Abstract
We have evaluated blood samples of chronic and congenital Trypanosoma cruzi-infected patients from the city of Reconquista located in the northeast of Argentina where no information was previously obtained about the genotype of infecting parasites. Fourteen samples of congenital and 19 chronical patients were analyzed by hybridization with DNA probes of minicircle hypervariable regions (mHVR). In congenital patients, 50% had single infections with TcIId, 7% single infections with TcIIe, 29% mixed infections with TcIId/e, and 7% had mixed infections with TcIId/b and 7% TcIId/b, respectively. In Chronical patients, 52% had single infections with TcIId, 11% single infections with TcIIe, 26% had mixed infections with TcIId/e, and 11% had non-identified genotypes. With these samples, we evaluated the minicircle lineage-specific polymerase chain reaction assay (MLS-PCR), which involves a nested PCR to HVR minicircle sequences and we found a correlation with hybridization probes of 96.4% for TcIId and 54.8% for TcIIe.
Introduction
American trypanosomiasis is a zoonosis caused by the protozoan Trypanosoma cruzi, extended throughout Latin America, where 16 to 18 million individuals are estimated to be infected and more than 50 million inhabitants are in a health hazard.1 Chagas disease is characterized by a broad range of clinical signs, which may be explained in part by the genetic heterogeneity of T. cruzi.2 In fact, a great diversity of T. cruzi clones with different biological, immunological, genetic, molecular, and pharmacological response characteristics have been isolated from different sources.3,4 In the last years, virulence and tropism differences have been reported in genetically distant strains.5–8 Furthermore, different T. cruzi genotypes have been correlated with different virulence and antibiotic resistance.9–13 These findings indicate that the genotype identification may be relevant to provide a prognosis of disease progression and/or differential treatment decisions based on the infective strain.
Currently, two major lineages are described in T. cruzi species, named T. cruzi I and T. cruzi II, and the T. cruzi II is subdivided into five sublineages, T. cruzi IIa, IIb, IIc, IId, and IIe.14,15 This classification is mainly based on methods requiring parasite isolation and culture. However, as the isolation of the parasites in cultures or mice could lead to strain selection, only direct characterization from insects and mammalian samples allow strains to be typed without bias.16 On this basis, and given the high sensitivity of kinetoplastid minicircle amplification by polymerase chain reaction (PCR), the polymorphism of minicircle hypervariable regions (mHVRs) has been used in many studies for the direct molecular characterization of parasitic populations from biological samples.8,17–22
Beyond the high variability of mHVR, the comparison of several minicircle regions within and among strains, has allowed identification of some sequences more represented in certain sublineages.23 In a previous work,24 we have described that a predominant minicircle sequence present in TcIIe and two others predominant minicircle sequences present in TcIId, are indeed present in reference strains from other T. cruzi lineages and sublineages.
However, the amount of these sequences is at least a thousand times more represented in the respective sublineage than in the others. Supported by this fact, we have developed a semiquantitative PCR directly from biological samples, using GrpI primers, designed by Velázquez and others,24 and primers (GrO-GrP) that amplify O and P sequences,23 to discriminate TcIIe and TcIId sublineages, respectively, and to differentiate them from the other lineages and sublineages.
In this work, we have evaluated this assay by extending the number of reference strains analyzed and testing biological samples by hybridization with a panel of genotype-specific minicircle DNA probes. This new method is highly useful for genotyping directly from blood of infected individuals because it does not require microorganism isolation, avoiding bias in the strain typing. Moreover, it uses kinetoplast DNA PCR products as templates, which is one of the most sensitive assays to detect parasite genetic material in biological samples.21,25–29 On the other hand, this assay requires only a PCR amplification and products are visualized by a simple agarose gel stained with ethidium bromide.
The biological samples analyzed in this work were obtained from acute and chronic chagasic patients from a high endemic region of northeastern Argentina. The current sampling extends epidemiological findings obtained by several groups of Latin America,8,22,30–32 and particularly of Argentina,21,33–36 showing the predominance of TcIId infection in human and mammalian hosts.
Materials and Methods
Reference strains.
Reference strains used in this work were: TcI strains, X10cl1; TcIIa strains, CANIIIcl1, TcIIb strains, TU18 cl93, IVV cl4, MCV, CBB cl3; TcIIc strains, M5631cl5 and P109/2; TcIId strains, JGG, XHCH56; NR cl3, SC43 cl1 and MN cl2; TcIIe strain, CL Brener and V195 cl1.
Patients and blood samples.
This study was carried out with 19 chronically and 14 congenitally infected individuals of Reconquista, Santa Fe province, Argentina who were positive by conventional serology enzyme-linked immunosorbent assay (ELISA) and indirect hemaglutination and/or microhematocrit in the congenital cases. Seven milliliters of blood (1 mL in neonates) were mixed with an equal volume of buffer Guanidine HCl/EDTA (6 M/0.2 M), boiled in a water bath for 15 minutes to shear and decatenate the minicircle DNA molecules from the kinetoplast network, and kept at −20°C until used for PCR assays.
Informed consent.
Informed consent was obtained from all human adult participants and neonate parents. The project was approved by the Ethical Committee of the Biochemistry Faculty, Nacional University of Litoral, Argentina.
DNA extraction.
Samples kept in polypropylene tubes containing guanidine/EDTA lysates were processed using 200 µL aliquots with the phenol-chloroform extraction, ethanol precipitation, and DNA resuspension in 200 µL sterile distilled water. Each set of purifications were performed with the respective positive and negative controls.
Hybridization with specific radiaoactive probes.
All PCR-amplified DNA from blood samples were analyzed by Southern blot assay with a panel of four genotype-specific probes from T. cruzi clones: CBB c13 (TCIIb), NR cl3 (TCIId), sp104 cl1 (TCI), and v195 cl1 (TCIIe). Because these probes derive from the whole kinetoplast, they are composed of all minicircle sequences present in a parasite clone. Clone sp104 was first isolated from Mepraia spinolai (sylvatic cycle). Meantime, the CBB and NR clones were isolated from chronically infected individuals, and clone v195 from Triatoma infestans (domestic cycle). The probes were radiolabelled with P32 and membranes were exposed and analyzed in a Personal Molecular Imager-FX (Bio-Rad, Hercules, CA). Moreover, probes were hybridized against a panel containing minicircle amplicons of T. cruzi clones as described.27
Minicircle lineage-specific PCR (MLS-PCR).
The assay consists in a nested PCR, in which the first reaction amplifies all mHVRs sequences, according to the classic T. cruzi detection.29 Briefly, amplification was performed in 50 µL reaction mixtures containing 5 µL of the total DNA isolated, 250 µM of each deoxynucleotide triphosphate, 100 pmol of each specific oligonucleotide for minicircle DNA S121 (5′-AAATAATGTACGGG[T/G]GAGATGCATGA-3′) and S122(5′-GGTTCGATTGGGGTTGGTGTAATATA-3′), 1.25 UI of Taq polymerase. The reaction mixtures were overlaid with 50 µL of mineral oil and subjected to 30 cycles of amplification in a thermocycler. Denaturation, annealing, and extension steps were performed for 60 sec each at 94, 60, and 72°C, respectively, with an initial denaturation at 94°C for 180 sec, and a final extension at 72°C for 5 minutes. Negative and positive DNA purification controls were included in each PCR reaction. Nested PCR were performed using quantified 330 bp PCR products as template. These products were quantified using a standard DNA curve according to protocol previously described.24 Briefly, pGEM-T easy vector DNA containing an insert of a CL–Brener–mHVR sequence was quantified by densitometry, at 260 nm. The proportional quantity of the released 330-bp PCR product was calculated, after digestion with EcoRI. The PCR products from each sample were then quantified by comparison of the electrophoresis gel images intensity obtained by digital capture with the DNA insert intensity taken as reference, using the Scion Image analysis software (Scion Corp., Frederick, MD). Dilutions were then performed to obtain 0.3 pg of DNA template, as previously standardized for minicircle lineage-specific polymerase chain reaction (MLS-PCR).24 To carry out this second PCR, three oligonucleotide primer pairs were used, two of them amplify two major TcIId sublineage minicircle sequences (GrO1- GrO2 and GrP1- GrP2) and the third amplify one major TcIIe sublineage minicircle sequence (Grp I 1- Grp I 2).
PCR conditions were the same as the first one, except for oligonucleotide primers (described below), with an annealing temperature of 55°C.
GrO1 5′-GGCTTAGGGTGTGGATAGG-3′
GrO25′-ATCGCGAAACCCATACAA-3′
GrP1 5′-GGGATTAGGGTATACTTAGTTGC-3′
GrP2 5′-TTCCAACCCCACAAATGATA-3′
Grp I 1 5′-ATCCAGACCCCAATTTTACTAC-3′
Grp I 2 5′-ATGTGATTGGATAGGTGATAGAT-3′
The MLS-PCR was performed on each sample with each of the three oligonucleotide primer pairs. Amplified products of first PCR negative and positive controls were included as negative and positive controls, respectively.
When the sensitivity of first PCR was below the established detection limit for the DNA quantification, the products were re-amplified using oligonucleotides complementary to minicircle regions, CV1 5′-GATTGGGGTTGGAGTACT-3′, and CV2 5′-TTGAACGGCCCTCCGAAAA-3′, obtaining an internal product of 290 bp. As template, it used a 10-fold dilution of first PCR amplification products, as described.37 The reaction mixtures were overlaid with 50 µL of mineral oil and subjected to a hot start at 95°C for 5 min and a first cycle at 48°C, 2 min and 70°C, 2 min; afterward, 28 cycles at 95°C for 5 sec, 48°C for 30 sec, and 70°C for 2 min; and a final cycle repeating previous steps with a final extension at 70°C for 15 min.
Statistics.
The χ2 test was used to assess the influence of the product concentration of the first PCR in the sensitivity of the MLS-PCR done with GrO or GrP primers, and Kappa index was used to analyze the concordance of hybridization probes with MLS-PCR.
Results
Evaluation of MLS-PCR using reference strains.
Fifteen strains, belonging to different lineages and sublineages of T. cruzi were analyzed by MLS-PCR using 0.3 pg of the first amplification product as reported in Materials and Methods. Typing of CAN III, M5631, MN, and CL Brener strains were performed twice, starting from parasite DNA obtained from collections of two different laboratories. As expected, the belonging to TcIIe sublineage amplified only with GrpI primers and strains belonging to TcIId sublineages amplified exclusively with GrO and GrP primers, indicating complete matching (Figure 1).
Figure 1.
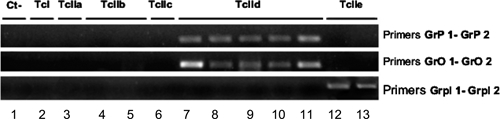
Agarose gel of minicircle lineage-specific polymerase chain reaction (MLS-PCR) performed with some reference strains of all Trypanosoma cruzi sublineages. 1.5% agarose gel Etidium Bromide-stained. Lanes: 1) Negative Control, 2) X10 cl1 (TcI), 3) CANIII cl1 (TcIIa), 4) CBB (TcIIb), 5) IVV cl4 (TcIIb), 6) M5631 cl5 (TcIIc), 7) MNcl2 (TcIId), 8) XHCH56 (TcIId), 9) NR cl3(TcIId), 10) JGG (TcIId), 11) SC43cl1(TcIId), 12) V195 cl1(TcIIe), 13) CL Brener (TcIIe).
For all TcIId strains, when the same amount of DNA template (0.3 pg) was used, the intensity of the bands was lower using GrO primers, except for SC43cl1 and MN cl2 strains, which displayed intense bands in the amplification using both GrO and GrP primers (Figure 1).
Genotyping of human samples by hibridization assay.
Until now, the typing of T. cruzi genotypes responsible for Chagas disease in northeastern Argentina was not boarded. In this work, we have done a preliminary analysis of genotypes present in this region using minicircle DNA probes and hybridization tests, which is a widely used method to approach this task.11,22,38 These results not only gave information about genotypes present in our region but they allowed us to make a genotyped panel of human samples to test with the MLS-PCR method. From the 14 congenitally infected individuals analyzed, 7 had single infections with TcIId sublineage, 1 with TcIIe, 4 mixed infections with TcIId/e, 1 with TcIId/b, and 1 with TcIIb/e. For two congenital patients (2- and 4-month-old samples) were taken: The results for the four samples indicated single infection with TcIId. For the 19 chronic-infected patients, we observed 10 single infections with TcIId, 2 with TcIIe, and 5 were mixed infections with TcIId/e. Two samples amplified with S121-122 primers but they were not recognized by any of the four probes. Infections with different genotypes are presented as percentages (Figure 2).
Figure 2.
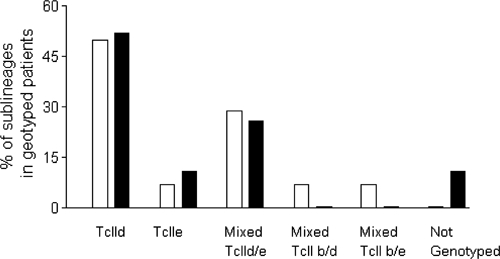
Infections with different Trypanosoma cruzi genotypes analyzed in the congenital and chronic samples performed with hybridization probes. Black bar indicates chronic samples and white bar indicates congenital samples. TcIId and TcIIe correspond to single infections with these sublinage, Mixed TcIId/e or TcIIb/e correspond to combined infections and Not Genotyped correspond to samples of patients that did not hybridize with any probe tested.
Genotyping of patient samples using MLS-PCR assay.
To test MLS-PCR directly from human-infected patients, all samples genotyped by hybridization tests were subjected to this nested PCR. In the first PCR, we used generic primers to amplify total mHVR sequence population and the second PCR was carried out using specific primers to detect predominant minicircle sequences of TcIId and TcIIe sublineages. In the congenital group, 16 samples from 14 congenitally infected individuals amplified with GrP primers, 10 of which amplified with GrO, which would indicate a minor detection with these primers. From these samples, which specifically amplified for TcIId sublineage two amplified also with GrpI primers, showing TcIId/e mixed infection. On the other hand, only one sample amplified exclusively with GrpI primers, indicating a unique sample belonging to the TcIIe sublineage.
In the chronic cases, 12 samples were amplified exclusively with GrP primers. Within these samples, only one was amplified with GrO-specific TcIId primers, and three of them shared a mixed amplification with Grp1 TcIIe primers. A single TcIIe infection was evidenced by a GrpI amplified sample. Finally, three of the samples that were amplified with S121-122 generic primers did not amplify with any pair of specific primers.
Results showed that all the samples (chronic and acute cases) genotyped by hybridization as TcIId, amplified with GrP primers, whereas only some of them amplified with GrO. When we evaluated MLS-PCR sensitivity to amplify O and P sequences, we found that it was related, with high significance (X2; P < 0.001), to the amount of amplified generic PCR product in the first PCR performed with generic primers (Table 1). Taking into account the higher sensitivity of GrP primer and its independence from the amount of the first PCR product, we used only these primers to genotype TcIId strains.
Table 1.
Evaluation of minicircle lineage-specific polymerase chain reaction (MLS-PCR) sensitivity to amplify specific O and P sequences for Trypanosoma cruzi TcIId sublineages in infected patient samples
| DNA template (ng) | Number of samples amplified with O or P primers (n/nt) | |
|---|---|---|
| Primers O (%) | Primers P (%)* | |
| > 56 | 8/8 (100) | 8/8 (100) |
| 11–56 | 10/11 (90) | 11/11 (100) |
| < 11 | 12/22 (55) | 22/22 (100) |
Higher than primers O (X2; P < 0.001).
Comparison between hybridization and MLS-PCR assay.
To compare the TcIId and TcIIe sublineages discrimination performance of the MLS-PCR assay with the hybridization one, we classified mixed infections with two sublineages as two individual results and single sublineage infection as a single result. The results obtained for each assay were then analyzed by Kappa index. By these criteria, the hybridization results of samples were classified as 27 TcIId (17 single infections and 9 TcIId/e mixed infections plus 1 TcIId/b mixed infection) and 13 TcIIe (3 single infections and 9 TcIId/e mixed infections plus 1 TcIIb/e mixed infections). The distribution of the number of TcIId, TcIIe, and no TcIId/e-infected patients is presented in a contingency table (Table 2). Three patient samples were non-genotyped by hybridization. On the other hand, the MLS-PCR results of samples were classified as 28 TcIId (24 single infections and 4 TcIId/e mixed infections) and 7 TcIIe (2 single infections and 5 TcIId/e mixed infections). Eight samples were not genotyped by MLS-PCR. The concordance between the hybridization test and MLS-PCR was 96.4% for TcIId sublineages (GrP primers) and 54.8% for TcIIe sublineages (GrpI primers). These results would indicate low sensitivity of GrpI primers to genotype TcIIe strains, whereas P primers were highly sensitive to genotype TcIId strains. Although a low sensitivity of GrpI primers was observed, the total concordance of the MLS-PCR assay using both GrP and Grp1 primers to genotype TcIId, TcIIe, or no TcIId/e had a statistical Kappa index of 0.69, indicating a good agreement.39
Table 2.
Contingency table to analyze the relationship between minicircle lineage-specific polymerase chain reaction (MLS-PCR) and hybridization assay to genotype TcIId and TcIIe Trypanosoma cruzi sublineages*
| Probes | MLS-PCR | |||
|---|---|---|---|---|
| TcIId | TcIIe | No TcIId/ No TcIIe | Total | |
| TcIId | 27 | 0 | 0 | 27 |
| TcIIe | 0 | 7 | 6 | 13 |
| No TcIId/ No TcIIe | 1 | 0 | 2 | 3 |
| Total | 28 | 7 | 8 | 43 |
Kappa index: 0.69.
Discussion
Human T. cruzi infecting strains have begun to be genotyped recently by different groups in different endemic regions of Argentina.21,33–36 Direct genotyping from blood samples is highly difficult because of the poor parasitaemia, mainly in the chronic phase of the infection. Consequently, most of the protocols have resorted to previous parasite isolation in culture medium or mammalian host. However, this approach may produce a selection from the original mixed infecting population,16 therefore direct PCR from sample would be a valuable method for accurate genotyping. Moreover, as the minicircle PCR amplification from minicircles is one of the most sensitive methods to obtain parasite's genetic material from biological samples, the use of mHVR can be successfully used to genotype T. cruzi.
Although the hybridization and MLS-PCR assays used here are based on the characterization of the same mHVR-PCR product, the last technique uses a second PCR, which identify unique minicircle sequences. This approach could increase the reliability and reproducibility with respect to hybridization with total minicircle sequences as the composition of DNA probes is complex and undefined.
Until now, most of the T. cruzi infections studied in mammalians and humans from Argentina, TcIId sublineages are highly prevalent, with a lower frequency of TcIIe.33–36 Keeping in mind these previous results, in this work we have focused on the evaluation of a new assay to characterize these sublineages. For this purpose, three pairs of primers were used. One primer amplifies GrpI, a major minicircle sequence in CL-Brener strain of TcIIe sublineage, which cover 40% of sequences found in that strain.24 On the other hand, two pairs of primers amplify predominant sequences in TcIId sublineage, covering 80% of sequences found in that sublineage.23 In a previous work, using specific probes for two TcIId predominant sequences (O and P), Virreira and others proposed a subclassification within this sublineage. In this way, TcIId were subdivided in MN-like, Bug2148-like, and TPK-like, if amplified PCR products hybridized mainly with O sequence, mainly with P, or equally with both of them, respectively. Recently, using this criterion the three subgroups of TcIId sublineage were described in northwestern Argentina.36 In our work, we have amplified these two predominant sequences by PCR in biological samples, however we could not amplify exclusively O sequences, although we have always obtained amplification for single P sequence, or O–P mixed sequences. Therefore, if we correlate our PCR results with Virreira's classification based on a hybridization test performed with the same oligonucleotids, these results have allowed us to type Bug 2148-like and TPK-like genotypes, although we did not find MN-like genotypes. Indeed, when reference strains were amplified using 0.3 pg of template, SC43 cl1 and MN cl2 were highly amplified with both sets of primers, whereas JGG, XHCH56, and NR cl3, were highly amplified with P primers but not with O primers. As MN cl2 belongs to MNcl2-like and SC43 cl1 belongs to TPK-like (previously classified by Virreira and others40), but rendered similar results by MLS-PCR, these results could indicate that the PCR discriminates two variants within TcIId sublineage instead of three.
The difference between the hybridization test performed by Virreira and others40 and MLS-PCR assay may be a result of a different ratio of O and P sequences in the purified DNA from samples as O sequence obtained from S121-122 generic PCR is less amplified than P sequence according to Table 1. Therefore, probably the amount of O sequences present in DNA template is insufficient to be amplified by means of this amount of template. Because of the higher sensitivity observed in PCR amplifications with P primers, we proposed only the use of these primers to genotype TcIId sublineage by MLS-PCR. Through these criteria a 96.4% of concordance was observed with the hybridization assay.
On the other hand, when TcIIe sublineage was genotyped using GrpI primers, a total concordance was observed when the strain reference panel was evaluated. However, a lower sensitivity was obtained when blood samples were compared with the hybridization test (54.8%). These observations would indicate that the use of Grp1 primers are not enough to type all the samples of this sublineage and other predominant sequences should be added to genotype misclassified samples.Nevertheless, we should not discard that v195 probe specific for TcIIe crossreacts with other sublineages.
Therefore, the PCR assay herein described allows identification of TcIId and TcIIe sublineages and TCIId/e mixed infections directly from blood samples. The assay is simpler to do and to standardize than hybridization assay because it uses only PCR amplification technique instead of radioactive or immunolabeled probes. When TcIId genotype was tested, the results were 96.4% coincident with hybridization assay. However, when TcIIe genotype was tested, the sensitivity of the new assay was 54.8% compared with hybridization assay. Currently, we are identifying new sets of primers to increase the sensitivity to type TcIIe genotype and to classify other genotypes. Because of the high sensitivity and specificity to detect TcIId genotype, the new tool described previously may be highly useful to assess the prevalence of TcIId sublineage directly from blood samples in wide geographic regions, and it would be very useful to support the results obtained by other methods.
When we analyzed the genotyping results of the overall infected samples from the Northeastern Argentina region, we detected TcIId predominance both in children and adults, and in a minor proportion, TcIIe. These results are compatible with those obtained from other regions of Argentina.33–36 Indeed, PCR-based techniques were recently applied to genotype T. cruzi samples from vector, animal, and human samples from Gran Chaco region, Santiago del Estero Province, Argentina, where a predominance of TcIId sublineage was detected in humans, and TcIIe in domestic mammals and vectors.21,35 The prevalence of TcIId was also found in congenital cases and their respective mothers, from different regions of Argentina and Bolivia.33 Similar results were obtained among congenitally infected newborns in nothwesten Argentina.36
However, a previous work, focused in a restricted endemic area of Chaco Province of Argentina, had reported for the first time the presence of TcIIe associated with dogs, and TcIId associated with human infection.36 The same sublineage was reported in peripheral blood of chagasic patients co-infected with human immunodeficiency virus (HIV), whereas other sublineages (TcIIb and TcIIe) were found in heart, brain, and skin lesions of these patients.33,41 Applying a serological diagnosis method, Di Noia and others42 showed that 95.4% of confirmed chagasic patients from Argentina were reactive to TSSA-II, a TcII-specific antigen, whereas only 3% of these sera displayed a mixed recognition to TSSA-II and TSSA-I, the latter specific for TcI. None of these patients showed exclusively TcI infection. Because we could not detect TcI in none of our patients our findings are in agreement with this work, although this lineage has been reported in the earliest studies done in our country. Indeed, the first studies using multilocus enzyme electrophoresis, showed that the more abundant (sub) lineages circulating in Argentina were TcI, TcIId, and TcIIb.43–45 Overall, TcII was more prevalent in the domestic area, whereas TcI and TcII infections were found in sylvatic mammals.46 A recent work suggests that TcI may be involved in infections of immunosuppressed patients,41 since this genotype was found in tissues different from blood. However, the cited work is a case report from a congenital acquired infection and no information about the geographical origin of the mother is available; then epidemiological data cannot be obtained about this patient.
We have also found 30.6% of mixed infections, predominantly TcIId/e, although one TcIIb/d and one TcIIb/e were also detected. It is important to clarify that mixed infections were observed in a minor proportion with MLS-PCR than hybridization assay, which may be a result of the lower detection of TcIIe sublineage by Grp1 primers used as previously discussed.
Interestingly, we found a similar distribution of sublineages in both congenital and chronicle patients. Similar results were obtained in northwestern Argentina36 and in a previous study performed in Bolivia.40 These results would indicate that congenital transmission is not caused by a specific genotype as was yet suggested by Virreira and others.40
In conclusion, our data about T. cruzi genotypes present in the Reconquista region of Santa Fe province, in coincidence with reports of other regions of our country cited previously, support the assumption that human infection by T. cruzi in Argentina is due principally to T. cruzi II. In that context, a correlation between clinical manifestation and the genotypes could be hypothesized as the cardiac sign that Chagas disease is predominant in Santa Fe province and in all of the country of Argentina.47–50 This hypothesis would be supported by the correlation of cardiac signs with TcII strains described by other authors,51,52 even though more evidences of this assumption should be obtained.
Footnotes
Financial support: This work received financial support from CAI+D 2009 PI 15-86, Universidad Nacional del Litoral and FONDECYT-Chile 1085154.
Disclosure: ISM and VG are researchers from the CONICET (Argentina).
Authors' addresses: Cristina Diez, Virginia Lorenz, Verónica Gonzalez, Andrea Racca, Iván Bontempi, and Iván Marcipar, Laboratorio de Tecnología Inmunológica, Facultad de Bioquímica y Cs Biológicas, Universidad Nacional del Litoral, Santa Fe, Argentina, E-mail: imarcipr@fbcb.unl.edu.ar. Silvia Ortiz and Aldo Solari, Program of Cellular and Molecular Biology, Biomedical Sciences Institute, Faculty of Medicine, University of Chile, Santiago, Chile, E-mail: asolari@med.uchile.cl. Silvia Manattini, Hospital Central de Reconquista, Santa Fe, Argentina, E-mail: equiroz@arnet.com.ar.
References
- 1.World Health Organization Control of Chagas disease. Report of a WHO expert committee. World Health Organ Tech Rep Ser. 2002;905:1–109. [PubMed] [Google Scholar]
- 2.Moncayo A. Chagas disease: current epidemiological trends after the interruption of vectorial and transfusional transmission in the southern Cone countries. Mem Inst Oswaldo Cruz. 2003;98:577–591. doi: 10.1590/s0074-02762003000500001. [DOI] [PubMed] [Google Scholar]
- 3.Andrade SG, Magalhaes JB, Pontes AL. Evaluation of chemotherapy with benznidazole and nifurtimox in mice infected with Trypanosoma cruzi strains of different types. Bull World Health Organ. 1985;63:721–726. [PMC free article] [PubMed] [Google Scholar]
- 4.Filardi LS, Brener Z. Susceptibility and natural resistance to Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Trans R Soc Trop Med Hyg. 1987;81:755–759. doi: 10.1016/0035-9203(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 5.Risso MG, Garbarino GB, Mocetti E, Campetella O, Gonzalez Cappa SM, Buscaglia CA, Leguizamon MS. Differential expression of a virulence factor, the trans-sialidase, by the main Trypanosoma cruzi phylogenetic lineages. J Infect Dis. 2004;189:2250–2259. doi: 10.1086/420831. [DOI] [PubMed] [Google Scholar]
- 6.Vago AR, Macedo AM, Adad SJ, Reis DD, Corrêa-Oliveira R. PCR detection of Trypanosoma cruzi DNA in oesophageal tissues of patients with chronic digestive Chagas' disease. Lancet. 1996;348:891–892. doi: 10.1016/S0140-6736(05)64761-7. [DOI] [PubMed] [Google Scholar]
- 7.Vago AR, Macedo AM, Oliveira RP, Andrade LO, Chiari E, Galvão LM, Reis D, Pereira ME, Simpson AJ, Tostes S, Pena SD. Kinetoplast DNA signatures of Trypanosoma cruzi strains obtained directly from infected tissues. Am J Pathol. 1996;149:2153–2159. [PMC free article] [PubMed] [Google Scholar]
- 8.Vago A, Andrade L, Leite A, Reis D, Macedo A, Adad S, Tostes S, Moreira M, Filo G, Pena S. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am J Pathol. 2000;156:1805–1809. doi: 10.1016/s0002-9440(10)65052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenière SF, Bosseno MF, Telleria J, Bastrenta B, Yacsik N, Noireau F, Alcazar JL, Barnabé C, Wincker P, Tibayrenc M. Different behavior of two Trypanosoma cruzi major clones: transmission and circulation in young Bolivian patients. Exp Parasitol. 1998;89:285–295. doi: 10.1006/expr.1998.4295. [DOI] [PubMed] [Google Scholar]
- 10.de Ornelas Toledo MJ, Bahia MT, Carneiro CM, Martins-Filho OA, Tibayrenc M, Barnabe C, Tafuri WL, de Lana M. Chemotherapy with benznidazole and itraconazole for mice infected with different Trypanosoma cruzi clonal genotypes. Antimicrob Agents Chemother. 2003;47:223–230. doi: 10.1128/AAC.47.1.223-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coronado X, Zulantai I, Rozas M, Apt W, Sanchez G, Rodriguez J, Ortiz S, Solari A. Dissimilar distribution of Trypanosoma cruzi clones in humans after chemotherapy with allopurinol anad itraconazole. J Antimicrob Chemother. 2006;58:216–219. doi: 10.1093/jac/dkl192. [DOI] [PubMed] [Google Scholar]
- 12.Martins HR, Silva RM, Valadares HM, Toledo MJ, Veloso VM, Vitelli-Avelar DM, Carneiro CM, Machado-Coelho GL, Bahia MT, Martins-Filho OA, Macedo AM, Lana M. Impact of dual infections on chemotherapeutic efficacy in BALB/c mice infected with major genotypes of Trypanosoma cruzi. Antimicrob Agents Chemother. 2007;51:3282–3289. doi: 10.1128/AAC.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins HR, Figueiredo LM, Valamiel-Silva JC, Carneiro CM, Machado-Coelho GL, Vitelli-Avelar DM, Bahia MT, Martins-Filho OA, Macedo AM, Lana M. Persistence of PCR-positive tissue in benznidazole-treated mice with negative blood parasitological and serological tests in dual infections with Trypanosoma cruzi stocks from different genotypes. J Antimicrob Chemother. 2008;61:1319–1327. doi: 10.1093/jac/dkn092. [DOI] [PubMed] [Google Scholar]
- 14.Brisse S, Dujardin JC, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol Biochem Parasitol. 2000;111:95–105. doi: 10.1016/s0166-6851(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 15.Brisse S, Verhoef J, Tibayrenc M. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol. 2001;31:1218–1226. doi: 10.1016/s0020-7519(01)00238-7. [DOI] [PubMed] [Google Scholar]
- 16.Devera R, Fernandes O, Coura JR. Should Trypanosoma cruzi be called “cruzi” complex? A review of the parasite diversity and the potential of selecting population after in vitro culturing and mice infection. Mem Inst Oswaldo Cruz. 2003;98:1–12. doi: 10.1590/s0074-02762003000100001. [DOI] [PubMed] [Google Scholar]
- 17.Britto C, Cardoso MA, Ravel C, Santoro A, Pereira JB, Coura JR, Morel CM, Wincker P. Trypanosoma cruzi: parasite detection and strain discrimination in chronic chagasic patients from northeastern Brazil using PCR amplification of kinetoplast DNA and nonradioactive hybridization. Exp Parasitol. 1995;81:462–471. doi: 10.1006/expr.1995.1139. [DOI] [PubMed] [Google Scholar]
- 18.Britto CM, Lima MM, Sarquis O, Pires MQ, Coutinho CF, Duarte R, Pacheco RS. Genetic polymorphism in Trypanosoma cruzi I isolated from Brazilian northeast triatomines revealed by low-stringency single specific primer-polymerase chain reaction. Parasitol Res. 2008;103:1111–1117. doi: 10.1007/s00436-008-1102-5. [DOI] [PubMed] [Google Scholar]
- 19.Morel C, Chiari E, Camargo EP, Mattei DM, Romanha AJ, Simpson L. Strains and clones of Trypanosoma cruzi can be characterized by pattern of restriction endonuclease products of kinetoplast DNA minicircles. Proc Natl Acad Sci USA. 1980;77:6810–6814. doi: 10.1073/pnas.77.11.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sturm N, Degrave W, Morel C, Simpson L. Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas' disease. Mol Biochem Parasitol. 1989;33:205–214. doi: 10.1016/0166-6851(89)90082-0. [DOI] [PubMed] [Google Scholar]
- 21.Marcet PL, Duffy T, Cardinal MV, Burgos JM, Lauricella MA, Levin MJ, Kitron U, Gürtler RE, Schijman AG. PCR-based screening and lineage identification of Trypanosoma cruzi directly from faecal samples of triatomine bugs from northwestern Argentina. Parasitology. 2006;132:57–65. doi: 10.1017/S0031182005008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Triana O, Ortiz S, Dujardin JC, Solari A. Trypanosoma cruzi: variability of stocks from Colombia determined by molecular karyotype and minicircle Southern blot analysis. Vet Parasitol Exp Parasitol. 2006;113:62–66. doi: 10.1016/j.exppara.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Telleria J, Lafay B, Virreira M, Barnabe C, Tibayrenc M, Svoboda M. Tr(i) panosoma cruzi: sequence analysis of the variable region of kinetoplast minicircles. Exp Parasitol. 2006;114:279–288. doi: 10.1016/j.exppara.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Velázquez M, Diez CN, Mora C, Diosque P, Marcipar IS. Trypanosoma cruzi: an analysis of the minicircle hypervariable regions diversity and its influence on strain typing. Exp Parasitol. 2008;120:235–241. doi: 10.1016/j.exppara.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Avila HA, Pereira JB, Thiemann O, De Paiva E, DeGrave W, Morel CM, Simpson L. Detection of Trypanosoma cruzi in blood specimens of chronic chagasic patients by polymerase chain reaction amplification of kinetoplast minicircle DNA: comparison with serology and xenodiagnosis. J Clin Microbiol. 1993;31:2421–2426. doi: 10.1128/jcm.31.9.2421-2426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wincker P, Brito C, Borges Pereira J, Cardoso MA, Oelemann W, Morel CM. Use of a simplified polymerase chain reaction procedure to detect Trypanosoma cruzi in blood samples from chronic chagasic patients in a rural endemic area. Am J Trop Med Hyg. 1994;51:133–139. doi: 10.4269/ajtmh.1994.51.771. [DOI] [PubMed] [Google Scholar]
- 27.Rozas M, Botto-Mahan C, Coronado X, Ortiz S, Cattan PE, Solari A. Coexistence of Trypanosoma cruzi genotypes in wild and periodomestic mammals in Chile. Am J Trop Med Hyg. 2007;77:647–653. [PubMed] [Google Scholar]
- 28.Diez M, Favaloro L, Bertolotti A, Burgos JM, Vigliano C, Lastra MP, Levin MJ, Arnedo A, Nagel C, Schijman AG, Favaloro RR. Usefuness of PCR strategies for early diagnosis of Chagas disease reactivation and treatment follow-up in heart transplantation. Am J Transplant. 2007;7:1633–1640. doi: 10.1111/j.1600-6143.2007.01820.x. [DOI] [PubMed] [Google Scholar]
- 29.Diez CN, Manattini S, Zanuttini JC, Bottasso O, Marcipar I. The value of molecular studies for the diagnosis of congenital Chagas disease in northeastern Argentina. Am J Trop Med Hyg. 2008;78:624–627. [PubMed] [Google Scholar]
- 30.Solari A, Campillay R, Ortíz S, Wallace A. Identification of Trypanosoma cruzi genotypes circulating in Chilean chagasic patients. Exp Parasitol. 2001;97:226–233. doi: 10.1006/expr.2001.4607. [DOI] [PubMed] [Google Scholar]
- 31.Salazar A, Schijman AG, Triana-Chávez O. High variability of Colombian Trypanosoma cruzi lineage I stocks as revealed by low-stringency single primer-PCR minicircle signatures. Acta Trop. 2006;100:110–118. doi: 10.1016/j.actatropica.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Lisboa CV, Pinho AP, Herrera HM, Gerhardt M, Cupolillo E, Jansen AM. Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) genotypes in neotropical bats in Brazil. Vet Parasitol. 2008;156:314–318. doi: 10.1016/j.vetpar.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares HM, Seidenstein ME, Piccinali R, Freitas JM, Levin MJ, Macchi L, Macedo AM, Freilij H, Schijman AG. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Diosque P, Barnabé C, Padilla AM, Marco JD, Cardozo RM, Cimino RO, Nasser JR, Tibayrenc M, Basombrío MA. Multilocus enzyme electrophoresis analysis of Trypanosoma cruzi isolates from a geographically restricted endemic area for Chagas disease in Argentina. Int J Parasitol. 2003;33:997–1003. doi: 10.1016/s0020-7519(03)00139-5. [DOI] [PubMed] [Google Scholar]
- 35.Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, Levin MJ, Kitron U, Gürtler RE, Schijman AG. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38:1533–1543. doi: 10.1016/j.ijpara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrales RM, Mora MC, Negrette OS, Diosque P, Lacunza D, Virreira M, Brenière SF, Basombrio MA. Congenital Chagas disease involves Trypanosoma cruzi sub-lineage IId in the northwestern province of Salta, Argentina. Infect Genet Evol. 2009;9:278–282. doi: 10.1016/j.meegid.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Veas F, Breniere SF, Cuny G, Brengues C, Solari A, Tibayrenc M. General procedure to construct highly specific kDNA probes for clones of Trypanosoma cruzi for sensitive detection by polymerase chain reaction. Cell Mol Biol. 1991;37:73–84. [PubMed] [Google Scholar]
- 38.Torres JP, Ortiz S, Muñoz S, Solari A. Trypanosoma cruzi isolates from Chile are heterogeneous and composed of mixed populations when characterized by schizodeme and Southern analyses. Parasitology. 2004;128:161–168. doi: 10.1017/s0031182003004475. [DOI] [PubMed] [Google Scholar]
- 39.Arimone Y, Miremont-Salamé G, Haramburu F, Molimard M, Moore N, Fourrier-Réglat A, Bégaud B. Inter-expert agreement of seven criteria in causality assessment of adverse drug reactions. Br J Clin Pharmacol. 2007;64:482–488. doi: 10.1111/j.1365-2125.2007.02937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virreira M, Alonso-Vega C, Solano M, Jijena J, Brutus L, Bustamante Z, Truyens C, Schneider D, Torrico F, Carlier Y, Svoboda M. Congenital Chagas disease in Bolivia is not associated with DNA polymorphism of Trypanosoma cruzi. Am J Trop Med Hyg. 2006;75:871–879. [PubMed] [Google Scholar]
- 41.Burgos JM, Begher S, Silva HM, Bisio M, Duffy T, Levin MJ, Macedo AM, Schijman AG. Molecular identification of Trypanosoma cruzi I tropism for central nervous system in Chagas reactivation due to AIDS. Am J Trop Med Hyg. 2008;78:294–297. [PubMed] [Google Scholar]
- 42.Di Noia J, Buscaglia C, De Marchi C, Almeida I, Frasch A. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas' disease is due to a single parasite lineage. J Exp Med. 2002;195:401–413. doi: 10.1084/jem.20011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montamat EE, Arauzo S, Cazzulo JJ, Subias E. Characterization by electrophoretic zymograms of 19 Trypanosoma cruzi clones derived from two chronic chagasic patients. Comp Biochem Physiol. 1987;87:417–422. doi: 10.1016/0305-0491(87)90161-1. [DOI] [PubMed] [Google Scholar]
- 44.Montamat EE, De Luca d'Oro GM, Perret B, Rivas C. Characterization of Trypanosoma cruzi from Argentina by electrophoretic zymograms. Acta Trop. 1992;50:125–133. doi: 10.1016/0001-706x(91)90005-5. [DOI] [PubMed] [Google Scholar]
- 45.Montamat EE, De Luca d'Oro GM, Gallerano RH, Sosa R, Blanco A. Characterization of Trypanosoma cruzi populations by zymodemes: correlation with clinical picture. Am J Trop Med Hyg. 1996;55:625–628. doi: 10.4269/ajtmh.1996.55.625. [DOI] [PubMed] [Google Scholar]
- 46.De Luca d'Oro GM, Gardenal CN, Perret B, Crisci JV, Montamat EE. Genetic structure of Trypanosoma cruzi populations from Argentina estimated from enzyme polymorphism. Parasitology. 1993;107:405–410. doi: 10.1017/s0031182000067755. [DOI] [PubMed] [Google Scholar]
- 47.Fabbro De Suasnábar D, Arias E, Streiger M, Piacenza M, Ingaramo M, Del Barco M, Amicone N. Evolutive behavior towards cardiomyopathy of treated (nifurtimox or benznidazole) and untreated chronic chagasic patients. Rev Inst Med Trop Sao Paulo. 2000;42:99–109. doi: 10.1590/s0036-46652000000200007. [DOI] [PubMed] [Google Scholar]
- 48.Dubner S, Schapachnik E, Riera AR, Valero E. Chagas disease: state-of-the-art of diagnosis and management. Cardiol J. 2008;15:493–504. [PubMed] [Google Scholar]
- 49.Abello M, González-Zuelgaray J, López C, Labadet C. Initiation modes of spontaneous monomorphic ventricular tachycardia in patients with Chagas heart disease. Rev Esp Cardiol. 2008;61:487–493. [PubMed] [Google Scholar]
- 50.Storino R, Auger S, Caravello O, Urrutia MI, Sanmartino M, Jörg M. Chagasic cardiopathy in endemic area versus sporadically infected patients. Rev Saude Publica. 2002;36:755–758. doi: 10.1590/s0034-89102002000700016. [DOI] [PubMed] [Google Scholar]
- 51.Venegas J, Coñoepan W, Pichuantes S, Miranda S, Apt W, Arribada A, Zulantay I, Coronado X, Rodriguez J, Reyes E, Solari A, Sanchez G. Differential distribution of Trypanosoma cruzi clones in human chronic chagasic cardiopathic and non-cardiopathic individuals. Acta Trop. 2009;109:187–193. doi: 10.1016/j.actatropica.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Carranza JC, Valadares HM, D'Avila DA, Baptista RP, Moreno M, Galvão LM, Chiari E, Sturm NR, Gontijo ED, Macedo AM, Zingales B. Trypanosoma cruzi maxicircle heterogeneity in Chagas disease patients from Brazil. Int J Parasitol. 2009;39:963–973. doi: 10.1016/j.ijpara.2009.01.009. [DOI] [PubMed] [Google Scholar]


