Abstract
• Background and Aims For rare endemics or endangered plant species that reproduce both sexually and vegetatively it is critical to understand the extent of clonality because assessment of clonal extent and distribution has important ecological and evolutionary consequences with conservation implications. A survey was undertaken to understand clonal effects on fine-scale genetic structure (FSGS) in two populations (one from a disturbed and the other from an undisturbed locality) of Echinosophora koreensis, an endangered small shrub belonging to a monotypic genus in central Korea that reproduces both sexually and vegetatively via rhizomes.
• Methods Using inter-simple sequence repeats (ISSRs) as genetic markers, the spatial distribution of individuals was evaluated using Ripley's L(d)-statistics and quantified the spatial scale of clonal spread and spatial distribution of ISSR genotypes using spatial autocorrelation analysis techniques (join-count statistics and kinship coefficient, Fij) for total samples and samples excluding clones.
• Key Results A high degree of differentiation between populations was observed (ΦST(g) = 0·184, P < 0·001). Ripley's L(d)-statistics revealed a near random distribution of individuals in a disturbed population, whereas significant aggregation of individuals was found in an undisturbed site. The join-count statistics revealed that most clones significantly aggregate at ≤6-m interplant distance. The Sp statistic reflecting patterns of correlograms revealed a strong pattern of FSGS for all four data sets (Sp = 0·072–0·154), but these patterns were not significantly different from each other. At small interplant distances (≤2 m), however, jackknifed 95 % CIs revealed that the total samples exhibited significantly higher Fij values than the same samples excluding clones.
• Conclusion The strong FSGS from genets is consistent with two biological and ecological traits of E. koreensis: bee-pollination and limited seed dispersal. Furthermore, potential clone mates over repeated generations would contribute to the observed high Fij values among genets at short distance. To ensure long-term ex situ genetic variability of the endangered E. koreensis, individuals located at distances of 10−12 m should be collected across entire populations of E. koreensis.
Keywords: Clonal structure, conservation, Echinosophora koreensis, monotypic genus, Fabaceae, fine-scale genetic structure, genets, ISSRs, sampling strategies
INTRODUCTION
Clonal reproduction is a common fact in many plant species. Mechanisms of clonal reproduction include vegetative spread, production of bulbils, and apomictic seeds. For rare, endemic and/or endangered plant species that reproduce both sexually and vegetatively, it is important to characterize clonality because clonal structure could affect overall population fitness. In self-compatible plant species, for example, it has been suggested that increasing levels of clonal structure will increase the likelihood of self-fertilization (inbreeding) by pollination among different ramets of the same genets (i.e. geitonogamy; Handel, 1985). Thus, the spatial distribution of clonal vs. that of sexual reproduction generally has differing effects on the levels of biparental inbreeding and apparent selfing rate, produced via mating by proximity through limited pollen dispersal (Peakall and Beattie, 1991; Eckert and Barrett, 1993). In outcrossing plant species, extensive clonality could reduce reproductive potential, overall population fitness, and effective population size, probably due to decreased probability of mating with different genets. Thus, knowledge of clonal structure within populations is crucial for understanding evolutionary processes in clonal plants.
Spatial statistical methods provide powerful tools for measuring the structure of genetic diversity within populations of target plants. Fine-scale spatial genetic structure (FSGS) can be quantified using spatial autocorrelation analysis to investigate population genetic processes (Sokal and Oden, 1978; Epperson, 1990; Heywood, 1991). When FSGS is analysed within populations of clonal plants, it is important to separate the number of genets (genetic individuals established via seeds) from the total samples including clonal ramets because inferences for micro-evolutionary processes and for conservation purposes from the pattern and magnitude of FSGS (such as the extent of pollen and seed dispersal, degree of inbreeding and genetic patch sizes) must be addressed from the number of genetic individuals (Chung and Epperson, 1999; Reusch et al., 1999; Hämmerli and Reusch, 2003; Alberto et al., 2005; Chung et al., 2005a, b). Therefore, it is essential to analyse fine-scale clonal structure using genetic markers a priori to understand better micro-evolutionary processes in clonal plant species. This point is relatively understudied among plant population geneticists, though there have been many studies on clonal diversity and structure within plant populations (e.g. Lynch et al., 1998; Sydes and Peakall, 1998; Suzuki et al., 1999; Burke et al., 2000; Rossetto et al., 2004; Torimaru and Tomaru, 2005; many references therein).
Inter simple sequence repeats (ISSR) are useful markers for estimating genetic variation within populations (Wolfe and Liston, 1998). Recent studies on clonal diversity of plant species have shown the great discriminating power of ISSR makers for separating genets from their clonal ramets within populations (Zietkiewicz et al., 1994; Esselman et al., 1999; Li and Ge, 2001; Xie et al., 2005).
In plant populations, limited seed dispersal generates, in the same way as clonality, spatial clustering of full- and half-sib cohorts (Hamrick and Nason, 1996; Kalisz et al., 2001). For clonal plants, if pollen dispersal is also spatially restricted within populations, then this sibling structure would further enhance biparental inbreeding or selfing via mating by proximity (Wright, 1969). Under this scenario, it is predicted that, repeated over generations, genetically distinct individuals would form a significant spatial clustering, and high levels of genetic similarity (strong spatial autocorrelation) between pairs of genets would exist over short distances.
Echinosophora koreensis was chosen as a study species for two reasons: (1) it reproduces both sexually and vegetatively; (2) it is endemic to Korea and belongs to a monotypic genus restricted to only a few localities in central Korea and may be at risk of extinction. To test the genetic prediction described above, two isolated populations of E. koreensis in central Korea were selected. The area (13 200 m2) covering the two populations has been designated as a Natural Monument of Korea (#372) since 1992. Then spatial distribution of individuals and their genotypes at the ramet and genet levels were quantified using ISSRs as genetic markers. To do this, spatial statistics [Ripley's L(d)-statistics] and spatial autocorrelation methods (join-count statistic and kinship coefficient, Fij) were used. With the information obtained on clonal and genetic structure, sampling strategies for ex situ conservation were provided.
MATERIALS AND METHODS
Study species
Echinosophora koreensis (Nakai) Nakai (Fabaceae) is a small shrub (<1 m tall) which belongs to a monotypic genus endemic to Korea (Park, 1973; Yim, 1993). In South Korea, populations of the species are known only in a few locations in Yanggu-gun and Injae-gun, in Kangwon-do Province (K. Huh, pers. comm.). Populations in Yanggu-gun and Injae-gun are considered to be at the southern limit of its distribution in Korea. Due to its rarity, the species is listed among the 58 endangered plant species proposed by the Ministry of Environment to be targeted for conservation in South Korea (http://www.me.go.kr). According to herbarium records, the species was also known in a few locations in North Korea (Myongsan, Pyangannam-do Province, and Bukchung, Hamgyeongnam-do Province). At present, there are no available samples from North Korea, and North Korean populations, if they still exist, have no protection measures in place. Based on available data concerning the total number of mature individuals (population size) for South Korea at the regional level (Gärdenfors et al., 2001), E. koreensis can be classified as endangered (EN) following criteria of the IUCN Red List C2a(i) (IUCN, 2001).
Echinosophora koreensis is a pioneer species that can potentially invade open, sunny forest gaps, and disturbed areas (Yim, 1993). The natural habitats are xeric, consisting of small granite gravel, where shoots are interconnected by rhizomes (a 180-cm-long rhizome was identified in one population), indicating that the species propagates both sexually and vegetatively. Ten to twenty yellow flowers (3–5 cm long) per inflorescence bloom in May and are frequently visited by the bumblebee Bombus diversus diversus (M. Y. Chung and M. G. Chung, personal observation). The species was found to be self-compatible under greenhouse conditions (M. Y. Chung and M. G. Chung, unpubl. res.). Pods (approx. 7 cm long) ripen from July to September and contain two or three seeds. There are apparently no specialized mechanisms of seed dispersal.
Studied sites and sampling procedure
Two study sites were selected in the centre of each of two populations in Yanggu-gun, where 286 visually identified shoots with fruits were mapped and leaf samples were collected. The first population (hereafter referred to HAN, 30 × 40 m in area, approx. 135 m a.s.l., N(r) = 150) is located on a north-east-facing hillside in Hanjeon-ri (Yanggu-eup), occupied by dead trunks of Pinus densiflora remaining from a fire in the area in December 2000. The second population (hereafter referred to YIM, 30 × 40 m in area, approx. 210 m a.s.l., N(r) = 136), 2·8 km south of HAN, is located on a north-facing hillside in Imdang-ri (Yanggu-eup) under a low-density old stand of Pinus densiflora. In YIM, other shrubs were cleared by government officials in an attempt to give more sun to E. koreensis. One leaf was collected from each flowering shoot of E. koreensis and stored at 4 °C until DNA was extracted.
DNA isolation and polymerase chain reaction (PCR) amplification
For ISSR–PCR, a total of 24 ISSR primers [University of British Columbia (UBC), Canada] were screened using four representatives from each of the two populations. Among them, six primers that gave clear polymorphisms were used in this study (Table 1). Total genomic DNA was extracted from fresh foliage by QIAGEN Plant Mini Kit. The amount of DNA was directly quantified with DynaQuantTM 200 (HOEFER Phamacia Biotech Inc.). The total volume of reaction mixtures for PCR amplification was 20 μL, which contained 5 ng of template DNA, 0·2 mm of each dNTP, 0·0025 % of BSA (Boehringer Manheim, Germany), 520 μL of 1·5 μm primers (UBC, Canada), 1·2 μL of 25 mm MgCl2 and 0·6 units of Taq DNA polymerase (Advanced Biotechnique, UK). Amplifications were performed in a PTC-200 thermocycler (MJ Research, USA) using a period of 5 min for one cycle for initial denaturation at 94 °C, followed by 45 cycles of 30 s of denaturation at 94 °C, 30 s annealing at 50 °C (for UBC #807 and #810) and 52 °C (for UBC #808, #824, #829 and #830), 1 min of extension at 72 °C, and a final extension step of 10 min at 72 °C. Amplification products of PCRs were fractioned on 1·5 % agarose gel in 1× TBE (Tris–boric acid–ethylendiamine tetraacetic acid) buffer at pH 8·0, stained with ethidium bromide, and photographed over a UV transilluminator. DNA size was calculated by comparing samples with a 100-bp DNA ladder (GIPCO BRL, USA) which was loaded in two separate lanes on the same gel for electrophoresis. To score ISSR fragments, it was assumed that each marker fragment or band represented the phenotype at a single biallelic distinct locus (Williams et al., 1990; Dawson et al., 1995). Smeared and weak bands of ISSRs were excluded. The majority of plant-primer combinations were run more than once to ensure reproducibility. ISSR fragments were scored as presence (1) and absence (0) of putative homologous bands, and were then transformed into a binary matrix.
Table 1.
Inter-simple sequence repeat (ISSR) primers used in this study
| Primer no. | Sequence |
|---|---|
| 807 | AGAGAGAGAGAGAGAGT |
| 808 | AGAGAGAGAGAGAGAGC |
| 810 | GAGAGAGAGAGAGAGAT |
| 824 | TCTCTCTCTCTCTCTCG |
| 829 | TGTGTGTGTGTGTGTGC |
| 830 | TGTGTGTGTGTGTGTGG |
Data analysis
Clonal structure
Since E. koreensis reproduces both sexually and vegetatively, it is important to determine whether shoots with identical marker genotypes are clones (Berg and Hamrick, 1994; Chung and Epperson, 1999; Chung et al., 2004; Torimaru and Tomaru, 2005). To carry out these analyses, the available genetic markers must have enough statistical power to discriminate clonal genotypes from identical sexually produced genotypes. In order to determine the probability (PG) that two random, sexually produced multilocus genotypes will be identical under the assumption of random mating, the following calculation was performed (e.g. Sydes and Peakall, 1998; Rossetto et al., 2004): PG = Π pi, where pi is the frequency of presence or absence of each band i in the multilocus dominant ISSR genotype. The discriminating power of the markers used in the present study was measured for each population as 1 – PG.. Since power was high and similar for both populations (1 – PG ≈ 1·0, Table 2), identical multilocus genotypes were considered as clones. Hereafter, reference is made to subscripts (r) and (g) for total samples [N(r)] and samples restricted to genets [N(g)], in order to distinguish between total samples and samples excluding clones for analysing levels of genetic diversity and FSGS.
Table 2.
Summary of clonal and genetic diversity estimates of two populations of Echinosophora koreensis
| No. of genets per population |
||
|---|---|---|
| No. of ramets per genet | HAN | YIM |
| 1 | 21 | 21 |
| 2 | 17 | 11 |
| 3 | 8 | 9 |
| 4 | 4 | 6 |
| 5 | 3 | 1 |
| 6 | 3 | 1 |
| 7 | 2 | 0 |
| 8 | 1 | 1 |
| 10 | 0 | 1 |
| 13 | 0 | 1 |
| N(r) | 150 | 136 |
| N(g) | 59 | 52 |
| PG | 3·16 × 10−18 | 4·76 × 10−18 |
| %P(r) | 80·5 % | 87·8 % |
| %P(g) | 80·5 % | 87·8 % |
| h(r) | 0·181 | 0·226 |
| h(g) | 0·164 | 0·208 |
N(r), number of ramets (shoots); N(g), number of genets (genotypes); PG, probability that two random, sexually produced multilocus genotypes will be identical; %P, percentage of polymorphic loci; h, genetic diversity. Subscripts (r) and (g) refer to total samples (including clonal ramets) and samples restricted to genets, respectively.
Join-count statistic
It is expected that putative clonal genotypes would be spatially clustered as expected for spreading via rhizomes. To test this hypohesis, spatial autocorrelation statistics (Sokal and Oden, 1978) were calculated for the total number of ‘unlike’ joins among multilocus genotypes (e.g. Chung and Epperson, 1999; Chung et al., 2004). Standard normal deviates (SND) were calculated using the program JCSP (B. K. Epperson, Michigan State University, East Lansing). An SND has an asymptotically standard normal distribution under the null hypothesis of random dispersion. An SND < −1·96 indicates a significant (P < 0·05) deficit of pairs of unlike (and excess of like joins) genotypes separated by a given range of Euclidean distances (Epperson, 1993). Hence, significant negative values at short distance intervals are indicative of clonal structure.
Spatial distribution of individuals
To assess the spatial distribution of individuals, Ripley's L(d)-statistics (Ripley, 1976, 1977) were used. Ripley's L(d) is calculated from the number of point pairs within concentric circles of increasing radii (d) around each plant. Since the use of circles with a radius greater than half the shortest plot side introduces excessive bias due to edge effects, radial distances of 1–15 m with 1-m lag were selected for each population (e.g. Parker et al., 1997; Burke et al., 2000; Cruse-Sanders and Hamrick, 2004; Ng et al., 2004). Values of L(d) = 0, L(d) > 0 and L(d) < 0 indicate spatial randomness, spatial clustering and spatial repulsion (hyperdispersal), respectively, up to distance d. Confidence intervals (CIs; 95 %) about the null hypothesis of spatial randomness were estimated by Monte Carlo simulation (199 replicates) with a value of L(d) outside of this envelope judged to be a significant departure from the null hypothesis. A univariate analysis was also conducted for samples that excluded clonal ramets [N(g)]. In this latter case the x and y co-ordinates of each ramet were placed at the genet's centre of mass. All calculations and simulations were performed using a program developed by P. Aldrich (Smithsonian Institution, National Museum of Natural History, USA) and E. Berg (Kenai National Wildlife Refuge, USA).
ISSR diversity
To estimate levels of genetic diversity, the percentage of polymorphic loci (P) was calculated. In order to estimate Nei's genetic diversity (h) in populations (Nei, 1973), an allozyme analysis was conducted on the same populations and inbreeding coefficients (FIS = 0·090 and 0·120 at HAN and YIM, respectively: M. Y. Chung and M. G. Chung, unpubl. res.) were calculated. The estimates of h were calculated on the basis of FIS values using the program POPGENE version 1·31(Yeh et al., 1999) for total samples and samples excluding clones. The program GenAlEx (Peakall and Smouse, 2001) was used to assess the partitioning of the genetic variance within and between populations by an analysis of molecular variance (AMOVA; Excoffier et al., 1992) in which analogues of F-statistics (so-called Φ statistics) are extracted. Estimation of these genetic parameters was based both on total samples and samples excluding clones.
Fine-scale genetic structure
To quantify the effects of clonal structure on the FSGS within E. koreensis populations, FSGS were analysed for both total samples and samples excluding clonal ramets. The program SPAGeDi (Hardy and Vekemans, 2002) was used to analyse FSGS, since (a) this program provides 95 % CIs of the slope (blog) of each correlogram and jackknifed means of blog of correlograms and their standard errors across loci, and (b) it allowed FSGS on the binary dominant data set (presence or absence of a certain fragment) to be analysed. The analysis of the pairwise kinship coefficients Fij, (Loiselle et al., 1995; Kalisz et al., 2001) with dominant markers in diploids was outlined in Hardy (2003). The estimation of Fij with ISSR markers (i.e. dominant markers) requires that FIS is known (Hardy, 2003). Therefore, FIS estimated on the basis of allozyme markers described above was used.
To visualize FSGS, values of Fij located within the distance classes (2-m intervals) were averaged and plotted against the distance. To assess statistical significance of the average Fij, each Fij value was compared with 95 % CIs generated under the null hypothesis of no spatial genetic structure (Fij = 0). Sample multilocus binary data were drawn at random with replacement and assigned to occupied map locations within the study population. Re-sampling was repeated 999 times, and the observed Fij values represented the 1000th statistic, for each distance class. The 95 % CIs were constructed as the interval from the 25th to the 976th ordered permutation estimates. Values of Fij that fall above or below the 95 % bootstrapped CIs are interpreted as showing significantly greater or lesser genetic structure (P < 0·05), respectively, than expected at random.
Under an isolation-by-distance model, Rousset (1997, 2000) showed that the probability of identity in state between two neutral genes decreases approximately linearly with the logarithm of spatial distance in two dimensions at a rate proportional to 1/Dσ2, where D is the effective population density and σ2 is half the average squared axial parent–offspring distance. Similarly, pairwise kinship coefficients, F(rij), between individuals i and j separated by distance r decrease linearly with the logarithm of distance r in a two-dimensional space (Vekemans and Hardy, 2004). Following the Vekemans and Hardy's method, F(rij) values were regressed on the logarithm of spatial distance rij to obtain the regression slope (blog) for each data set (total samples and excluding clones in HAN and YIM, respectively). To test for FSGS (i.e. isolation by distance), each blog was evaluated using a Mantel test with 1000 permutations under null hypothesis (blog = 0). Since blog depends to some extent on sampling scale used and it is negative, the Sp statistic reflecting the rate of decrease of pairwise kinship with distance was estimated (Vekemans and Hardy, 2004). The statistic allowed within-population FSGS patterns within the present study to be compared among each other and to patterns observed in other taxa (e.g. Alberto et al., 2005; reviewed in Vekemans and Hardy, 2004). The Sp statistic is equal to –blog/[1 – F(i=1)] (Vekemans and Hardy, 2004), where F(i=1) is the mean Fij at the smallest distance interval [hence F(i=1) = F(2 m)]. To determine whether a significant difference of the Sp values exists between four comparisons, t-tests were conducted, assuming that the distribution of the Sp values per each locus was approximately normal.
Finally, to test whether mean values of Fij at ≤2 m between total samples and samples excluding clones are significantly different each other, the approximation of a 95 % CI (±1·96 times the standard error) was used. All analyses, including estimation of jackknifed standard errors, were conducted using the program SPAGeDi.
RESULTS
Clonal structure
Given the available marker variation, the power to discriminate clonal genotypes from sexually produced genotypes identical by chance alone was 1 − PG ≈ 1·0 for each population (PG = 4·76 × 10−18 to 3·16 × 10−18; Table 2). Given this power, ramets sharing the same genotype were treated as putative clones, finding that within populations 85–86 % of genets formed clones consisting of two or more ramets (Table 2). Clones ranged in size from one to eight ramets at HAN and one to 13 ramets at YIM. However, the distribution of clone sizes was not significantly different between populations (contingency χ2-test: χ2 = 7·33, d.f. = 9, P = 0·603). Join-count statistics revealed a statistically significant deficit of joins between unlike multilocus genotypes (i.e. excesses of pairs of identical genotypes) compared with random distribution expectations, only at distances of 0 m < d < 2 m (standard normal deviate test statistic, SND = −51·3 for HAN and −38·9 for YIM) and for 2 m < d < 6 m (SND = −19·4 for HAN and −11·3 for YIM), but not at greater distances within the two populations (Fig. 1). Together, these results indicate the positive spatial clustering of identical multilocus genotypes, consistent with the expectations of clonal structure.
Fig. 1.
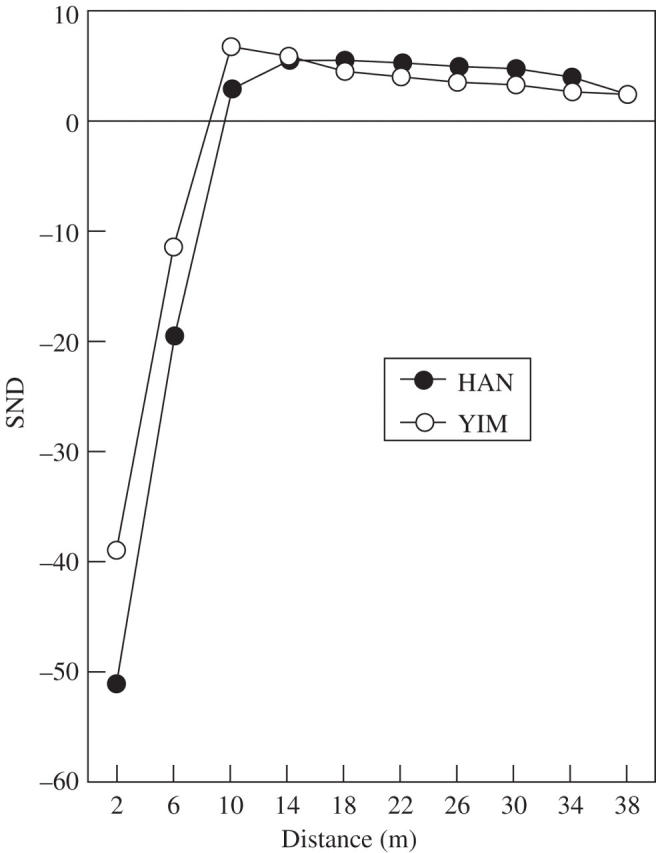
Correlograms for populations HAN and YIM of Echinosophora koreensis showing the relationships between pairs of the total number of unlike joins among multilocus genotypes (SND, standard normal deviate).
Spatial distribution of individuals
Visual inspection of the spatial arrangement of plants in HAN suggested a less clumped distribution, whereas a more clumped distribution of shoots was observed in YIM. Ripley's L(d)-statistics indicated a somewhat patchy distribution of shoots at <6 m and a hyperdispersal distribution beyond this distance to 15 m in HAN. However, when clones are excluded, a near random distribution of genets is evident at 2–13 m (Fig. 2). In contrast to HAN, significant aggregation both of total shoots and genets was found at YIM for all distance intervals (Fig. 2).
Fig. 2.
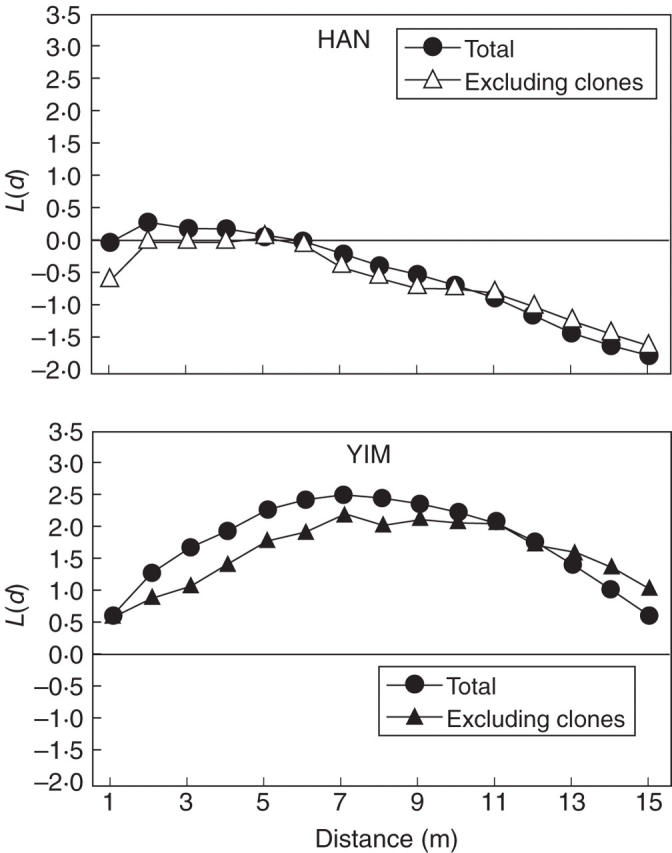
Results of Ripley's L(d)-statistics, observed L(d) estimates for univariate Ripley analysis in two populations (HAN and YIM) of Echinosophora koreensis for total samples (including clonal ramets; circles) and for sample genets (excluding clones; triangles). Estimates that depart significantly from the null hypothesis [L(d) = 0] are indicated by closed symbols.
ISSR diversity
Of the 41 putative ISSR loci examined, 36 were polymorphic (100 % criterion) across two populations (%P = 87·8 %). Genetic diversity calculated from both total population samples and samples excluding identical multilocus genotypes (and hence clones) was slightly higher for YIM [h(r) = 0·226 at YIM vs. 0·181 at HAN] but with similar estimates between total samples and those excluding clones (Table 2). AMOVA revealed moderate levels of genetic variability among individuals within populations and significant and high degrees of differentiation between populations (ΦST(r) = 0·226 for total samples, P < 0·001; ΦST(g) = 0·184 for samples excluding clones, P < 0·001).
Fine-scale genetic structure
Thirty-three (HAN) and 36 (YIM) loci were chosen for spatial autocorrelation analysis according to the criteria described in Materials and methods. Overall slopes (blog) of all four correlograms were significantly different from the null hypothesis of no spatial genetic structure (blog = 0): blog(r) of −0·114 (95 % CI = −0·005, 0·004) for total samples and blog(g) of −0·076 (95 % CI = −0·012, 0·010) for genets at HAN; blog(r) of −0·142 (95 % CI = −0·006, 0·004) for total samples and blog(g) of −0·080 (95 % CI = −0·012, 0·010) for genets at YIM (Fig. 3).
Fig. 3.
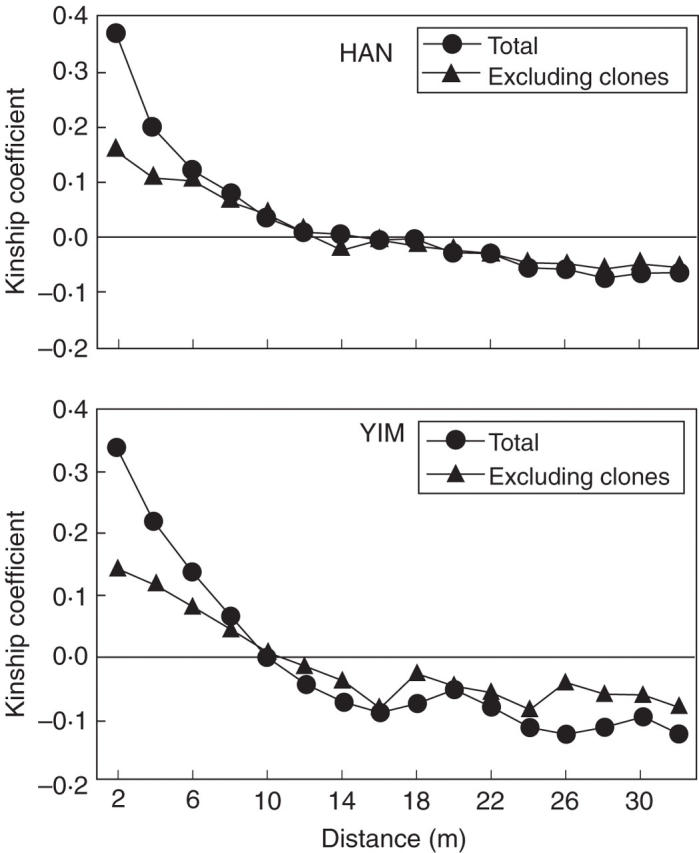
Correlograms of kinship coefficients (Fij) in two populations (HAN and YIM) of Echinosophora koreensis for total samples (including clonal ramets; circles) and for genets (excluding clones; triangles). Estimates that depart significantly from the null hypothesis (Fij = 0) are indicated by closed symbols.
Sp values revealed strong patterns of FSGS for total samples and they were very close between the two populations [Sp = 0·148 ± 0·040 (s.e.) at HAN; 0·154 ± 0·040 at YIM]. Values decreased about 2-fold were found in samples excluding clones (0·072 ± 0·033 at HAN; 0·093 ± 0·034 at YIM). These patterns, however, were not significantly different from each other.
In contrast, the magnitude of Fij at ≤2 m differed significantly between total samples and samples excluding clones at HAN (mean: 0·372, 95 % CI = 0·310, 0·437 for total; mean: 0·160, 95 % CI = 0·085, 0·237 for genets) and YIM (mean: 0·339, 95 % CI = 0·262, 0·424 for total; mean: 0·142, 95 % CI = 0·042, 0·241 for genets). Differences between populations for total samples and genets, however, were not significantly different at this spatial scale.
The distance at which the mean Fij value first intercepts the x co-ordinate may represent the length of a genetic patch size (Sokal and Wartenberg, 1983; Smouse and Peakall, 1999; Epperson, 2003; Escudero et al., 2003). Although x is sensitive to the spatial scale of sampling and is not a characteristic of populations (Fenster et al., 2003; Vekemans and Hardy, 2004), it can be used to compare similarly sampled populations. The average intercept was similar between total samples and samples excluding clones in both populations. The approximate patch size is 10–12 m (Fig. 3).
DISCUSSION
Levels of genetic diversity
Moderate levels of genetic diversity found within the total samples were similar to those found in samples excluding clones. This indicates that clones do not significantly affect levels of genetic diversity within populations of Echinosophora koreensis. Mean genetic diversity within populations was h(g) = 0·204, and the percentage of polymorphic loci was %P = 87·8 %. These values are higher than those found in other endemic species (Li and Ge, 2001; Ge et al., 2005; Sheng et al., 2005; Xie et al., 2005). This comparison suggests that the marginal populations of E. koreensis are not genetically depauperate. However, genetic differentiation between the two populations of E. koreensis was high for total samples [ΦST(r) = 0·226]. This differentiation was somewhat lower when considering only genets [ΦST(g)], with estimates decreasing to 0·184. This level of differentiation may reflect occasional pollen flow through bumblebees but restricted seed dispersal between populations separated by 2·8 km.
Spatial distribution and fine-scale genetic structure
Join-count statistics revealed that most clones aggregate significantly at ≤6-m interplant distance, which reflects the spatial distribution of ramets at short distances. Significant aggregation of both total shoots and genets was found at YIM. In contrast to this, HAN showed a near random distribution of genets but a slight aggregation of shoots at <6 m. This suggests different population histories and/or ecological conditions between the two sites. At YIM many distinct patches grow in gaps under high density of Pinus densiflora overstorey. At HAN, in contrast, only a few P. densiflora grew before a recent fire. The entire area surrounding and including HAN had been disturbed by domesticated animals (e.g. cows and goats) before designation of Natural Monument in 1980s. The relatively open habitat of HAN provides suitable light for E. koreensis individuals. Clones may, therefore, be more likely to proliferate at HAN.
Significant evidence of FSGS was detected in two populations both at the ramet and genet levels, and a similar pattern of correlograms between treatments (ramets vs. genets) and populations was found. The pronounced FSGS found in samples excluding clones (genets) of two populations (Sp = 0·072 and 0·093) suggests restricted dispersal of E. koreensis. The Sp values estimated here are within the range of those reported in predominantly selfing herbaceous species, particularly in leguminous species (Sp = 0·055–0·263; Zoro Bi et al., 1997; Bonnin et al., 2001; reviewed in Vekemans and Hardy, 2004). The finding that HAN showed a near random distribution of genets but a significantly positive autocorrelation of genotypes at <10 m may need explanation. Disturbance (e.g. fire) and open habitats at HAN may have encouraged colonization and establishment at the HAN site with the initially established genets spreading in various directions over <6 m distance. With a relatively high density of clonal ramets, many of these may have died randomly via thinning, resulting in a weak aggregation of shoots and a near random distribution of genets. Genotypes, however, still showed significant clumping in short-distance intervals as seen in a population of Pinus clausa in Florida, USA (Parker et al., 2001).
FSGS within populations is primarily determined by the effects of limited seed and pollen dispersal, isolation in small demographic patches, and microhabitat requirements (Wright, 1943; Linhart et al., 1981; Slatkin and Arter, 1991; Hamrick and Nason, 1996; McCauley, 1997; Parker et al., 2001; Chung et al., 2003). Among these factors, probably the most widely studied influence on patterns of FSGS is seed dispersal (Hamrick and Nason, 1996). If, at the scale of investigation, seed dispersal is localized, it will result in spatial clustering of genetically related individuals (full-sibs, half-sibs, first cousins, etc.) and will result in the development of significant FSGS (Wright, 1943; Sokal and Wartenberg, 1983; Barbujani, 1987). In E. koreensis no specialized seed or pod dispersal mechanism is known. Thus, limited seed dispersal may be a primary factor responsible for the observed spatial genetic structuring in this species. Nonetheless, the strong (magnitude of Fij at ≤ 2 m) FSGS observed in genets may also be related to limited pollen dispersal. The observed pollinators of E. koreensis in the HAN populations were bumblebees which exhibited a large proportion of short flight distances [2·54 ± 1·34 (s.d.) m, n = 54] within patches (M. Y. Chung and M. G. Chung, unpubl. res.), though bumblebees may travel between patches as well [a leptokurtic dispersal of pollen; Roubik (1989)]. The effect of the mating system on the level of genetic structure (i.e. FIS) will primarily influence between-individual kinship (Fij; Cockerham, 1969) values of the correlograms at the shortest distances, because at short distances Fij values are highly correlated with inbreeding (Crow and Kimura, 1970; Hardy and Vekemans, 1999). Under random mating, Fij between individuals is a measure of the inbreeding coefficient of their hypothetical offspring with expected values of 0·25 for full-sibs, 0·125 for half-sibs, and 0·0625 for first cousins (Crow and Kimura, 1970). Mean Fij(g) of 0·160 (HAN) and 0·142 (YIM) were estimated from the data set excluding clones at ≤2-m interplant distance, which corresponds to Fij values expected for half-sibs (Crow and Kimura, 1970). Considering this high level of relatedness, it is further suggested that, since clonal ramets aggregate at ≤6 m, the potential of selfing via geitonogamy over generations in E. koreensis might lead to the high Fij values observed among genets within a distance of 2 m.
To sum up, there were no statistically significant differences in the Sp values of the four treatments (total samples and excluding clones from each of two populations), although approx. 2-fold stronger patterns were detected in total samples. In addition, similar x-intercepts were also found when comparing total samples and samples excluding clones. Thus, clones appear to have little effect on patch size as inferred from the correlogram intercepts. However, at ≤2-m interplant distance, the total samples exhibited significantly higher Fij values than population samples excluding clones, primarily due to the aggregation of clonal ramets at ≤6 m distance. This indicates that clones themselves enhance the magnitude but not patterns of spatial autocorrelation. In terms of evolutionary processes, the strong pattern of FSGS found in genets of E. koreensis would be due to limited pollen and seed dispersal, probably coupled with clonal effects on breeding structure.
Implications for conservation
Although local populations of E. koreensis currently maintain moderate levels of genetic diversity within populations, the two populations are at risk due to illegal collectors and high human frequentation. These activities could decrease N(g) in the two studied sites. Furthermore, only 45 flowering shoots were located in another isolated population in Wolmyeong-ri (M.Y.C. and M.G.C., personal observation), suggesting that N(g) in this population is small, and random genetic drift could be significant. Long-term genetic variability of E. koreensis can only be ensured by implementing appropriate conservation and management strategies. The distribution of genetic variation in space is the prime factor to take into account in the conservation and management of natural plant populations (McCue et al., 1996). For example, sampling methods for seed stocks and decisions on the size of an area for population preservation may greatly benefit from this information. The estimation of the average intercept across alleles has important implications for ex situ and in situ conservation. Based on the correlogram intercepts of average Fij in local populations of E. koreensis, individuals closer than 10–12 m tend to be quite similar genetically, representing ‘pseudoreplication’ in terms of genetic variability. To optimize sampling design and avoid pseudoreplication, only individuals at greater distances should be collected (Diniz-Filho and Telles, 2002), and they should be sampled from each location since the two populations are significantly differentiated.
Acknowledgments
The authors thank J. Stireman, J. López-Pujol and E. R. Myers for making helpful suggestions, J. D. Nason for discussion of the Sp statistic, and B. K. Epperson for providing the program JCSP.
LITERATURE CITED
- Alberto F, Gouveia, L, Arnaud-Haond S, Pérez-Lloréns JL, Duarte CM, Serrão EA. 2005. Within-population spatial genetic structure, neighbourhood size and clonal subrange in the seagrass Cymodocea nodosa. Molecular Ecology 14: 2669–2681. [DOI] [PubMed] [Google Scholar]
- Barbujani G. 1987. Autocorrelation of gene frequencies under isolation by distance. Genetics 117: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg EE, Hamrick JL. 1994. Spatial and genetic structure of two sandhills oaks: Quercus laevis and Quercus margaretta (Fagaceae). American Journal of Botany 81: 7–14. [Google Scholar]
- Bonnin I, Ronfort J, Wonzniak F, Olivieri I. 2001. Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae). Molecular Ecology 10: 1371–1384. [DOI] [PubMed] [Google Scholar]
- Burke JM, Bulger MR, Wesselingh RA, Arnold ML. 2000. Frequency and spatial patterning of clonal reproduction in Louisiana iris hybrid populations. Evolution 54: 137–144. [DOI] [PubMed] [Google Scholar]
- Chung MG, Epperson BK. 1999. Spatial genetic structure of clonal and sexual reproduction in populations of Adenophora grandiflora (Campanulaceae). Evolution 53: 1068–1078. [DOI] [PubMed] [Google Scholar]
- Chung MY, Epperson BK, Chung MG. 2003. Genetic structure of age classes in Camellia japonica (Theaceae). Evolution 57: 62–73. [DOI] [PubMed] [Google Scholar]
- Chung MY, Nason JD, Chung MG. 2004. Implication of clonal structure for effective population size and genetic drift in a rare terrestrial orchid, Cremastra appendiculata. Conservation Biology 18: 1515–1524. [Google Scholar]
- Chung MY, Nason JD, Chung MG. 2005a. Patterns of hybridization and population genetic structure in the terrestrial orchids Liparis kumokiri and Liparis makinoana (Orchidaceae) in sympatric populations. Molecular Ecology 14: 4389–4402. [DOI] [PubMed] [Google Scholar]
- Chung MY, Suh Y, López-Pujol J, Nason JD, Chung MG. 2005b.. Clonal and fine-scale genetic structure in populations of a restricted Korean endemic, Hosta jonesii (Liliaceae) and its implications for conservation. Annals of Botany 96: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham CC. 1969. Variance in gene frequencies. Evolution 23: 72–84. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. 1970. An introduction to population genetic theory. New York, NY: Harper and Row.
- Cruse-Sanders JM, Hamrick JL. 2004. Spatial and genetic structure within populations of wild American ginseng (Panax quinquefolius L., Araliaceae). Journal of Heredity 95: 309–321. [DOI] [PubMed] [Google Scholar]
- Dawson IL, Simons AJ, Waugh R, Powell W. 1995. Diversity and genetic differentiation among subpopulations of Gliricidia sepium revealed by PCR-based assays. Heredity 74: 10–18. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, Telles MPC. 2002. Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conservation Biology 16: 924–935. [Google Scholar]
- Eckert CG, Barrett SCH. 1993. Clonal reproduction and patterns of genotypic diversity in Decodon verticillatus (Lythraceae). American Journal of Botany 80: 1175–1182. [Google Scholar]
- Epperson BK. 1990. Spatial patterns of genetic variation within plant populations. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding, and genetic resources. Sunderland, MA: Sinauer, 229–253.
- Epperson BK. 1993. Spatial and space–time correlations in systems of subpopulations with genetic drift and migration. Genetics 133: 711–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson BK. 2003. Geographical genetics. Princeton, NJ: Princeton University Press.
- Escudero A, Iriondo JM, Torres ME. 2003. Spatial analysis of genetic diversity as a tool for plant conservation. Biological Conservation 113: 351–365. [Google Scholar]
- Esselman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD. 1999. Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Molecular Ecology 8: 443–451. [Google Scholar]
- Excoffier L, Smouse PE, Quattro M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Vekemans X, Hardy OJ. 2003. Quantifying gene flow from spatial genetic structure data in a metapopulation of Chamaecrista fasciculate (Leguminosae). Evolution 57: 995–1007. [DOI] [PubMed] [Google Scholar]
- Gärdenfors U, Hilton-Taylor C, Mace GM, Rodríguez JP. 2001. The application of IUCN Red List criteria at regional levels. Conservation Biology 15: 1206–1212. [Google Scholar]
- Ge X-J, Yu Y, Yuan Y-M, Huang H-W, Yan C. 2005. Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of north-west China as revealed by ISSR analysis. Annals of Botany 95: 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerli A, Reusch TBH. 2003. Inbreeding depression influences genet size distribution in a marine angiosperm. Molecular Ecology 12: 619–629. [DOI] [PubMed] [Google Scholar]
- Hamrick JL, Nason JD. 1996. Consequences of dispersal in plants. In: Rhodes Jr OE, Chesser RK, Smith MH, eds. Population dynamics in ecological space and time. Chicago, IL: University of Chicago Press, 203–236.
- Handel SN. 1985. The intrusion of clonal growth patterns on plant breeding systems. American Naturalist 125: 367–384. [Google Scholar]
- Hardy O. 2003. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Molecular Ecology 12: 1577–1588. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. 1999. Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity 83: 145–154. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618. [Google Scholar]
- Heywood JS. 1991. Spatial analysis of genetic variation in plant populations. Annual Review of Ecology and Systematics 22: 335–355. [Google Scholar]
- IUCN. 2001. IUCN red list categories: version 3.1. Prepared by the IUCN Species Survival Commission. IUCN, Gland, Switzerland and Cambridge, UK (available at http://www.iucn.org/themes/ssc).
- Kalisz S, Nason JD, Hanzawa FA, Tonsor SJ. 2001. Spatial genetic structure in Trillium grandiflorum: the roles of dispersal, mating, history and selection. Evolution 55: 1560–1568. [DOI] [PubMed] [Google Scholar]
- Li A, Ge S. 2001. Genetic variation and clonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers. Annals of Botany 87: 585–590. [Google Scholar]
- Linhart YB, Mitton JB, Sturgeon KB, Davis ML. 1981. Genetic variation in space and time in population of ponderosa pine. Heredity 46: 407–426. [Google Scholar]
- Loiselle BA, Sork VL, Nason JD, Graham C. 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82: 1420–1425. [Google Scholar]
- Lynch AJJ, Barnes RW, Cambecèdes J, Vaillancourt RE. 1998. Genetic evidence that Lomatia tasmanica (Proteaceae) is an ancient clone. Australian Journal of Botany 46: 25–33. [Google Scholar]
- McCauley DE. 1997. The relative contribution of seed and pollen movement to the local genetic structure of Silene alba. Journal of Heredity 88: 257–263. [Google Scholar]
- McCue KA, Buckler ES, Holtsford TP. 1996. A hierarchical view of genetic structure in the rare annual plant Clarkia springvillensis. Conservation Biology 10: 1425–1434. [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided population. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KKS, Lee SL, Koh CL. 2004. Spatial structure and genetic diversity of the two tropical tree species with contrasting breeding systems and different ploidy levels. Molecular Ecology 13: 657–669. [DOI] [PubMed] [Google Scholar]
- Park M-K. 1973. Keys to the herbaceous plants in Korea (Dicotyledoneae). Seoul: Jeongumsa [in Korean].
- Parker KC, Parker AJ, Beaty RM, Fuller MM. 1997. Population structure and spatial pattern of two coastal populations of Ocala sand pine (Pinus clausa (Chapm. ex Engelm.) Vasey ex Sarg. var. clausa D. B. Ward). Journal of the Torrey Botanical Society 124: 22–33. [Google Scholar]
- Parker KC, Hamrick JL, Parker AJ, Nason JD. 2001. Fine-scale genetic structure in Pinus clausa (Pinaceae) populations: effects of disturbance history. Heredity 87: 99–113. [DOI] [PubMed] [Google Scholar]
- Peakall R, Beattie AJ. 1991. Genetic consequences of worker ant pollination in a self-compatible, clonal orchid. Evolution 45: 1837–1848. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse P. 2001. GenAlEx V5: Genetic Analysis in Excel. Population genetic software for teaching and research. Canberra: Australian National University. [DOI] [PMC free article] [PubMed]
- Reusch TBH, Hukriede W, Stam WT, Olsen J. 1999. Differentiating between clonal growth and limited gene flow using spatial autocorrelation of microsatellites. Heredity 83: 120–126. [DOI] [PubMed] [Google Scholar]
- Ripley BD. 1976. The second-order analysis of stationary point processes. Journal of Applied Probability 13: 965–981. [Google Scholar]
- Ripley BD. 1977. Modelling spatial patterns. Journal of the Royal Statistical Society, Series B 39: 172–192. [Google Scholar]
- Rossetto M, Gross CL, Jones R, Hunter J. 2004. The impact of clonality on an endangered tree (Elaeocarpus williamsianus) in a fragmented rainforest. Biological Conservation 117: 33–39. [Google Scholar]
- Roubik DW. 1989. Ecology and natural history of tropical bees. Cambridge: Cambridge University Press. [DOI] [PubMed]
- Rousset F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. 2000. Genetic differentiation between individuals. Journal of Evolutionary Biology 13: 58–62. [Google Scholar]
- Sheng Y, Zheng W, Pei K, Ma K. 2005. Genetic variation within and among populations of a dominant desert tree Haloxylon ammodendron (Amaranthaceae) in China. Annals of Botany 96: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Arter HE. 1991. Spatial autocorrelation methods in population genetics. American Naturalist 138: 499–517. [Google Scholar]
- Smouse PE, Peakall R. 1999. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82: 561–573. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Oden NL. 1978. Spatial autocorrelation in biology. I. Methodology. Biological Journal of the Linnean Society 10: 199–228. [Google Scholar]
- Sokal RR, Wartenberg DE. 1983. A test of spatial autocorrelation analysis using an isolation-by-distance model. Genetics 105: 219–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki JI, Herben T, Krahulec F, Hara T. 1999. Size and spatial pattern of Festuca rubra genets in a mountain grassland: its relevance to genet establishment and dynamics. Journal of Ecology 87: 942–954. [Google Scholar]
- Sydes M, Peakall R. 1998. Extensive clonality in the endangered shrub Haloragodendron lucasii (Haloragaceae) revealed by allozyme and RAPDs. Molecular Ecology 7: 87–93. [Google Scholar]
- Torimaru T, Tomaru N. 2005. Fine-scale clonal structure and diversity within patches of a clone-forming dioecious shrub, Ilex leucoclada (Aquifoliaceae). Annals of Botany 95: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans X, Hardy O. 2004. New insights from fine-scale structure analyses in plant populations. Molecular Ecology 12: 921–935. [DOI] [PubMed] [Google Scholar]
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research 18: 6531–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Liston A. 1998. Contributions of the polymerase chain reaction to plant systematics. In: Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants. II. DNA sequencing. New York: Kluwer, 203–236.
- Wright S. 1943. Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1969. Evolution and genetics of populations. Vol. 2. The theory of gene frequencies. Chicago, IL: University of Chicago Press.
- Xie G-W, Wang D-L, Yuan Y-M, Ge X-J. 2005. Population genetic structure of Monimopetalum chinense (Celastraceae), an endangered endemic species of eastern China. Annals of Botany 95: 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Boyle T, Yang RC, Ye Z, Xiyan JM. 1999. POPGENE, the user friendly shareware for population genetic analysis, version 1.31. University of Alberta and Centre for International Forestry Research.
- Yim K.-B. 1993. Botanical treasures (natural monuments) of Korea. Seoul: Daewonsa [in Korean].
- Zietkiewicz E, Rafalski A, Labuda D. 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20: 176–183. [DOI] [PubMed] [Google Scholar]
- Zoro Bi I, Maquet A, Baudoin J-P. 1997. Spatial patterns of allozyme variants within three wild populations of Phaseolus lunatus L. from the central valley of Costa Rica. Belgian Journal of Botany 129: 149–155. [Google Scholar]


