pheromone biosynthetic enzymes during the functional development of the T. ni pheromone gland showed that the amount of Δ11-desaturase activity in the pheromone gland increases by two to three orders of magnitude during the first 2 days after adult eclosion (31). The Δ11-desaturase activity of T. ni is, thus, coincident with the tissue-specific occurrence, rapid induction, and high steady state levels of PDesat-TnΔ11Z transcript in the pheromone glands of posteclosion females.
Figure 2.
Northern blot of T. ni poly(A)+ RNAs isolated from various tissues and hybridized with the PDesat-TnΔ11Z digoxygenin-labeled probe. Lanes: 1, adult (48 hours posteclosion) pheromone glands; 2, 4, and 6, no RNA; 3, in vitro synthesized PDesat-TnΔ11Z transcript; 5, adult (48 hours posteclosion) fat bodies; 7, third instar larval fat bodies. RNA loadings were 3 μg/lane except for the positive control in lane 3, which was <50 ng. Exposure time for the film shown was 90 minutes. The position of the arrow corresponds to the mobility of an RNA of ≈1,250 nt and was interpolated from the relative mobilities of RNAs present in a molecular mass ladder (not shown).
Functional Expression of PDesat-TnΔ11Z in Yeast.
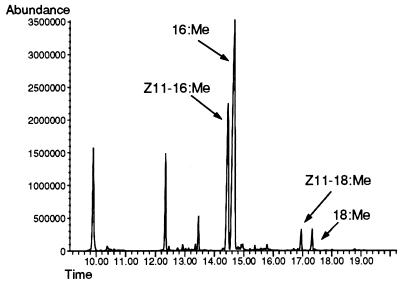
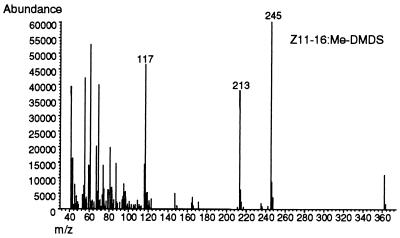
The demonstration that the UFA auxotrophy of a desaturase-deficient ole1 strain of S. cerevisiae can be complemented genetically with a plasmid encoding a rat stearoyl-CoAΔ9- desaturase cDNA (19) indicates the functional conservation of the interactions between acyl-CoA desaturases and the electron transport protein components of the functional desaturase complex (i.e., cytochrome b5 reductase and cytochrome b5). This, in conjunction with the experimental finding that the ole1 strain’s UFA auxotrophy can be rescued by supplementation of the growth medium with Δ11-UFAs (18), led us to predict that transformation of the ole1 strain with plasmids expressing the T. ni Δ11- desaturase also would meet the strain’s nutritional requirement for UFAs. To test this prediction, we constructed the plasmid YEpOLEX-CLR7 and transformed the L8–14C strain containing a nonreverting ole1 mutation with it. The YEpOLEX-CLR7 plasmid encodes a chimeric desaturase comprising a 27-aa amino terminal sequence fused via a short linker to the PDesat-TnΔ11Z protein. Transcription of the mRNA is under the regulation of the OLE1 promoter, which is repressed by oleic acid and palmitoleic acid but not by Δ11-UFAs (44, 45). When URA+ transformant colonies were transferred to complete (yeast extract/peptone/dextrose) medium lacking supplemental UFAs, the transformants grew, indicating complementation of the ole1 mutation by the yeast/PDesat-TnΔ11Z fusion protein. Analysis of the UFAs present in the YEpOLEX-CLR7-transformed strain revealed both Z11–16 and Z11–18 fatty acids in a ratio of ≈10-to-1 and no detectable Z9–16 or Z9–18 fatty acids (Fig. 3). The structural identities of the UFAs of the transformants were verified by making dimethyl disulfide adducts and analyzing their distinct mass spectra. The dimethyl disulfide adduct of Z11–16 methyl ester produces three diagnostic fragments: two methyl sulfide fragments at m/z 117 and 245 and a degradation product of the latter at m/z 213 (Fig. 4), compared with two fragments at m/z 145 and 217 and a degradation product of the latter at m/z 185 for the Z9 isomer (data not shown).
Figure 3.
GC/MS total ion spectrum of fatty acid methyl esters of the YEpOLEX-CLR7-transformed ole1 strain. The profile of the whole cell lipid extract shows the resolution of the identified monounsaturated products by capillary GLC.
Figure 4.
Mass spectral confirmation of the double bond position of the 16-carbon unsaturated product of the YEpOLEX-CLR7-transformed ole1 strain. The diagnostic m/z values in the MS scan of the methyl sulfide degradation products of the dimethyl disulfide adduct of Z11–16:Me are labeled.
Taken together, these findings unequivocally demonstrate that the PDesat-TnΔ11Z protein is homologous to acyl-CoA Δ9-desaturases occurring in species representing phylogenetically distant lineages and that it is the apoprotein component of the functional acyl-CoA Δ11-desaturase complex of the T. ni pheromone gland. The diversity of catalytic specificities that occur among the pheromone desaturases provides a unique opportunity to investigate the mechanistic basis of the desaturation reaction. The present investigation provides the technical precedent for the isolation and characterization of additional pheromone desaturases, which, in conjunction with functional studies, should permit the identification of specific mutational events that gave rise to additional catalytic specificities.
Acknowledgments
We thank Drs. Philipp Strittmatter and Charles Martin for gifts of cDNA clones, Drs. Claude Wicker-Thomas and Jean-Marc Jallon for sharing DNA sequence data before publication, Ms. Patricia Marsella-Herrick for technical assistance with molecular biology, and Ms. Kathy Poole for insect rearing and dissections. This work was supported by a grant to W.L.R. from the National Science Foundation (IBN-9514211) and by grants to D.C.K. from the National Science Foundation (IBN-9004979), the U.S. Department of Agriculture (97-35302-4345), and the Environmental Protection Agency/National Science Foundation (BES-9728367).
ABBREVIATION
- UFA
unsaturated fatty acid
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF035375).
References
- 1.Jeffcoat R. Essays Biochem. 1979;15:1–36. [PubMed] [Google Scholar]
- 2.Vigh L, Los D A, Horvath I, Murata N. Proc Natl Acad Sci USA. 1993;90:9090–9094. doi: 10.1073/pnas.90.19.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiku P E, Gracey A Y, Macartney A I, Beynon R J, Crossins A R. Science. 1996;271:815–818. doi: 10.1126/science.271.5250.815. [DOI] [PubMed] [Google Scholar]
- 4.Raju P K, Reiser R. J Biol Chem. 1967;242:379–384. [PubMed] [Google Scholar]
- 5.Jeffcoat R, Brawn P R, Safford R, James A T. Biochem J. 1977;161:431–437. doi: 10.1042/bj1610431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strittmatter P, Spatz L, Corcoran D, Rogers M J, Setlow B, Redline R. Proc Natl Acad Sci USA. 1974;71:4565–4569. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad M R, Joshi V C. J Biol Chem. 1979;254:6362–6369. [PubMed] [Google Scholar]
- 8.Holloway P W. Biochemistry. 1971;10:1556–1560. doi: 10.1021/bi00785a008. [DOI] [PubMed] [Google Scholar]
- 9.Rogers M J, Strittmatter P. J Biol Chem. 1973;248:800–806. [PubMed] [Google Scholar]
- 10.Shimakata T, Mihara K, Sato R. J Biochem (Tokyo) 1972;72:1163–1174. doi: 10.1093/oxfordjournals.jbchem.a130004. [DOI] [PubMed] [Google Scholar]
- 11.Spatz L, Strittmatter P. Proc Natl Acad Sci USA. 1971;68:1042–1046. doi: 10.1073/pnas.68.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spatz L, Strittmatter P. J Biol Chem. 1971;248:793–799. [PubMed] [Google Scholar]
- 13.Thiede M A, Ozols J, Strittmatter P. J Biol Chem. 1986;261:13230–13235. [PubMed] [Google Scholar]
- 14.Ntambi J M, Buhrow S A, Kaestner K H, Christy R J, Sibley E, Kelly T J, Jr, Lane M D. J Biol Chem. 1988;263:17291–17300. [PubMed] [Google Scholar]
- 15.Kaestner K H, Ntambi J M, Kelly T J, Lane M D. J Biol Chem. 1989;264:14755–14761. [PubMed] [Google Scholar]
- 16.Li J, Ding S-F, Habib N A, Fermor B F, Wood C B, Gilmore R S. Int J Cancer. 1994;57:348–352. doi: 10.1002/ijc.2910570310. [DOI] [PubMed] [Google Scholar]
- 17.Wicker-Thomas C, Henriet C, Dallerac R. Insect Biochem Mol Biol. 1997;27:963–972. doi: 10.1016/s0965-1748(97)00077-5. [DOI] [PubMed] [Google Scholar]
- 18.Stukey J E, McDonough V M, Martin C E. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 19.Stukey J E, McDonough V M, Martin C E. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 20.Mitchell A G, Martin C E. J Biol Chem. 1995;270:20144–20149. [Google Scholar]
- 21.Meesters P A E P, Eggink G. Yeast. 1996;12:723–730. doi: 10.1002/(sici)1097-0061(19960630)12:8<723::aid-yea963>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 22.Shanklin J, Whittle E, Fox B G. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 23.Bjostad L B, Wolf W A, Roelofs W L. In: Pheromone Biochemistry. Prestwich G D, Blomquist G J, editors. New York: Academic; 1987. pp. 77–120. [Google Scholar]
- 24.Roelofs W L, Wolf W A. J Chem Ecol. 1988;14:2019–2031. doi: 10.1007/BF01014247. [DOI] [PubMed] [Google Scholar]
- 25.Wolf W A, Roelofs W L. In: Biocatalysis in Agricultural Biotechnology. Whitaker J R, Sonnet P E, editors. Washington, DC: Am. Chem. Soc.; 1989. pp. 323–331. [Google Scholar]
- 26.Foster S P, Roelofs W L. Arch Insect Biochem Physiol. 1988;8:1–9. [Google Scholar]
- 27.Zhao C, Löftsedt C, Wang X. Arch Insect Biochem Physiol. 1990;15:57–65. [Google Scholar]
- 28.Wolf W A, Roelofs W L. Arch Insect Biochem Physiol. 1986;3:45–52. [Google Scholar]
- 29.Rodriguez F, Hallahan D L, Pickett J A, Camps F. Insect Biochem Mol Biol. 1992;22:143–148. [Google Scholar]
- 30.Tang J, Wolf W A, Roelofs W L, Knipple D C. Insect Biochem. 1991;21:573–581. [Google Scholar]
- 31.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 32.Tang J D, Roelofs W L, Knipple D C. In: Molecular Insect Science. Hagedorn H, Hildebrand J, Kidwell M, Law J, editors. New York: Plenum; 1990. p. 368. [Google Scholar]
- 33.Gould S J, Subramani S, Scheffler I E. Proc Natl Acad Sci USA. 1989;86:1934–1938. doi: 10.1073/pnas.86.6.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamb A, Weir M, Rudy B, Varmus H, Kenyon C. Proc Natl Acad Sci USA. 1989;86:4372–4376. doi: 10.1073/pnas.86.12.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engler-Blum G, Meier M, Frank J, Müller G A. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 36.Mounier N, Prudhomme J C. Biochimie (Paris) 1986;68:1053–1061. doi: 10.1016/s0300-9084(86)80179-1. [DOI] [PubMed] [Google Scholar]
- 37.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buser H R, Arn H, Guerin P, Rauscher S. Anal Chem. 1983;55:818–822. [Google Scholar]
- 39.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 41.Gillot C. Entomology. New York: Plenum; 1980. pp. 32–41. [Google Scholar]
- 42.Boore J, Collins T M, Stanton D, Daehler L L, Brown W M. Nature (London) 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 43.Friedrich M, Tautz D. Nature (London) 1995;376:165–167. doi: 10.1038/376165a0. [DOI] [PubMed] [Google Scholar]
- 44.McDonough V M, Stukey J E, Martin C E. J Biol Chem. 1992;267:5931–5936. [PubMed] [Google Scholar]
- 45.Choi J Y, Stukey J E, Hwang S Y, Martin C E. J Biol Chem. 1996;271:3581–3589. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]