Abstract
• Background and Aims Representatives from Papaver, Roemeria, Stylomecon and Meconopsis were studied to elucidate phylogenetic relationships between Papaver and these closely allied genera.
• Methods Two molecular data sets were used individually and combined and included sequences from the internally transcribed spacer region (ITS) of 18S–26S nuclear ribosomal DNA and the trnL intron and the trnL–trnF intergenic spacer region of plastid DNA.
• Key Results Parsimony analysis demonstrated that the genus is not monophyletic unless the closely related Roemeria, Stylomecon and Meconopsis cambrica are included in a revised circumscription of Papaver. Three distinct clades are resolved in a combined ITS and trnL–F analysis. Clade 1 consists of Papaver sect. Meconella and Asian Meconopsis. Clade 2 contains a group here identified as Papaver s.s., comprising sections Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium. Clade 3 consists of Papaver sect. Argemonidium and Roemeria refracta. A number of diagnostic indels support these groupings. Within clade 2, sects. Papaver and Rhoeadium are either not monophyletic or lack evidence supporting their monophyly.
• Conclusions The results of this molecular analysis indicate that a number of morphological characters such as valvate capsule dehiscence, dark or light filaments and sessile stigmatic discs have arisen in parallel. The phylogenetic trees are incongruent with the existing taxonomy of Papaver, and a revised classification is suggested.
Keywords: DNA, Meconopsis, nrITS, Papaver, phylogenetics, Roemeria, Stylomecon, trnL–F
INTRODUCTION
Papaver L. is the largest genus of subfamily Papaveroideae of Papaveraceae sensu Kadereit (1993a). Based on morphological considerations, subfamily Papaveroideae, including subfamily Platystemonoideae, can be divided into two major clades (Kadereit et al., 1997; Schwarzbach and Kadereit 1999). These are a New World clade containing Arctomecon Torr. et Frem., Argemone L., Romneya Harv., Canbya Parry, Platystemon Benth., Meconella Nutt. and Hesperomecon Greene, and a largely Old World clade with Papaver, Meconopsis Vig., Stylomecon Benth. and Roemeria Medic. The latter group shares characters such as semicampyltropous ovules and a seed coat with a fine layer of crystals. Additionally, meconic acid is found only in species of these four genera (Cordell, 1981). Determining relationships among these four genera of Papaveroideae is the primary focus of this paper.
Papaver consists of approximately 80 annual, biennial and perennial herbs distributed in central and south-western Asia, central and southern Europe and northern Africa (Kadereit, 1988a). Papaver sect. Meconella has a panarctic–alpine distribution that includes north-eastern North America. Papaver aculeatum Thunb. (sect. Horrida) is indigenous to South Africa, and P. californicum A. Gray (sect. Californicum) is indigenous to western North America. Papaver is characterized by the absence of a style and the possession of stigmatic tissue arranged radially on a sessile stigmatic disc crowning the ovary. The latest taxonomic revision of Papaver (Kadereit, 1988a) recognized 11 sections (Argemonidium Spach.; Carinatae Fedde; Californicum Kadereit; Horrida Elk.; Oxytona Bernh.; Meconidium Bernh.; Meconella Spach; Papaver L.; Pilosa Prantl; Pseudopilosa Gunther; Rhoeadium Bernh.). Detailed taxonomic accounts of many of the sections have been published (Goldblatt, 1974; Kadereit, 1986a, b, 1987, 1988b, c, 1989, 1993b, 1996). The separation of species into sections is based on a combination of characters, including mode of capsule dehiscence (through valves or pores), colour of anthers and filaments (pale or dark), and general capsule characteristics such as size, shape and indumentum. Based on these characters, Kadereit (1988a) recognized four groups of sections within Papaver. The first group consists of sects. Californicum, Meconella and Meconidium and is characterized by pale filiform filaments and anthers, and valvate capsule dehiscence. The second group consists of sect. Argemonidium alone and is characterized by dark clavate filaments and anthers and poricidal capsule dehiscence. The third group comprises sects. Horrida, Pilosa and Pseudopilosa and is characterized by pale filiform filaments and anthers and poricidal capsule dehiscence. Finally, group four comprises sects. Carinatae, Oxytona, Papaver and Rhoeadium and is characterized by dark (sometimes pale) filiform (sometimes clavate) filaments and always dark anthers and poricidal capsule dehiscence.
Meconopsis comprises approximately 50 perennial monocarpic or polycarpic herbs, distributed primarily in southern central Asia. Meconopsis cambrica (L.) Vig. is the only European representative of the genus. Meconopsis is considered to be distinct from Papaver based on the possession of stigmatic tissue borne on top of a style (although species without styles do exist). Roemeria comprises three annual species distributed mainly in south-western and central Asia and Europe. It has long, linear, bristly capsules with sessile stigmas borne directly on top of the ovary. Stylomecon is a monotypic genus comprising the annual S. heterophylla (Benth.) G. Taylor native to western North America and is characterized by the possession of stigmatic tissue borne on top of a style. Although it is similar to Meconopsis in capsule characteristics, it is recognized as a distinct genus primarily based on its annual habit and geographical distribution (Taylor, 1930; Kadereit et al., 1997).
Delimitation of taxa into their respective genera (Papaver, Meconopsis, Stylomecon, Roemeria) seems straightforward based on the distinction of capsule characteristics. Previous molecular phylogenetic analyses of these genera (Kadereit and Sytsma, 1992; Kadereit et al., 1997), however, demonstrated that they form a monophyletic group within Papaveroideae (Kadereit, 1993a) and provided evidence that Papaver sensu Kadereit (1988a) is not monophyletic. These molecular analyses included a restriction site analysis of plastid DNA (Kadereit and Sytsma, 1992) and an RFLP analysis of the plastid trnK region (Kadereit et al., 1997). It was demonstrated that Roemeria was sister to P. sect. Argemonidium and Stylomecon was sister to P. sect. Californicum, indicating that Papaver was monophyletic only if these genera were included in Papaver. In addition, the European Meconopsis cambrica did not group with the Asian species of this genus. Meconopsis cambrica resolved as sister to a group of sections of Papaver including Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium, leading Kadereit et al. (1997) to view these sections as Papaver s.s. Determining the interrelationships of these sections was limited by the small number of species sampled in their study. Generally, only a single species was used to represent sections, and single individuals were used to represent species. The non-monophyly of Papaver s.l. indicates that the stigmatic disc typical for the genus may have arisen several times independently. To define Papaver based on a single character that has multiple origins would be taxonomically and phylogenetically unsound. The results of these molecular analyses also demonstrated that some of the infrageneric taxonomic groupings suggested by Kadereit (1988a) were artificial.
The objective of this paper is to examine phylogenetic relationships within Papaver and allied genera by comparing nucleotide sequences obtained from plastid and nuclear ribosomal sequences. The two molecular regions used were the internally transcribed spacer region (ITS) of 18S–26S nuclear ribosomal DNA (Sun et al., 1994; Baldwin et al., 1995) and the trnL intron and the trnF intergenic spacer region of plastid DNA (Taberlet et al., 1991). All regions are relatively small in size (i.e. trnL–trnF ∼500–900 bp; and ITS ∼700 bp), which facilitates successful amplification and sequencing (Taberlet et al., 1991; Baldwin et al., 1995; Kelchner, 2000).
Combining sequences from different genomes (nuclear, plastid and mitochondrial) is common in molecular phylogenetics, as long as they produce congruent results, and has resulted in greater understanding of relationships within a wide range of plant groups (see Savolainen and Chase, 2003). Both DNA regions used here have proven useful at similar taxonomic levels in other plant groups (e.g. Gielly et al., 1994; Sun et al., 1994; Baldwin et al., 1995; Wendel et al., 1995; Gielly and Taberlet, 1996; Wendel and Doyle, 1998; Kelchner, 2000; Hodkinson et al., 2002). However, these DNA regions and their subsequent combination have not been applied to Papaver phylogenetics. The topology of the trees obtained here from the comparative analysis of the ITS and trnL–F regions is interpreted in terms of morphological, chemotaxonomic and geographical similarities.
MATERIALS AND METHODS
Specimens
Material was obtained from various botanical gardens and commercial sources and grown to maturity either at the National Botanic Garden, Glasnevin, Ireland, or in the glasshouse of the Department of Pharmacognosy, University of Dublin, Trinity College, Ireland. DNA obtained from herbarium material was also used. Voucher specimens were kept for each accession and stored in the Herbarium of the Department of Botany, Trinity College Dublin, Ireland (TCD). DNA was stored at the Department of Botany, TCD, DNA Bank. Voucher specimens for each accession and sequences obtained from GenBank are listed in Table 1.
Table 1.
Species and associated voucher specimens used in the study
| Taxon | GenBank number ITS: trnL–F | ID number* | Voucher or reference |
|---|---|---|---|
| Argemone mexicana L. | AY328303·1; AY328248·1 | Y. M. Yuan et al., unpubl. | |
| Chelidonium majus L. | AY328251·1; AY328308·1 | Y. M. Yuan et al., unpubl. | |
| Eomecon chionantha Hance | AY328254·1; AY328306·1 | Y. M. Yuan et al., unpubl. | |
| Meconopsis aculeata Royle | AY328263·1; AY328227·1 | Y. M. Yuan et al., unpubl. | |
| Meconopsis betonicifolia Franch. | DQ250323; DQ251174 | 032 | 1998·0451 |
| Meconopsis betonicifolia Franch | AY328236·1; AY328292·1 | Y. M. Yuan et al., unpubl. | |
| Meconopsis cambrica L. | DQ250277; DQ251128 | 001 | 2000·0001 |
| Meconopsis cambrica L. | DQ250278; DQ251129 | 048 | 1992·0611 |
| Meconopsis delavayi Franch. ex Prain | AY328211·1; AY328285·1 | Y. M. Yuan et al., unpubl. | |
| Meconopsis lancifolia Franch. ex Prain | AY328212·1; AY328282·1 | Y. M. Yuan et al., unpubl. | |
| Papaver aculeatum Thunb. | DQ250317; DQ251168 | 131 | 2000·0131 |
| Papaver aculeatum Thunb | DQ250316; DQ251167 | 151 | 2000·0659 |
| Papaver alpinum spp. rhaeticum Mgf. | DQ250261; DQ251112 | 150 | 2000·0150 |
| Papaver alpinum spp. alpinum L. | DQ250268; DQ251119 | 102 | 2000·0568 |
| Papaver anomalum Fedde | DQ250263; DQ251116 | 106 | 2000·1321 |
| Papaver anomalum ‘album’ Fedde | DQ250264; DQ251115 | 078 | 2000·1806 |
| Papaver apulum Ten. | DQ250300; DQ251151 | 084 | 2000·0601 |
| Papaver argemone L. | DQ250298; DQ251149 | 153 | 2000·0153 |
| Papaver armeniacum ssp. armeniacum L. | DQ250302; DQ251153 | 154 | 2000·1717 |
| Papaver armeniacum ssp. armeniacum L. | DQ250297; DQ251148 | 095 | 2000·0604 |
| Papaver armeniacum ssp. armeniacum L. | DQ250311; DQ251162 | 087 | 2000·0651 |
| Papaver armeniacum ssp. armeniacum L. | DQ250312; DQ251163 | 082 | 2000·0795 |
| Papaver armeniacum ssp. armeniacum L. | DQ250259; DQ251110 | 103 | 2000·0793 |
| Papaver armeniacum ssp. armeniacum L. | DQ250294; DQ251145 | 107 | 2000·0107 |
| Papaver atlanticum Ball et Cross. | DQ250307; DQ251158 | 092 | 2000·0603 |
| Papaver atlanticum Ball et Cross. | DQ250315; DQ251166 | 099 | 2000·0615 |
| Papaver atlanticum Ball et Cross. | DQ250293; DQ251144 | 077 | 2000·0472 |
| Papaver atlanticum Ball et Cross. | DQ250303; DQ251154 | 156 | 2000·0156 |
| Papaver bracteatum Lindl. | DQ250286; DQ251137 | 028 | 2000·0028 |
| Papaver bracteatum Lindl. | DQ250287; DQ251138 | 031 | 2000·0031 |
| Papaver californicum A.Gray | DQ250318; DQ251169 | 170 | 2000·0170 |
| Papaver croceum Ledeb. | DQ250258; DQ251109 | 015 | 1999·00340 |
| Papaver croceum Ledeb. | DQ250257; DQ251108 | 104 | 2000·0104 |
| Papaver croceum Ledeb. | DQ250266; DQ251117 | 105 | 2000·0653 |
| Papaver croceum Ledeb. | DQ250284; DQ251135 | 148 | AS 95/23 Kadereit et al., 1996 |
| Papaver commutatum Fisch et Mey. | DQ250313; DQ251164 | 102 | 2000·0605 |
| Papaver dubium ssp. dubium L. | DQ250270; DQ251121 | 024 | 2000·0024 |
| Papaver dubium ssp. dubium L. | DQ250319; DQ251170 | 162 | 2000·1706 |
| Papaver dubium L. ssp. erosum (Litv.) Kadereit | DQ250271; DQ251122 | 168 | 2000·0610 |
| Papaver dubium ssp. lecoquii Syme | DQ250322; DQ251173 | 174 | 2000·0606 |
| Papaver dubium L. ssp. lecoquii (Lamotte) Syme var. albiflorum Besser | DQ250267; DQ251118 | 100 | 2000·0600 |
| Papaver glaucum Boiss. et Hausskn | DQ250310; DQ251161 | 089 | 2000·0609 |
| Papaver glaucum Boiss. et Hausskn | DQ250309; DQ251160 | 177 | 2000·0177 |
| Papaver glaucum Boiss. et Hausskn | DQ250308; DQ251159 | 189 | 2000·0189 |
| Papaver hybridum L. | DQ250301; DQ251152 | 167 | 1999·1723 |
| Papaver miyabeanum Tatew. | DQ250276; DQ251127 | 019 | 1999·0339 |
| Papaver miyabeanum Tatew. | DQ250265; DQ251116 | 186 | 2000·0186 |
| Papaver macrostomum Boiss. et Huet | DQ250275; DQ251126 | 160 | RBGE 34139 |
| Papaver nudicaule ssp. nudicaule L. | DQ250260; DQ251111 | 086 | 2000·0598 |
| Papaver orientale L. | DQ250292; DQ251143 | 011 | 2000·0011 |
| Papaver orientale L. | DQ250289; DQ251140 | 035 | 2000·0035 |
| Papaver orientale L. | DQ250290; DQ251141 | 135 | 2000·0135 |
| Papaver orientale L. | DQ250291; DQ251142 | 179 | RBGE 19880542A |
| Papaver pavonium Fischer & Meyer ssp. pavonium | DQ250283; DQ251134 | 138 | 2000·0138 |
| Papaver pilosum ssp. strictum Wendt ex Kadereit | DQ250321; DQ251172 | 081 | 2000·0768 |
| Papaver pilosum Sibth. & Sm. ssp. pilosum Wendt | DQ250320; DQ251171 | 182 | 2000·0182 |
| Papaver pseudo-orientale (Fedde) Medv. | DQ250269; DQ251120 | 014 | 2000·0014 |
| Papaver pseudo-orientale (Fedde) Medv. | DQ250288; DQ251139 | 039 | 2000·0039 |
| Papaver pseudo-orientale (Fedde) Medv. | DQ250296; DQ251147 | 093 | 2000·0632 |
| Papaver pseudo-orientale (Fedde) Medv. | DQ250285; DQ251136 | 139 | 2000·0794 |
| Papaver radicatum Rottb. | DQ250262; DQ251113 | 094 | 2000·0769 |
| Papaver rhoeas L. | DQ250272; DQ251123 | 090 | 2000·0090 |
| Papaver rhoeas L. | DQ250273; DQ251124 | 147 | KJ93/7 Kadereit et al., 1996 |
| Papaver rupifragum Boiss et Reut. | DQ250314; DQ251165 | 017 | 1999·0342 |
| Papaver somniferum ssp. setigerum (DC.) L.Corb | DQ250279; DQ251130 | 016 | 2000·0016 |
| Papaver somniferum L. | DQ250281; DQ251132 | 052 | 2000·0052 |
| Papaver somniferum L. | DQ250305; DQ251156 | 063 | 2000·0063 |
| Papaver somniferum L. | DQ250280; DQ251131 | 130 | 2000·0130 |
| Papaver somniferum L. | DQ250304; DQ251155 | 166 | 2000·0166 |
| Papaver somniferum ssp. somniferum L. | DQ250282; DQ251133 | 068 | 2000·0068 |
| Papaver somniferum ssp. somniferum L. | DQ250306; DQ251157 | 142 | 2000·0142 |
| Roemeria refracta DC. | DQ250299; DQ251150 | 171 | 2000·0171 |
| Stylomecon heterophylla Taylor | DQ250295; DQ251146 | 183 | 2000·0183 |
Vouchers are deposited in the Herbarium of Trinity College Dublin (TCD).
*The ID number represents the identification number used in this study and to differentiate taxa with the same name.
Outgroup selection
Outgroup taxa were selected on the basis of the plastid DNA restriction site analysis of Kadereit and Sytsma (1992) and the morphological work of Kadereit (1993a). Eomecon chionantha Hance (Papaveraceae subfamily Chelidonioideae) was chosen as an outgroup taxon owing to its position as given in previous studies. Argemone mexicana L. (Papaveraceae subfamily Papaveroideae) and Chelidonium majus L. (Papaveraceae subfamily Chelidonioideae) were also included as outgroups.
DNA extraction
DNA was extracted from 0·5–1·0 g of fresh leaf material using a modified 2 % CTAB procedure of Doyle and Doyle (1987), precipitated using 100 % ethanol or isopropanol for at least 48 h at −20 °C, pelleted and washed with 70 % ethanol and purified via the Concert™ Rapid PCR Purification System (Life Technologies, Gaithersburg, MD, USA). DNA was then stored in TE buffer (10 mm Tris/HCl, 1 mm EDTA, pH 8·0) at −80 °C until required.
DNA sequencing
For amplification and sequencing of the ITS region the forward and reverse primers of Sun et al. (1994) were used. The trnL intron and the trnL–trnF spacer (hereafter the trnL–F region) were amplified and sequenced as one segment using primers ‘c’ and ‘f’ of Taberlet et al. (1991). Difficulties were encountered when attempting to amplify certain sequence regions from herbarium specimens. Their successful amplification and sequencing was achieved using the internal primers (2 and 3) for ITS of Baldwin et al. (1995) and the internal primers (d and e) for the trnL intron and the trnL–F region (Taberlet et al., 1991). PCRs for both regions were carried out in 50-µL reactions using 1 % PCR buffer (Promega, Madison, WI, USA), 2·5 mm MgCl2, 0·2 µm of each primer, 0·2 mm of each dNTP, 1 U Taq polymerase (Promega) and approximately 50 ng template DNA. Reaction conditions for the trnL–F region were: denaturation at 94 °C for 3 min followed by 30 cycles of 1 min at 94 °C, 1 min at 51 °C, 1 min at 72 °C and a final extension at 72 °C for 7 min in a Peltier thermal cycler (PTC 200; MJ Research). PCR amplification of the ITS region was achieved using a touchdown PCR strategy involving denaturation at 94 °C for 3 min followed by 30 cycles of 1 min at 94 °C, 1 min at 60–52 °C (over the first eight cycles with the remaining cycles at 52 °C), 1 min at 72 °C and a final extension at 72 °C for 7 min. Successfully amplified DNA fragments were purified using the Concert™ Rapid PCR Purification System (Life Technologies) and sequenced using Big Dye Terminator Cycle Sequencing Kits v1.1 (Applied Biosystems, Foster City, CA, USA) on an Applied Biosystems 310 or 377 automated DNA sequencer, all according to the manufacturer's protocols and with the same primers as used for the initial amplification.
Sequence analysis and phylogenetic reconstruction
Forward and reverse sequence reads were assembled using Sequencher™ version 3.1 (Gene Codes Corporation, 1998) to obtain a contiguous sequence for the target DNA region. Consensus sequences for all accessions were imported into SE-Al v2.0 (sequence-alignment, Rambaut, 2001) in which sequences were aligned by inserting gaps manually within the data matrix following the guidelines of Kelchner (2000). The aligned matrix was imported into PAUP v4.0b for phylogenetic analysis (Swofford, 2003). Gaps were treated as missing data. Regions of the sequence alignment that contained a substantial number of alignment gaps were omitted from the analyses because the positional homology within these regions is uncertain (Swofford et al., 1996). Omitted regions included 12-, 53- and 38-bp hypervariable regions of the ITS aligned matrix (corresponding to positions 138–150, 240–293 and 478–516, respectively) and the initial 14 and final 65 nucleotides of the trnL–F aligned matrix. Independent phylogenetic analysis of the trnL intron and the trnL–trnF spacer regions yielded broadly congruent trees (results not shown). If incongruence was found it was not supported by bootstrap analysis (soft incongruence; Seelanen et al., 1997). For the purposes of this study both regions were combined for parsimony analysis. These are part of the non-recombining plastid genome and are frequently combined for phylogenetic reconstruction (e.g. Hopper et al., 1999; Chase et al., 2000; Hodkinson et al., 2002) because they should have the same phylogenetic history.
Maximum parsimony (MP) trees were obtained from the resulting matrices using heuristic search options. Searches included 1000 replicates of random addition sequence (saving no more than 30 trees per replicate to reduce time spent swapping large islands of trees) with the tree bisection reconnection (TBR) branch-swapping algorithm and MulTrees on (keeping multiple equally most-parsimonious trees). Internal support was assessed using 1000 bootstrap replicates (Felsenstein, 1985), simple addition sequence, TBR swapping and MulTrees on (holding 30 trees per replicate; see Salamin et al., 2003). Groups with bootstrap percentages (BP) of 90–100 were considered to be strongly supported, 80–89 moderately supported and 50–79 weakly supported. Only groups with BP >50 that are consistent with the strict consensus tree are shown.
No major conflicts (hard incongruence) between the separate trees were identified between single-region analyses. Accordingly, the trnL–F data were joined with the ITS data in a combined analysis. The incongruence length difference (ILD) test (or similar congruence tests) was not applied as this can be ineffective in identifying combinability of data and in some cases has been shown to be misleading (Yoder et al., 2001). Our decision to combine was based on the pattern of major clades and their respective bootstrap percentages. The combined analysis of trnL–F and ITS data was also performed using the same parameters as stated above for the single gene region analyses.
RESULTS
Analysis of ITS
The lengths of ITS1, ITS2 and 5·8S were confirmed using a comparative alignment of Papaver rhoeas ITS1, ITS2 and 5·8S obtained from GenBank (Schwarzbach and Kadereit, 1999; accession no. AF098920). The 5·8S region ranged from 157 to 169 bp across all accessions used in this study. Relatively little variation was encountered within the 5·8S region, with only 10 of the 171 nucleotides (5·8 % of the final aligned matrix) being variable, but all were potentially parsimony-informative. The ITS1 and ITS2 spacers ranged in length from 218 to 260 bp and from 216 to 257 bp, respectively. A considerable proportion of both regions were variable. Of the 189 variable sites within the aligned ITS1 (66·5 % of the aligned ITS1 region), 152 were potentially parsimony-informative. Of the 171 variable sites found within the aligned ITS2 (50·4 % of the ITS2 region) 128 were potentially parsimony-informative. The G+C content of both ITS1 and ITS2 ranged from 50·5 to 60·2 % and 55·3 to 63·8 %, respectively. Independent analysis of the ITS1 and ITS2 spacers yielded broadly congruent trees (results not shown; Carolan, 2004). For the purposes of this study both regions plus the 5·8S gene were combined for parsimony analysis. The entire aligned ITS matrix (ITS1, 5·8S and ITS2) was 705 bp long; 324 sites were variable, and 235 of these were potentially parsimony-informative. Figure 1 shows one of 151 equally most-parsimonious trees from the ITS analysis. It has 826 steps, with a consistency index (CI) of 0·57 and a retention index (RI) of 0·83.
Fig. 1.
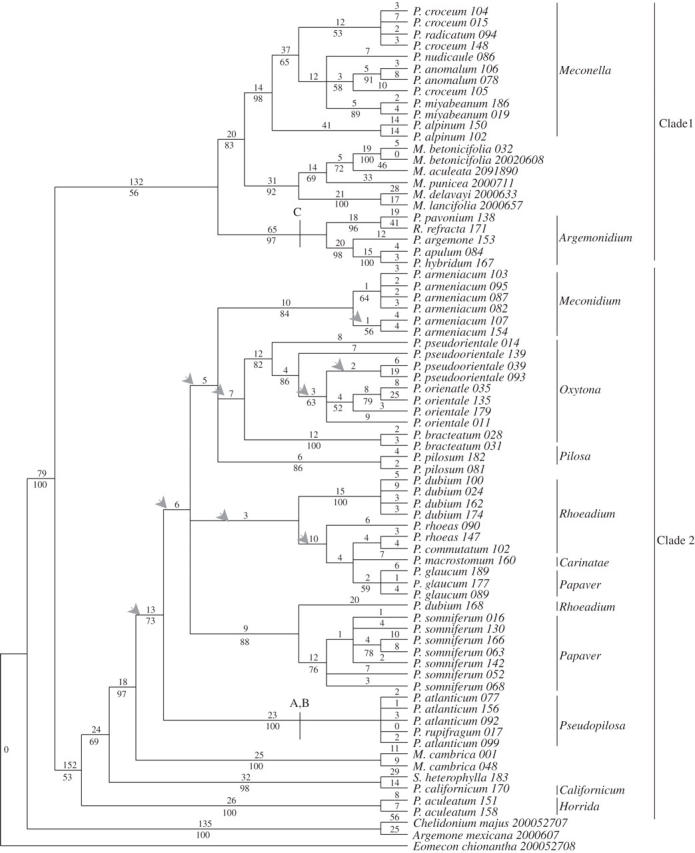
One of 151 equally most-parsimonious trees generated from the ITS sequences. Support for each node is represented by bootstrap percentages (BP) below the branch (shown only when >50 % and consistent with the strict consensus tree). An arrow indicates clades that did not appear in the strict consensus tree. Numbers above each branch indicate the numbers of character changes along each lineage (accelerated transformation, ACCTRAN, optimization). Groups that possess characteristic indels are indicated using letters (refer to Table 2).
Two distinct clades were found within the ITS tree (Fig. 1). Clade 1 comprises sects. Argemonidium, Meconella and the Asian representatives of Meconopsis. This clade is sister to all other sections of Papaver in all equally most-parsimonious trees but is itself weakly supported with only 56 % bootstrap support (bootstrap percentage; BP). Roemeria refracta groups with the species of sect. Argemonidium (97 BP). Within clade 1, sect. Meconella forms a well-supported group (98 BP). The Asian representatives of Meconopsis are resolved as sister to sect. Meconella in all equally most-parsimonious trees (83 BP).
Clade 2 (53 BP) comprises the remaining sections of Papaver, including Meconopsis cambrica and Stylomecon heterophylla. Papaver aculeatum (sect. Horrida) and a group comprising sect. Californicum and Stylomecon heterophylla resolve independently but sister to the remaining sections of clade 2 (53 and 69 BP, respectively). The positioning of Stylomecon heterophylla as sister to P. californicum is well supported (98 BP). Meconopsis cambrica and the remaining sections of Papaver form a well-supported group (97 BP). Sections Papaver and Rhoeadium are not monophyletic in this tree, as indicated by the grouping of Papaver glaucum (sect. Papaver) with representatives of sect. Rhoeadium (including sect. Carinatae; <50 BP) and the grouping of P. dubium ssp. erosum (sect. Rhoeadium) with Papaver somniferum (sect. Papaver; 88 BP). However, there is also little evidence contradicting their monophyly. Within clade 2, sect. Pseudopilosa is characterized by the possession of a number of unique indels. These include a 4-bp indel at positions 75–78 (A, Fig. 1; Table 2) and a 4-bp indel at positions 216–219 (B, Fig. 1; Table 2) of the aligned ITS matrix. Omitting these indels (gapped sites) from the analysis did not affect the sister group position of Pseudopilosa (with respect to the majority but not all of clade 2).
Table 2.
Insertions/deletions in the ITS and trnL–F regions for representatives used in this study
| Region and start points | Diagnostic indel or sequence | Tree annotation | Taxon |
|---|---|---|---|
| ITS 75 | TATA indel | A | Sect. Pseudo-pilosa |
| ITS 216 | TCTC indel | B | Sect. Pseudo-pilosa |
| ITS 695 | T indel | C | Sect. Argemonidium; Roemeria refracta |
| trnL–F 162 | TATA indel | D | Sects. Californicum; Horrida; Meconella; Asian Meconopsis; Stylomecon heterophylla |
| trnL–F 186 | TAGAG intel | E | Papaver commutatum; P. dubium ssp. erosum; P. glaucum; P. macrostomum; P. rhoeas |
| trnL–F 261 | GCCC indel | F | Sects. Californicum; Horrida; Meconella; Asian Meconopsis; Stylomecon heterophylla |
| trnL–F 643 | 10-bp deletion | G | Sect. Argemonidium (excluding P. pavonium) |
Start points of the indel are based on the aligned matrix for that given region. Characters mapped onto phylogenetic trees are given as letters.
Analysis of trnL–F
The total lengths of the trnL intron and the trnL–F spacer were confirmed using a comparative alignment of the Meconopsis betonicifolia trnL intron, trnL–F spacer and the 3′ trnL exon sequence obtained from GenBank (Y. M. Yuan et al., unpubl. data, Zhongshan University, P. R. China; accession AY328263). Little variation was encountered within the 3′ trnL exon region (50 bp long, including one parsimony informative character). The unaligned trnL intron and trnL–F spacer regions ranged in length from 467 to 505 bp and 384 to 422 bp, respectively. The final aligned matrix had a total length of 951 characters (539, 50 and 362 sites for the trnL intron, the 3′ trnL exon and the trnL–F spacer, respectively). The 128 variable sites found within the aligned trnL intron (representing 23·7 % of the trnL intron) consisted of 74 potentially parsimony-informative characters, and the trnL–F spacer contained 165 variable characters (representing 45·5 % of the spacer), of which 103 were potentially parsimony informative. The G+C content of both the trnL intron and the trnL–F spacer ranged from 32 to 36·5 % and 33·6 to 41·4 %, respectively. In total the aligned trnL–F matrix was 951 bp long; 294 sites were variable, and 176 of these were potentially parsimony informative. Phylogenetic analysis of the trnL–F matrix produced eight equally most-parsimonious trees (468 steps, CI = 0·77, RI = 0·91; Fig. 2).
Fig. 2.
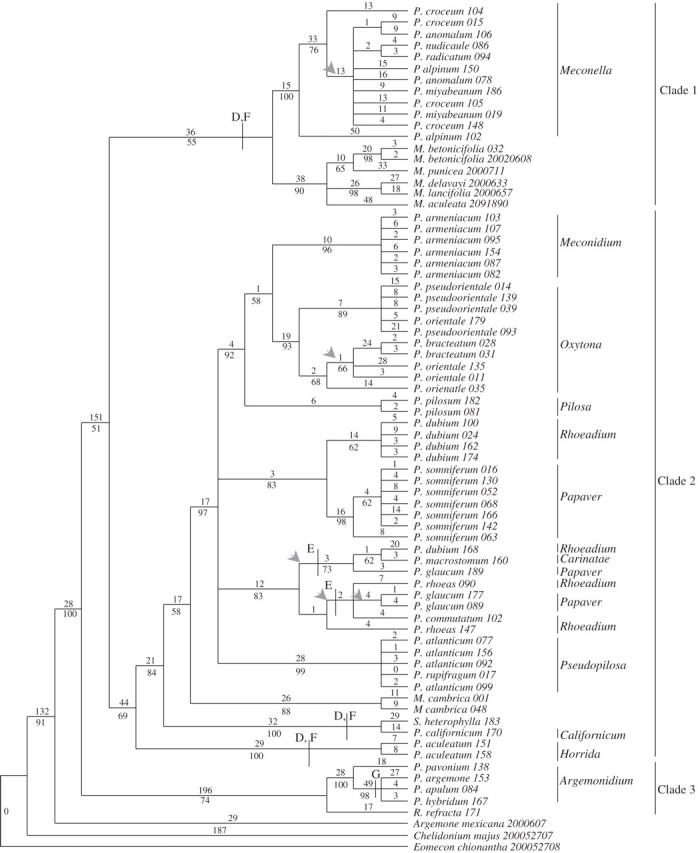
One of eight equally most-parsimonious trees generated from the trnL–F sequences using maximum parsimony. Support for each node is represented by bootstrap percentages (BP) below the branch (shown only when >50 % and consistent with the strict consensus tree). Numbers above each branch indicate the numbers of character changes along each lineage (ACCTRAN optimization). An arrow indicates branches that did not appear in the strict consensus tree. Groups that possess characteristic indels are indicated using letters (refer to Table 2).
Three main clades are present in the trnL–F trees, which (Fig. 2) are broadly congruent with the ITS analysis. The separation of P. sect. Argemonidium and Roemeria refracta (clade 3, 74 BP) and Meconella (clade 1, 100 BP) from the main group containing the remaining sections of Papaver (clade 2, 69 BP) is evident. Section Argemonidium is well supported (100 BP). The Asian representatives of Meconopsis (excluding M. aculeata) form a well-supported group (90 BP) and are sister to representatives of sect. Meconella (55 BP). Members of sect. Meconella possessed two characteristic 4-bp indels at positions 162 and 261 (D and F, Fig. 2; Table 2), which are also present in the outlying sections of clade 2, such as P. sects. Californicum, Horrida, Stylomecon heterophylla and Asian Meconopsis. Papaver argemone, P. apulum and P. hybridum share a 10-bp deletion at positions 644–653 (G, Fig. 2; Table 2), which is not found in P. pavonium.
The remainder of the sections form a weakly supported group (clade 2; 69 BP). Papaver sects. Horrida, Californicum and Meconopsis cambrica are resolved independently but sister to the main group in clade 2 with similar topologies to the ITS trees. Papaver californicum and Stylomecon heterophylla form a well-supported group (100 BP). Within clade 2, a group comprising sects. Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium was resolved (97 BP). Section Oxytona groups with sect. Meconidium (58 BP), with sect. Pilosa as sister to these (92 BP). The sampled species of section Pseudopilosa are well supported (99 BP). The members of sect. Rhoeadium do not form a monophyletic group; P. dubium (excluding P. dubium ssp. erosum) groups more closely with Papaver somniferum (sect. Papaver) than other members of sect. Rhoeadium (83 bp). In addition, P. dubium (excluding P. dubium ssp. erosum) does not possess a 5-bp indel at positions 186–190 (E, Fig. 2; Table 2) that the other members of sect. Rhoeadium share. Papaver glaucum groups within the main P. rhoeas clade (83 BP) and shares the indel (E, Fig. 2) with these Rhoeadium species.
Analysis of combined ITS and trnL–F
The combined trnL–F and ITS matrix was 1659 bp long. Parsimony analysis of the matrix generated eight equally most-parsimonious trees of 1332 steps with a CI of 0·63 and an RI of 0·84 (Fig. 3). The combination of the ITS and trnL–F data sets showed increased bootstrap support for the majority of groupings compared with those found in the individual analyses. Three clades are resolved. Clade 1 (90 BP) comprises P. sect. Meconella (100 BP) and Asian Meconopsis (99 BP). Clade 2 (81 BP) comprises the remaining sections of Papaver, Meconopsis cambrica and Stylomecon heterophylla. Section Horrida (100 BP) is sister to the rest of clade 2. The single representative of sect. Californicum (P. californicum) shares a close affinity with Stylomecon heterophylla (100 BP). Within clade 2, the main group of sections (Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium) is evident and well supported (99 BP). Of these, sect. Pseudopilosa is most divergent and monophyletic within Papaver (100 BP). Support for the positioning of Meconopsis cambrica as sister to the core sections of clade 2 and its separation from the other representatives of Meconopsis increased to 99 BP in comparison with 97 BP in the ITS tree and 58 BP in the trnL–F tree.
Fig. 3.
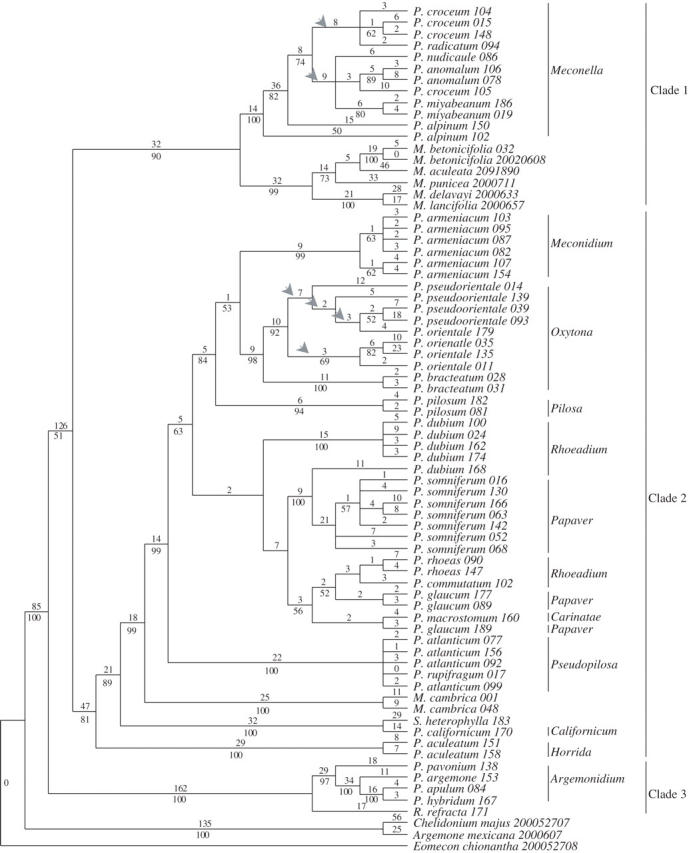
One of eight equally most-parsimonious trees generated from the combined ITS and trnL–F data sets using maximum parsimony. Support for each node is represented by bootstrap percentages (BP) below the branch (shown only when >50 % and consistent with the strict consensus tree). Numbers above each branch indicate the numbers of character changes along each lineage (ACCTRAN optimization). An arrow indicates branches that did not appear in the strict consensus tree.
Sections Meconidium (99 BP), Oxytona (98 BP) and Pilosa (94 BP) form a well-supported clade (84 BP). Sections Papaver and Rhoeadium are not monophyletic. Papaver glaucum (sect. Papaver) groups with species of sects. Rhoeadium and Carinatae (56 BP; P. commutatum, P. dubium ssp. erosum, P. macrostomum and P. rhoeas) and not with the other representatives of sect. Papaver. Finally, clade 3 comprises sect. Argemonidium plus Roemeria refracta (100 BP).
DISCUSSION
Phylogenetics of Papaver and related genera
The combination of nuclear ribosomal ITS and plastid trnL–F nucleotide sequences in a phylogenetic analysis resulted in well-resolved and well-supported trees. Three main lineages can be identified (clades 1, 2 and 3; Fig. 3). The results also show that Papaver is only monophyletic if Roemeria, Stylomecon heterophylla and Meconopsis cambrica are included in this genus. This is consistent with the molecular studies of Kadereit and Sytsma (1992) and Kadereit et al. (1997). Evidently, the topologies and major groupings of the phylogenetic trees produced in this analysis are incongruent with the generally accepted definitions of these closely interrelated genera. The major groupings found in this analysis are discussed below and interpreted in light of their morphology and biogeography.
Papaver sect. Argemonidium and Roemeria
Kadereit (1986a) revised Papaver sect. Argemonidium and concluded that it contains four annual, half-rosette species, P. apulum, P. argemone, P. hybridum and P. pavonium. Papaver apulum, P. argemone and P. pavonium are closely related and occur allopatrically from around the Adriatic Sea through Turkey–Iran to the Himalayas. The fourth species, P. hybridum, occupies a wide range from the Macaronesian Islands towards the Himalayas (Kadereit, 1986a, 1988a). The four species of this section are well differentiated in capsule and petal characters (Kadereit, 1986a) but are clearly closely related to each other as demonstrated by the groupings within the molecular phylogenetic trees (clade 3; 97 BP; Fig. 3). Within sect. Argemonidium, P. apulum and P. hybridum are sister species in both the ITS and the trnL–F analyses. In all analyses sect. Argemonidium is distinct from the other sections of Papaver and has characteristic indels (Table 2). The molecular distinctness of sect. Argemonidium is also supported by morphological differences (Fedde, 1909; Ernst, 1962; Cullen, 1965; Kadereit, 1986a; Markgraf, 1958), which include presence of an apical plug in the capsules, long internodes above the basal leaf rosette, polyporate pollen grains, bristly capsules and sepal morphology.
From a taxonomic point of view the most significant relationship involving sect. Argemonidium is the close grouping of its members with the genus Roemeria. In all analyses sect. Argemonidium and Roemeria are sister to each other. This affinity has been suggested by previous authors based on morphological observations (Günther, 1975; Morales Torres et al., 1988) and has been supported by previous molecular analyses (Kadereit and Sytsma, 1992; Kadereit et al., 1997). Some of the morphological characters that separate sect. Argemonidium from Papaver are shared with Roemeria, including polyporate pollen grains, sepal morphology and long internodes above the basal leaf rosette (shared with R. hybrida). Relationships within the Argemonidium–Roemeria group are unclear owing to incongruence between the ITS and trnL–F phylogenetic trees and also between these results and previous molecular analyses by Kadereit and Sytsma (1992) and Kadereit et al. (1997). The trees resulting from ITS sequences clearly show P. pavonium ssp. pavonium and Roemeria refracta as sister species (99 BP). This placement was also demonstrated by Kadereit and Sytsma (1992) and Kadereit et al. (1997). Based on the molecular similarity between P. pavonium and R. refracta and the fact that P. pavonium has a similar geographical distribution to R. refracta and R. hybrida, Kadereit and Sytsma (1992) and Kadereit et al. (1997) postulated that Roemeria had arisen from within sect. Argemonidium and most probably directly from Papaver pavonium or an ancestor of this species. However, in the analyses of the maternally inherited trnL–F region (Fig. 2), sect. Argemonidium and Roemeria are sister groups, indicating that Roemeria can be considered distinct from sect. Argemonidium but not distinct from Papaver. Incongruence of trees generated from these differently inherited DNA regions is sometimes attributed to hybridization. Given that the two species in question are both diploid (Podlech and Dieterle, 1969; Kadereit, 1986a), allopolyploidy cannot explain the different topologies of the ITS and trnL–F trees. However, hybridization or introgression could explain these differences. Divergence of ITS repeat types could also have occurred before the divergence of the Argemonidium–Roemeria group. Paralogy could therefore also explain the pattern, with one ITS repeat type retained in the P. pavonium–Roemeria group and an alternative type in the others.
The current taxonomy of Papaver and relatives does not take account of the distinctiveness of sect. Argemonidium and its close relationship to Roemeria. We here suggest a re-classification accommodating our molecular results. Elevation of sect. Argemonidium to genus level or a combination of sect. Argemonidium as a subgenus of Roemeria would be appropriate taxonomic treatments of these groups. We favour the former option because of the substantial morphological differences between sect. Argemonidium and Roemeria.
Papaver sect. Meconella and Meconopsis (excluding M. cambrica)
The scapose, perennial species of Papaver sect. Meconella (represented in this study by Papaver alpinum, P. anomalum, P. croceum, P. miyabeanum and P. radicatum) form a monophyletic group (100 BP in the trnL–F and combined analyses and 98 BP in the ITS analysis; Figs 1–3). Section Meconella is widely distributed across central, inner and eastern Asia, Siberia, Scandinavia through Greenland and northern Canada, with representatives found also in mountainous regions of Europe and the Rocky Mountains in North America (Rändel, 1974; Kadereit, 1988a). The species included in this study represent a limited sample from this distribution (five of 30 species; Rändel, 1974; Kadereit, 1988a).
The distinctness of this section from Papaver (excluding sect. Argemonidium) is also supported, as is its placement with Meconopsis excluding M. cambrica (Fig. 3; clade 1; 90 BP). A number of morphological characters have been used to define sect. Meconella (Hanelt, 1969; Rändel, 1974; Kadereit, 1988a). These include bristly, valvate capsules, simple or dissected pinnatisect leaves, pale anthers and filaments, and yellow, orange or white petals.
The species of sect. Meconella can be divided into two groups based on the degree of leaf dissection (finely dissected leaves: Papaver alpinum, P. miyabeanum and P. radicatum; broad leaf lobes: Papaver anomalum and P. croceum), but such a grouping is not supported by our molecular analysis. This morphological character has been discussed previously by Kadereit (1990) and Kadereit and Sytsma (1992) with reference to P. alpinum. The authors of these studies regarded finely dissected leaves to be a primitive character in sect. Meconella and suggested that this character may support a relationship to sect. Argemonidium with species of similar leaf morphology. Although P. alpinum is sister to the remaining Meconella species in the combined analysis, the other representatives of sect. Meconella with finely dissected leaves group more closely with species possessing broad leaf lobes. In addition, a molecular analysis of P. alpinum s.l. by Bittkau and Kadereit (2002) found that within this species broad leaf lobes are ancestral.
The position of sect. Meconella is not fully congruent with topologies obtained from earlier molecular analyses (Kadereit and Sytsma, 1992; Kadereit et al., 1997). The results from those analyses indicated that Meconella is sister to all sections of Papaver (Kadereit and Sytsma, 1992) or that sections Meconella and Argemonidium were resolved as sister to each other (Kadereit et al., 1997). However, bootstrap support for the sister-group relationship of these two sections (in the latter study) was low (<50 BP). Based on the topology of the major clades in our molecular trees, it can be concluded that sect. Meconella (and probably Meconopsis) is derived from a lineage that separated earlier from that giving rise to most other sections of Papaver (excluding Argemonidium).
The Asian representatives of Meconopsis were resolved as sister to sect. Meconella and share the diagnostic indels of sect. Meconella (D and F; Table 2). This grouping is incongruent with results of previous molecular analyses (Jork and Kadereit, 1995; Kadereit et al., 1997). The results of those analyses demonstrated that within Asian Meconopsis two distinct clades existed (based on an RFLP analysis of plastid DNA fragments). The first clade comprised species such as Meconopsis chelidonifolia and M. villosa that are sister to the other representatives of Asian Meconopsis (clade 2) plus the remaining Old World Papaveroideae (Meconopsis cambrica, Papaver, Roemeria, Stylomecon) used in that analysis. Only representatives of this second clade were included in the analyses reported here.
A significant amount of morphological difference exists between sect. Meconella and Papaver s.s. (clade 2). Although species of sect. Meconella possess a sessile stigmatic disc similar to the stigmatic discs typical of Papaver, it has been noted (Rändel, 1977; Kadereit et al., 1997) that the stigmatic discs of sect. Meconella may not be homologous to those found in other sections of Papaver (excluding Argemonidium). The stigmatic discs of Meconella consist in some cases of stigmatic tissue only, or there are deep incisions between the stigmatic rays. In addition, certain species of Meconella have polyporate instead of tricolpate pollen grains, a characteristic also found in some species of Meconopsis and Papaver sect. Argemonidium. No species of Meconella with polyporate pollen were included in this study.
If the current circumscription of Papaver is followed and sect. Meconella is retained within Papaver, a strict interpretation of the trees produced in this analysis would imply that Meconopsis should also be considered to be a member of Papaver. To retain Meconopsis as a genus would require a separation of sect. Meconella from Papaver. For example, it could either be raised to genus rank or included in Meconopsis. Meconella is therefore treated as a subgenus of Papaver, recognizing the distinction between Meconella and other Papaver subgenera but also recognizing that evidence exists for the amalgamation of Papaver and Meconopsis.
Papaver sects. Californicum and Horrida
Papaver sects. Californicum and Horrida are distributed outside the main geographical range of Papaver. Papaver aculeatum (sect. Horrida) is native to South Africa and is characterized by an indumentum of relatively long bristles, poricidal capsules, and pale filiform filaments and anthers. All green parts of the plant are covered with patent bristles (Kadereit, 1988c). Papaver californicum (sect. Californicum) is native to the west coast of North America and has a slender, ribbed, glabrous capsule, a many-flowered racemose inflorescence, pale anthers and filaments, and valvate capsule dehiscence (Kadereit, 1988b). Both species are annuals. In the ITS, trnL–F and combined trees both sections are attached to basal nodes within the main clade of Papaver (clade 2; Figs 1–3), and sect. Californicum is sister to the ‘core’ group of Papaver (sects. Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium) and Meconopsis cambrica. Papaver aculeatum shares morphological and cytological characteristics with sects. Pilosa and Papaver. Similarities between P. aculeatum and P. somniferum (sect. Papaver) include auriculate–amplexicaulous leaves and a chromosome base number of n = 11, both characteristics found only in these two species. However, both these characters appear to have evolved in parallel (Figs 1–3). Similarities between sects. Horrida and Pilosa include racemose inflorescences, pale filiform filaments and the possession of long capsules with flat stigmatic discs (Kadereit, 1988c). However, these two sections do not associate in the molecular analysis presented here, and convergence is therefore also implied to explain the similarity in morphology. Papaver californicum shares characteristics with sect. Meconidium, including valvate capsule dehiscence and pale filiform filaments, but species of these two groups are geographically widely separated and do not associate in the molecular trees.
The results from the molecular analysis support the view of Kadereit et al. (1997) that Stylomecon heterophylla arose from within Papaver and should not be considered a separate genus. Stylomecon heterophylla and P. californicum are both native to California and grow in similar habitats (Kadereit, 1988b). Morphological similarities between these species include leaf shape, glabrous/globose buds, orange petals, and pale anthers and filiform filaments (Ernst, 1962; Kadereit, 1988b). The two species are differentiated by capsular morphology, with S. heterophylla possessing a distinct style that is similar to those found in many representatives of Meconopsis. In the ITS, trnL–F and combined analysis, S. heterophylla and P. californicum form a well-supported group (100 BP in the trnL–F and combined trees, Figs 2 and 3; 96 BP in the ITS trees, Fig. 1). The two species appear to have diverged relatively recently. Stylomecon heterophylla possesses the 4-bp indel diagnostic for sects. Meconella, Californicum and Horrida (incl. Asian Meconopsis) at positions 261–265 in the trnL–F region (F, Fig. 2; Table 2). The separation of S. heterophylla from Papaver therefore is not justified based solely on differences in capsule characteristics.
The results of the molecular analysis can be interpreted in a number of ways for taxonomic conclusions concerning sects. Californicum and Horrida. Both sections are successively sister to the highly supported (99 BP; Fig. 3) core group of Papaver comprising sects. Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium. However, sects. Californicum and Horrida possess the characteristic 4-bp indel at positions 248–252 in the trnL–F region shared with Asian Meconopsis and representatives of sect. Meconella. The disjunct geographical distributions of sect. Californicum (North America) and sect. Horrida (South Africa) might indicate a wider distribution of Papaver at some point during its evolutionary history, with extinction occurring in North America and Africa leaving these two sections geographically isolated (Randel, 1974; Kadereit et al., 1997), or indicate long-distance dispersal. Taking into account the outlying positions of sects. Californicum and Horrida in the molecular trees, they seem to derive from a relatively ancient lineage of Papaver. The positions of Californicum and Horrida within the core Papaver clade in the analyses here are congruent with previous molecular analyses (Kadereit and Sytsma, 1992; Kadereit et al., 1997).
It is recommended that sects. Californicum and Horrida be elevated to the rank of subgenera within Papaver, i.e. subgen. Californicum and subgen. Horrida. The separation of Stylomecon heterophylla from Papaver is rejected. The clear relationship of this species to P. californicum, as indicated by similarities in morphology, geographical distribution, and nucleotide sequences within the ITS and trnL–F gene regions, favours its inclusion in subg. Californicum. Considering these differences in capsule morphology, subg. Californicum should contain two species, Papaver californicum and P. heterophylla (=Stylomecon heterophylla).
Meconopsis cambrica
The only European species of Meconopsis, M. cambrica, is well separated from the representatives of Asian Meconopsis in the molecular analysis here. Meconopsis cambrica occupies a well-supported (99 BP; Fig. 3) sister-group position to the remaining sections of Papaver (excluding Argemonidium, Californicum, Horrida and Meconella). This supports the view (Kadereit et al. 1997) that two distinct lineages within Meconopsis s.l. exist and that Meconopsis in its current circumscription is neither monophyletic nor distinct from Papaver. Meconopsis cambrica shares diagnostic trnL–F indels with the majority of Papaver (excluding Argemonidium, Californicum, Horrida and Meconella). Meconopsis cambrica could have arisen either in parallel with the Asian representatives of Meconopsis, clade 2, i.e. core Papaver (sects. Carinatae, Papaver, Pilosa, Pseudopilosa, Oxytona, Meconidium and Rhoeadium), or from within a lineage best recognized as members of an expanded Meconopsis. Both these views were proposed by Kadereit et al. (1997), who favoured the latter view based on geographical, phytochemical and morphological considerations. Topological considerations alone favour parallel evolution as M. cambrica is embedded in clade 2 in our ITS/trnL–F trees.
It is evident from the results of this analysis that incongruence exists with previous taxonomic classifications regarding the positioning of M. cambrica. If M. cambrica is recognized as Meconopsis, Papaver s.s. (i.e. after the exclusion of the groups discussed above) is not monophyletic. It is suggested to include M. cambrica (as Papaver cambrica L.) in Papaver. However, an appropriate treatment of this species is difficult owing to the lack of apparent morphological similarities with extant Papaver species. There is no obvious section or group of species with which to place Papaver cambrica. Although unsatisfactory from a taxonomic perspective it may be necessary to describe a new monotypic section for this species within Papaver. The alternative is to leave it as incertae sedis until further evidence is found regarding its placement.
Inter-sectional relationships in Papaver s.s.
Clade 2 contains a well-supported (99 BP; Fig. 3) group of sections including Carinatae, Meconidium, Oxytona, Papaver, Pilosa, Pseudopilosa and Rhoeadium (hereafter described as Papaver s.s.). This is the largest inclusive group of Papaver s.l. This group was described by Kadereit et al. (1997) as representing the typical species of Papaver. Within this group, inter-sectional relations are not fully resolved, but sections are generally well supported (Figs 1–3).
Section Pseudopilosa (represented in the combined analysis by P. atlanticum and P. rupifragum) forms a well-supported group in the combined analysis (100 BP; Fig. 3) and is sister to the remaining sections of Papaver s.s. Representatives of sect. Pseudopilosa are characterized by having unique 5- and 4-bp indels at positions 75–79 (A, Fig. 1) and 216–219 (B, Fig. 1) of the ITS region, respectively (Table 2). The species of this section are of subscapose to scapose habit and are found in south-western Asia, northern Africa and southern Spain.
Section Pilosa comprises a single perennial subscapose species with a number of subspecies found predominantly in western Turkey (Kadereit, 1996). The species is characterized by convolute leaf vernation, poricidal capsule dehiscence and pale filiform filaments. The separation of sect. Pilosa from sect. Pseudopilosa based on morphological and phytochemical differences (Popov, 1937; Günther, 1975; Kadereit, 1996) is supported by the results of the combined analysis here (Fig. 3). Papaver pilosum is sister to sects. Oxytona and Meconidium (86 BP). Section Oxytona comprises a polyploid series including diploid P. bracteatum (2n = 14), tetraploid P. orientale Fedde (2n = 28) and allohexaploid P. pseudo-orientale Fedde (2n = 42) and is found predominantly in the Caucasus Mountains, eastern Turkey and north-western Iran (Goldblatt, 1974). The group is characterized by their perennial habit, poricidal capsule dehiscence, and dark filaments and anthers. Section Meconidium, comprising four biennial species (represented in the analysis here by two subspecies of P. armeniacum), occupies a continuous geographical range in southern and eastern Turkey, the Caucasus Mountains, northern Iraq and north-western Iran and possesses glabrous or bristly capsules, valvate capsule dehiscence, and pale filaments and anthers. Sections Meconidium, Oxytona and Pilosa are heterogeneous morphologically, and identification of synapomorphies for this group is difficult. The three species of sect. Oxytona are clearly monophyletic (98 BP; Fig. 3). Genomic and fluorescence in situ hybridization studies (Carolan, 2004) have indicated that the diploid P. bracteatum was a parent of the hexaploid P. pseudo-orientale. The clear inter-relationship between these species has been demonstrated previously using AFLP fingerprinting (Carolan et al., 2002).
The remaining sections of Papaver s.s. are sects. Carinatae, Papaver and Rhoeadium. The results of the molecular analyses question whether these sections are monophyletic. Papaver sect. Rhoeadium consists of 17 predominantly annual species (Günther, 1975; Kadereit, 1989) and is represented in this study by Papaver commutatum, P. dubium and P. rhoeas. The centre of diversity of sect. Rhoeadium is south-western Asia and the Aegean area with some species found in the central or western Mediterranean, the Balkans and the western Himalayas (Kadereit, 1989). Characteristic morphological traits include poricidal capsules and dark (sometimes light) filaments. However, the section is extremely diverse in morphological characteristics. Kadereit (1989) recognized three species groups within sect. Rhoeadium based on geographical and morphological traits. The first group contains species with longer than broader capsules, such as P. dubium, and only tetraploid (2n = 28) and hexaploid (2n = 42) species. The second group contains diploid species (2n = 14), including P. arenarium and P. commutatum, and is diverse morphologically. The third group is morphologically more uniform than the P. arenarium group and consists of diploid species, including P. rhoeas. The similarity of the P. rhoeas and P. arenarium groups (the latter represented by P. commutatum) suspected by Kadereit (1989) is weakly supported (56 BP; Fig. 3) here. In addition, the representatives of these two groups possess a diagnostic 5-bp indel at positions 186–191 of the trnL–F region (E, Table 2). A separation exists in some analyses (Figs 2 and 3) between the P. rhoeas/P. arenarium groups and the P. dubium group. In the trnL–F trees, the P. dubium group is clearly allied to P. somniferum (83 BP; Fig. 2). However, in the ITS trees obtained (Fig. 1) the P. dubium group is weakly allied to the P. rhoeas group (BP < 50 %). Papaver dubium also lacks a characteristic 5-bp trnL–F indel (E, Table 2) unique to the other representatives of sect. Rhoeadium (including P. glaucum). In addition, some incongruence between the ITS and trnL–F topologies exists with respect to P. dubium ssp. erosum. In the ITS analysis P. dubium ssp. erosum groups with P. somniferum (sect. Papaver; 88 BP; Fig. 1), and in the trnL–F tree it groups within a subclade comprising P. commutatum, P. glaucum, P. macrostomum and P. rhoeas (83 BP; Fig. 2).
The single representative of sect. Carinatae (P. macrostomum) consistently fell within the P. rhoeas group and shares its diagnostic trnL–F indel (Figs 1–3; E, Table 2). Papaver macrostomum, distributed in Iran, Iraq and Turkey, possesses all the morphological characteristics of sect. Rhoeadium but has been separated into a separate section based on the possession of a deciduous stigmatic disc (Fedde, 1909; Kadereit, 1987). No support for the separation of P. macrostomum from sect. Rhoeadium is found in the ITS and trnL–F trees.
The four annual representatives of sect. Papaver (represented in this study by P. glaucum and P. somniferum) from the western Mediterranean and south-western Turkey to Cyprus, Iran, Afghanistan and Pakistan do not form a monophyletic group in our analyses of ITS and trnL–F. Species of this section are characterized by the possession of more or less strongly auriculate–amplexicaulous leaves, poricidal capsule dehiscence and dark (sometimes pale) filaments. Papaver glaucum shows more sequence similarity to sect. Rhoeadium. This division within sect. Papaver has previously been demonstrated (Kadereit and Sytsma, 1992). The study by these authors also demonstrated that P. glaucum and P. gracile (members of this section) are more closely related to P. rhoeas and P. dubium of sect. Rhoeadium. Many morphological and geographical similarities exist between the two sections (see Kadereit, 1988a). Phytochemically, P. glaucum differs from P. somniferum in not accumulating morphinane alkaloids but rather has some alkaloids similar to those found in P. rhoeas (Preininger et al., 1981; Preininger, 1986). Papaver gracile, P. glaucum and P. decaisnii, like the majority of Papaver, have a base chromosome number of n = 7. Papaver somniferum has a base chromosome number of n = 11 (Hammer and Fritsch, 1977). These differences in chromosome number and alkaloid spectra led Novak and Preininger (1980) and Preininger et al. (1981) to separate these three species into their new sect. Glauca. Reckin (1973) transferred these species to sect. Rhoeadium. The presence in P. glaucum of the diagnostic 5-bp indel at positions 186–191 of the trnL–F region (E, Table 2), characteristic for the Papaver rhoeas group, further questions the classification of P. glaucum in sect. Papaver.
Although our study could demonstrate the non-monophyly of sects. Papaver and Rhoeadium, limited sampling of species and limited support for some groups do not allow us to reclassify Papaver s.s. confidently into sections apart from the inclusion of M. cambrica just discussed. However, Papaver s.s. should be treated as Papaver subg. Papaver. It seems likely from the molecular results that subg. Papaver will contain sects. Meconidium, Oxytona, Papaver (including Rhoeadium and Carinatae), Pilosa and Pseudopilosa.
Evaluation of morphological characters previously viewed as diagnostic for Papaver
Papaver has been defined primarily by the possession of a capsule with a sessile stigmatic disc. The results of the molecular analyses presented here clearly demonstrate that a number of species with sessile stigmatic discs are close relatives of taxa that possess a style. This is demonstrated by S. heterophylla and P. californicum and P. sect. Meconella and Asian Meconopsis. Furthermore, the structure of the stigmatic disc in sect. Argemonidium is different from all other stigmatic discs due to the formation of a plug-like structure in the interior of the capsule. This can be regarded as evidence for its independent evolution from other species with a typical stigmatic disc.
Papaver has generally been considered to represent the most derived lineage of Papaveroideae, and hence the sessile stigmatic disc was deemed to be an advanced character. The results here are congruent with this view with respect to Papaver s.s. only. In light of the groupings generated in our phylogenetic analysis it is not inconceivable that the sessile stigmatic disc has arisen on a number of occasions from ancestors with a style. Independent origins of the stigmatic disc in Papaver have been suggested previously (Kadereit and Sytsma, 1992).
Two morphological characters were considered of primary significance for the evaluation of relationships within Papaver, particularly at the inter-sectional level. These are the mode of capsule dehiscence and the degree of pigmentation of filaments and anthers. The possession of pale filaments and anthers by the majority of genera of Papaveroideae and of dark filaments in part of Papaver s.s. indicates that pale filaments might be ancestral. Dark filaments seem to have evolved more than once, or there have been reversals to pale filaments in some sections (e.g. Meconidium and Pilosa). Molecular and morphological data separate sect. Argemonidium from the other sections with dark filaments (Carinatae, Oxytona, Papaver and Rhoeadium).
Sections Meconella and Californicum have valvate capsule dehiscence and an outlying position with respect to the other sections of Papaver. This indicates that valvate capsule dehiscence may be primitive. However, this character is also found in sect. Meconidium, which falls within Papaver s.s. Its presence here suggests that this character is a synapomorphy for the species of sect. Meconidium. Thus, the results of this analysis indicate that valvate capsule dehiscence has evolved independently at least three times within Papaver s.l.
The combination of morphological, biogeographical and molecular characters has made possible a novel interpretation of relationships in Papaver and allies, and allows for more useful taxonomies to be generated. A formal taxonomic revision of Papaver infrageneric groupings is in preparation.
Acknowledgments
We thank Matthew Jebb of the National Botanical Garden, Glasnevin, Ireland, for his assistance in the acquisition and cultivation of a considerable portion of the material used in this study. In addition, thanks are due to Brian S. Luther (Seattle, USA), Suzanne Cubey (Herbarium of the Royal Botanic Garden, Edinburgh, UK), Hans Hansen (University of Copenhagen, Botanic Gardens) and Rainer Greissl (Mainz, Germany) for the provision of material, and to Joe Reilly, Mary Deegan and Maureen Brunt for assistance in cultivating and maintaining the living collection at Trinity College, Dublin, Ireland.
LITERATURE CITED
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Cambell CS, Donoghue MJ. 1995. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden 82: 247–277. [Google Scholar]
- Bittkau C, Kadereit JW. 2002. Phylogenetic and geographical relationships in Papaver alpinum L. (Papaveraceae) based on RAPD data. Botanische Jahrbücher für Systematik, Pflanzengeschichte und Pflanzengeographie 123: 463–479. [Google Scholar]
- Carolan JC. 2004. Phylogenetic analysis of Papaver L. PhD thesis, University of Dublin, Trinity College, Ireland.
- Carolan JC, Hook ILI, Walsh JJ, Hodkinson TR. 2002. Using AFLP markers for species differentiation and assessment of genetic variability of in vitro cultured Papaver bracteatum (section Oxytona). In vitro cellular and developmental biology. Plant 38: 300–307. [Google Scholar]
- Chase MW, de Bruijn AY, Reeves G, Cox AV, Rudall PJ, Johnson MAT, Eguiarte LE. 2000. Phylogenetics of Asphodelaceae (Asparagales): an analysis of plastid rbcL and trnL–F DNA sequences. Annals of Botany 86: 935–956. [Google Scholar]
- Cordell GA. 1981. Alkaloids derived from phenylalanine and tyrosine. In: Cordell GA, ed. Introduction to alkaloids: a biogenetic approach. New York: John Wiley and Sons, 490–512.
- Cullen J. 1965. Papaveraceae. In: Davis PH, ed. Flora of Turkey and the east Aegean Islands, Vol. I. Edinburgh213–247.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletins 19: 11–15. [Google Scholar]
- Ernst WR. 1962. A comparative morphology of the Papaveraceae. PhD thesis, Stanford University, Stanford, California.
- Fedde F. 1909. Papaveraceae–Hypecoideae et Papaveraceae–Papaveroideae. In: Engler A, ed. Das Pflanzenreich. Leipzig: Englemann, 228.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gielly LP, Taberlet P. 1996. A phylogeny of the European gentians inferred from chloroplast trnL (UAA) intron sequences. Botanical Journal of the Linnaean Society 120: 57–75. [Google Scholar]
- Gielly LP, Yuan YM, Kupfer P, Taberlet P. 1994. Phylogenetic use of noncoding regions in the genus Gentiana L.: chloroplast trnL (UAA) intron versus nuclear ribosomal internal transcribed spacer sequences. Molecular Phylogenetics and Evolution 5: 460–466. [DOI] [PubMed] [Google Scholar]
- Goldblatt P. 1974. Biosystematic studies in Papaver section Oxytona. Annals of the Missouri Botanic Garden 61: 264–296. [Google Scholar]
- Günther KF. 1975. Beitrage zur Morphologie und verbreitung der Papaveraceae 2. Teil: Die Wuchsformen der Papaverae, Escoscholzieae und Platystemonoideae. Flora 164: 393–436. [Google Scholar]
- Hammer K, Fritsch R. 1977. Zur Frage nach der Ursprungsart des Kulturmohns (Papaver somniferum L.). Kulturpflanze 25: 113–124. [Google Scholar]
- Hodkinson TR, Chase MW, Takahashi C, Leitch IJ, Bennett MD, Renvoize SA. 2002. The use of DNA sequencing (ITS and trnL–F), AFLP and fluorescent in-situ hybridisation to study allopolyploid Miscanthus (Poaceae). American Journal of Botany 89: 279–286. [DOI] [PubMed] [Google Scholar]
- Hopper SD, Fay MF, Rossetto M, Chase MW. 1999. A molecular phylogenetic analysis of the bloodroot and kangaroo paw family, Haemodoraceae: taxonomic, biogeographic and conservation implications. Botanical Journal of the Linnean Society 131: 285–299. [Google Scholar]
- Jork KB, Kadereit JW. 1995. Molecular phylogeny of the Old World representatives of Papaveraceae subf. Papaveroideae with special emphasis on the genus Meconopsis Vig. In: Jensen U, Kadereit JW, eds. Systematics and evolution of the Ranunculiflorae. Plant Systematics and Evolution 9 (Suppl.): 171–180.
- Kadereit JW. 1986a. A revision of Papaver section Argemonidium. Notes of the Royal Botanic Garden Edinburgh 44: 25–43. [Google Scholar]
- Kadereit JW. 1986b. A revision of Papaver L. sect Papaver (Papaveraceae). Botanische Jahrbücher für Systematik, Pflanzengeschichte und Pflanzengeographie 108: 1–16. [Google Scholar]
- Kadereit JW. 1987. A revision of Papaver sect Carinatae (Papaveraceae). Nordic Journal of Botany 7: 501–504. [Google Scholar]
- Kadereit JW. 1988a. Sectional affinities and geographical distribution in the genus Papaver L. (Papaveraceae). Beiträge zur Biologie der Pflanzen 63: 139–156. [Google Scholar]
- Kadereit JW. 1988b. Papaver L. sect Californicum Kadereit, a new section of the genus Papaver. Rodora 90: 7–13. [Google Scholar]
- Kadereit JW. 1988c. The affinities of the south-hemispherical Papaver aculeatum Thunb. (Papaveraceae). Botanische Jahrbücher für Systematik, Pflanzengeschichte und Pflanzengeographie 109: 335–341. [Google Scholar]
- Kadereit JW. 1989. A revision of Papaver section Rhoeadium Spach. Notes of the Royal Botanic Garden Edinburgh 45: 225–286. [Google Scholar]
- Kadereit JW. 1990. Some suggestions on the geographical origin of the central, west and north European synantropic species of Papaver L. Botanical Journal of the Linnean Society 103: 221–231. [Google Scholar]
- Kadereit JW. 1993a. Papaveraceae. In: Kubitzki K, Rohweer JG, Bittrich V, eds. The families and genera of vascular plants, Vol. II. Berlin: Springer-Verlag, 20–33.
- Kadereit JW. 1993b. A revision of Papaver sect. Meconidium. Edinburgh Journal of Botany 50: 125–148. [Google Scholar]
- Kadereit JW. 1996. A revision of Papaver L. sects Pilosa Prantl and Pseudopilosa M. Popov ex Gunther (Papaveraceae). Edinburgh Journal of Botany 53: 285–309. [Google Scholar]
- Kadereit JW, Sytsma KJ. 1992. Disassembling Papaver: a restriction site analysis of chloroplast DNA. Nordic Journal of Botany 12: 205–217. [Google Scholar]
- Kadereit JW, Schwarzbach AE, Jork KB. 1997. The phylogeny of Papaver s.l. (Papaveraceae): polyphyly or monophyly? Plant Systematics and Evolution 204: 75–98. [Google Scholar]
- Kelchner SA. 2000. The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden 87: 182–198. [Google Scholar]
- Markgraf F. 1958. Familie Papaveraceae. In: Hegi P, ed. Illustrietrte Flora von Mittelurropaa Band IV, 2nd edn. Teil 1. Munich: Hanser, 15–49.
- Morales Torres C, Mendoza Castellon R, Romero Garcia AT. 1988. La posicion sistematica de Papaver argemone L.: interes evolutivo del orden Papaverales: 1. Lagascalia 15: 181–189. [Google Scholar]
- Novak J, Preininger V. 1980. Sect., Glauca-nova sekce rodu Papaver. Presila 52: 97–101. [Google Scholar]
- Podlech D, Dieterle A. 1969. Chromosomenstudien an Afghanischen Pflanzen. Candollea 24: 185–243. [Google Scholar]
- Popov MG. 1937. Papaveraceae. In: Komarov VL, ed. Flora of the USSR. Moscow-Leningrad: Nauka, 470–474.
- Preininger V. 1986. Chemotaxonomy of Papaveraceae and Fumariaceae. In: The alkaloids. London: Academic Press, 2–98.
- Preininger V, Novak J, Santavy F. 1981. Isolierung und Chemie der Alkaloide aus Pflanzen der Papaveraceae LXXXXI. Glauca, eine neue Sektion der Gattung Papaver. Planta Medica 41: 119–123. [DOI] [PubMed] [Google Scholar]
- Rändel U. 1974. Beitrage zur Kenntnis der Sippenstruktuir der Gattung Papaver L. Sectio Scapiflora Reihenb. Im vergleich mit P. alpinum L. (Papaveraceae). Feddes Repert 86: 19–37.
- Rändel U. 1977. Uber die grondlandischen Vertreter der section Lasiotrachyphylla Bernh. (Papaveraceae). Feddes Repert 88: 421–450. [Google Scholar]
- Reckin J. 1973. A contribution to the cytology of Papaver gracile Auch. Including proposals for a revision of the section Mecones. Caryologia 26: 245–251. [Google Scholar]
- Salamin N, Chase MW, Hodkinson TR, Savolainen V. 2003. Assessing internal support with large phylogenetic DNA matrices. Molecular Phylogenetics and Evolution 27: 528–539. [DOI] [PubMed] [Google Scholar]
- Seelanan T, Schnabel A, Wendel J F. 1997. Congruence and consensus in the cotton tribe (Malvaceae). Systematic Botany 22: 259–290. [Google Scholar]
- Savolainen V, Chase MW. 2003. A decade of progress in plant molecular phylogenetics. Trends in Genetics 12: 717–724. [DOI] [PubMed] [Google Scholar]
- Schwarzbach AE, Kadereit JW. 1999. Phylogeny of prickly poppies, Argemone (Papaveraceae), and the evolution of morphological and alkaloid characters based on ITS nrDNA sequence variation. Plant Systematics and Evolution 218: 257–279. [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. 1994. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics 89: 26–32. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*: Phylogenetic analysis using parsimony (* and other methods), version 4.0b 10. Sunderland, MA: Sinauer Associates.
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. 1996. Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK, eds. Molecular systematics, 2nd edn. Sunderland, MA: Sinauer Associates, 407–514.
- Taberlet PL, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Moleular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Taylor G. 1930. Stylomecon: a new genus of Papaveraceae. Journal of Botany 68: 138–140. [Google Scholar]
- Wendel JF, Doyle JJ. 1998. Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants II. London: Kluwer Academic Publishers, 265–296.
- Wendel JF, Schnabel A, Seelanan T. 1995. An unusual ribosomal DNA sequence from Gossypium gossypioides reveals ancient, cryptic, intergenomic introgression. Molecular Phylogenetics and Evolution 4: 298–313. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Irwin JA, Payseur, BA. 2001. Failure of the ILD to determine data combinability for slow Loris phylogeny. Systematic Biology 50: 108–121. [PubMed] [Google Scholar]


