Abstract
• Background and Aims Intraspecific genome size variability of Festuca pallens occurring on relict rocky steppes in Central Europe was studied on two ploidy levels and three geographical scales: (1) local scale of 24 populations, (2) landscape scale of three transects in river canyons or hill systems, and (3) global scale of 160 samples covering the whole distribution area.
• Methods DAPI flow cytometry of homogeneously cultivated samples (≥1 year), measured randomly with two internal standards, Lycopersicon esculentum and Pisum sativum. Differences in DNA content were confirmed (1) by the double peaks of simultaneously measured samples, (2) based on measurements carried out in different seasons, and (3) by additional measurements with propidium iodide.
• Key Results On a global scale, the relative DNA content ranged between 1·170-fold in diploids and 1·164-fold in tetraploids. A maximum difference of 1·088-fold between the mean relative DNA content of nearby populations was found. In 16 of 24 populations significant variability was shown (P < 0·001, 1·121-fold as maximum). For both ploidy levels, the relative genome size had the same range and geographical pattern, correlated with geographical coordinates (P < 0·01). Diploids with larger genomes occur on relict habitats (P < 0·01), and in areas of periglacial steppes (20 000 years ago; P < 0·02). In tetraploids, the relative DNA content differs among the three previously recognized geographical types (Alpine, Pannonian and Scabrifolia, P < 0·001). Tetraploids have a relative DNA content smaller than twice that of the diploids (P < 0·001). An influence of microhabitat on DNA content variation was not confirmed.
• Conclusions Genome size variability occurs over all spatial scales: intrapopulation, landscape and global. Correlation between geographical coordinates and palaeovegetation type, concomitant with diploids and tetraploids, and no influence of microhabitat were found. Genome size decreases in tetraploids. Lower CVs, and thus higher accuracy, resolution and reproducibility, favour DAPI measurements for the study of intraspecific genome size variability.
Keywords: Festuca subgen. Festuca, fescue, Gramineae, polyploidy, Poaceae, genome size, flow cytometry, Central Europe, infraspecific C-value variation, DAPI, PI, DAPI/PI correlation
INTRODUCTION
Festuca pallens is a narrow-leaved fescue of the type section (Festuca sect. Festuca, Poeae, Poaceae). Close relatives are found in the Mediterranean region and the mountains of south and south-west Europe, the assumed evolution centre of the group.
Festuca pallens is an allogamic, wind-pollinated species (Auquier, 1977), and is well separated morphologically from other Central European fescues. Setaceous tiller leaves are 0·6–1·3 mm in diameter, without remarkable ligulas and are usually apparently pruinose. The sub-epidermal sclerenchyma on a tiller leaf cross-section forms a complete to partly interrupted sclerenchyma ring. Two other Central European relatives (F. psammophila and F. vaginata) differ in awns usually being shorter than 1·1 mm.
Geographically, Festuca pallens is restricted to Central Europe, from France and Belgium to the Alps and Hercynian Mountains to the Carpathians and the Transylvanian Highlands of Romania (see Fig. 6). It grows on rocky steppes, open rocky outcrops, rocks, cliffs and promontories, with no obvious bedrock type preference, typically on slopes of river canyons, valleys and gorges in rocky highlands, karst areas and lower mountains from 100 to 1500 m a.s.l. (related F. psammophila and F. vaginata are psamophilous). Most petrophilous species frequently occurring together with F. pallens have a largely southern European distribution range that overlaps in Central Europe (Seseli osseum, Sedum album, Alyssum saxatile, A. montanum, Minuartia setacea, Galium glaucum, Sedum reflexum, Lactuca perennis, Fumana procumbens, Helianthemum canum, Dianthus gratianopolitanus). These rocky habitats were naturally treeless areas during glacial and interglacial periods of the last ice age. Their relict character is also evidenced by the frequent co-occurrence with many glacial relicts (Saxifraga paniculata, Primula auricula, Biscutella laevigata, Sesleria albicans, S. rigida). There is also a large group of endemic or relict species that entirely prefer habitats occupied by Festuca pallens: Daphne arbuscula (a Tertiary relict and endemic of the Muráňská planina Mountains of Slovakia), Festuca tatrae (a Tertiary relict, west Carpathian endemic), Ferrula sadleriana (a periglacial relict and Pannonian endemic), Dianthus moravicus (endemic to the rocky slopes of river canyons in the south-east of the Czech Republic), Helictotrichon decorum (endemic to the Romanian Carpathians), Seseli leucospermum (Pannonian endemic) and Viola jooi (endemic to the South Carpathians in Romania, Ukraine and Moldavia). In northernmost, isolated localities in the Central Europe, several petrophilous species of wider ecological niches obviously prefer F. pallens habitats, e.g. Stipa eriocaulis (Pálava Hills, Czech Republic), Arenaria grandiflora (Pálava Hills) and Notholaena marantae (Mohelno, Czech Republic). Besides its natural habitats, Festuca pallens occasionally invades quarries and secondary rocky sites along railways and roads.
Festuca pallens shows two ploidy levels (Pils, 1981; Šmarda and Kočí, 2003; Šmarda et al., 2005), which can be distinguished on the basis of several minute morphological and anatomical characters (leaf roughness, sclerenchyma structure; Šmarda, 2001). Diploids (2n = 14) are dispersed within the whole distribution area. Tetraploids (2n = 28) are restricted to several geographical regions and can be used to divide the population into three geographical and morphological types (Tracey, 1980; Pils, 1981; Šmarda and Kočí, 2003): (1) Alpine type occurring in the Eastern Alps in the area around Graz, Austria; (2) Pannonian type, growing in the calcareous hills of the Pannonian Lowland; and (3) Scabrifolia type, scattered in the hills of the Hercynian region and the Rhineland (Czech Republic, Germany). These types lack any taxonomical evaluation and are usually used independently on the subspecies of Festuca pallens in Flora Europaea. These subspecies, as recognized by Markgraf-Dannenberg (1980), seem to be of an ambiguous taxonomic value (Šmarda and Kočí, 2003; Šmarda et al., unpubl. data). Rarely, mixed ploidy populations, hybrids and triploid plants also occur (Šmarda and Kočí, 2003; Šmarda et al., 2005).
The history of the study of intraspecific genome size variability covers the last 40 years, since the first report of Evans et al. (1966). Despite this, the crucial question of whether variability really exists independently of chromosomal mutations remains controversial. Although genome size differences among related species with the same chromosome number are widely accepted as one of a species' evolutionary attributes, analogous variability within a species is often queried. This skepticism follows from: (1) instrumental or methodical errors of some authors reviewed in the critical reassessments of Greilhuber (1988, 1997, 1998, 2005), Greilhuber and Ebert (1994), Greilhuber and Obermayer (1997, 1998), Obermayer and Greilhuber (1999, 2005) and Temsch and Greilhuber (2000); (2) differences in the content of cytosolic compounds and seasonal intraspecific changes in the content of such secondary metabolites influenced the intensity of nuclear fluorescence (Price et al., 2000; Noirot et al., 2005); and (3) measurements from different laboratories are not completely comparable (Doležel et al., 1998). A comprehensive critical overview of opinions of intraspecific genome size variability was provided by Greilhuber (2005); the reader is referred also to Bennett (1985), Price (1988) and Greilhuber (1998).
Ecological and geographical differentiation of genome size has been documented either on interspecific or intraspecific levels (cf. overview in Knight et al., 2005). Intraspecific genome size variability may also indicate microevolutionary differentiation (Murray, 2005). Considerable intraspecific and intrapopulation variability was documented recently in several Festuca species from several populations in Romania (Šmarda, 2006). Considerable variation was found especially in Festuca pallens, where samples from two distant populations were found to differ maximally by 1·092-fold in relative DNA content (Šmarda, 2006). For an understanding of the causes of such intraspecific genome size variability, a comparison of patterns over various geographical scales (including the whole species distribution area) is required.
Among the recent methods of DNA content estimation, the most precise results are routinely produced with flow cytometry (Doležel and Bartoš, 2005). Flow cytometry with DAPI staining has enabled the detection of a 1·04-fold difference between samples as a clear double peak in simultaneous measurements (Doležel and Göhde, 1995).
The following key questions are herein. (1) What range and pattern of relative DNA content variability occurs in Festuca pallens on three geographical scales: local (within the population), landscape (in transects through isolated canyons or hill systems) and global (within the whole distribution area)? (2) Is there any microhabitat, ecogeographical or historical interpretation of relative DNA content variation on these scales? (3) Is there a geographically based relationship between relative genome sizes of diploids and tetraploids? (4) Is the relative DNA content of tetraploids twice that of their diploid progenitors? (5) Does the relative DNA content correlate with the delimitation of tetraploid geographical morphotypes (Alpine, Pannonian, Scabrifolia)? (6) Is the DAPI staining applicable to the study of intraspecific genome size as well as PI staining? (7) Are the DAPI measurements repeatable over long periods and in different seasons?
MATERIAL AND METHODS
Experimental material
On a global scale, 160 samples covering the whole distribution area of Festuca pallens Host were used. All three tetraploid geographical (morpho)types (26 samples of the Pannonian type, 25 of the Scabrifolia type and 11 of the Alpine type) and ploidy levels (94 diploids, three triploids, 62 tetraploids and one pentaploid) were included in these samples. Before measurements were taken, all samples were cultivated under standard homogeneous conditions of the experimental garden in the Faculty of Education, Masaryk University, Brno, for 1 year or more. Voucher specimens are deposited in the herbarium of the Masaryk University in Brno (BRNU). The chromosome number of 41 samples studied (29 diploids, two triploids, ten tetraploids) had been documented by Šmarda and Kočí (2003).
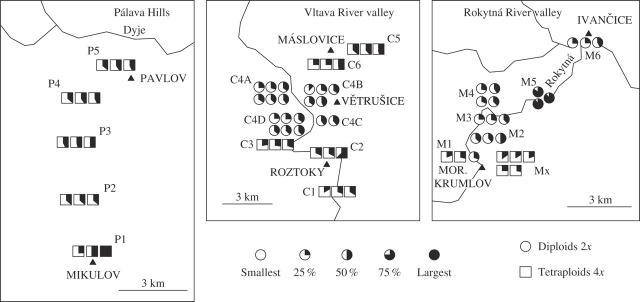
On a landscape scale, three transects (73 plants collected 6–9 October 2004) were analysed: (1) Rokytná River Canyon (basic agglomerate and gneiss)—a 13-km-long transect including five diploid populations, one mixed population and one tetraploid population of the Pannonian type previously karyologically analysed (Šmarda and Kočí, 2003); (ii) Pálava Hills (Jurassic limestone)—a 9-km-long transect that included five populations of the tetraploid Pannonian type; and (iii) Vltava River Canyon (spilite and gneiss bedrock)—a 13-km-long transect that included five populations of the tetraploid Scabrifolia type and four populations of diploids. In each transect, at least five populations homogeneous at the ploidy level, and three (to six) plants per population were sampled. Within each population, samples were collected from the most contrasting microhabitats (see below under Statistical treatment and variables) or to cover major morphological variability.
On the local (intrapopulation) scale, 24 populations (88 samples) were analysed. A second set of 73 samples from 21 populations of three landscape transects was also taken, with 15 samples collected at the same time from an additional three isolated populations: (1) Skalní mlýn in the Moravian Karst (Devonian limestone, diploids); (2) Horky Nature Reserve east of Brno (agglomerate rocky slope, tetraploid Pannonian type); and (3) Prokopské údolí valley south-west of Prague (Devonian limestone bedrock, tetraploid Scabrifolia type). Five plants per population were sampled using the strategy used for analysis on the landscape scale. For details of sample localities see Table 1 and Supplementary information at http://www.aob.oupjournals.org.
Table 1.
Population characteristics
| One-way ANOVA |
|||||||
|---|---|---|---|---|---|---|---|
| Locality | Code | Sample no. | Ploidy level | Sample/standard ratio (mean) | Maximum difference (fold) | F | P |
| Rokytná River* | |||||||
| M. Krumlov—plate | Mx | 5 | 4x | 3·017 | 1·026 | 41·88 | <0·001 |
| M. Krumlov—slopes† | M1 | 3 | 2x | 3·041 | 1·015 | 49·51 | <0·001 |
| M. Krumlov—Floriánek | M2 | 3 | 2x | 1·565 | 1·018 | 18·26 | <0·001 |
| Rokytná—Tábor | M3 | 3 | 2x | 1·543 | 1·022 | 49·79 | <0·001 |
| Rokytná—Slepen. skály | M4 | 4 | 2x | 1·551 | 1·024 | 59·88 | <0·001 |
| Budkovice | M5 | 3 | 2x | 1·670 | 1·011 | 1·95 | 0·177 |
| Ivančice | M6 | 3 | 2x | 1·535 | 1·032 | 26·62 | <0·001 |
| Pálava Hills* | |||||||
| Mikulov—Sv. Kopeček | P1 | 3 | 4x | 3·098 | 1·121 | 266·77 | <0·001 |
| Mikulov—Kočičí skála | P2 | 3 | 4x | 2·984 | 1·005 | 1·07 | 0·368 |
| Klentnice—Stolová hora | P3 | 3 | 4x | 3·044 | 1·008 | 2·31 | 0·134 |
| Pavlov—Děvín | P4 | 3 | 4x | 3·022 | 1·011 | 3·11 | 0·074 |
| Pavlov—Dívcí hrady | P5 | 3 | 4x | 3·024 | 1·010 | 6·07 | 0·012 |
| Vltava River* | |||||||
| Praha-Suchdol | C1 | 3 | 4x | 2·950 | 1·021 | 37·92 | <0·001 |
| Roztoky-Žalov | C2 | 3 | 4x | 3·012 | 1·051 | 30·53 | <0·001 |
| Podmoráň | C3 | 3 | 4x | 2·942 | 1·010 | 6·24 | 0·011 |
| Větrušice—North-1 | C4A | 6 | 2x | 1·542 | 1·022 | 9·12 | <0·001 |
| Větrušice—North-2 | C4B | 5 | 2x | 1·538 | 1·056 | 69·60 | <0·001 |
| Větrušice—Central-1 | C4C | 2 | 2x | 1·554 | 1·006 | 6·48 | 0·029 |
| Větrušice—Central-2 | C4D | 6 | 2x | 1·544 | 1·027 | 19·62 | <0·001 |
| Máslovice—East | C5 | 3 | 4x | 3·027 | 1·014 | 3·88 | 0·044 |
| Máslovice—West | C6 | 3 | 4x | 2·963 | 1·042 | 144·92 | <0·001 |
| Isolated populations | |||||||
| Bedřichovice—Horky | H | 5 | 4x | 2·914 | 1·021 | 15·90 | <0·001 |
| Adamov—Skalní mlýn | K | 5 | 2x | 1·554 | 1·018 | 15·52 | <0·001 |
| Praha—Prokopské údolí | S | 5 | 4x | 2·953 | 1·077 | 206·48 | <0·001 |
Transects
The only diploid sample from a mixed population was excluded from the analysis.
Flow cytometry
Relative DNA content was determined using DAPI-stained flow cytometry. Measurements were taken at the Institute of Botany and Zoology, Masaryk University, Brno, with a PA-I Partec ploidy analyser. A two-step procedure (Otto, 1990) was used. A sample of young, basal, just developing tiller leaf was chopped using a sharp razor blade together with two standards in a glass Petri dish containing 0·5 mL Otto I buffer (0·1 m citric acid, 0·5 % Tween 20). An additional 0·5 mL Otto I buffer was added. The crude nuclei suspension was filtered through a 50 µm nylon mesh. One millilitre of Otto II buffer (0·4 m Na2HPO4.12H2O) supplemented with 2 µg mL−1 4′,6-diamidino-2-phenylindole (DAPI) was then pipetted to the nuclei suspension. The youngest leaves of Lycopersicon esculentum ‘Stupické polní tyčkové rané’ and Pisum sativum ‘Ctirad’ were used simultaneously as internal standards. A total of 5000 cells were analysed in each measurement. Sample ratios to both internal standards were calculated. Consequently, the sample/Pisum ratio was converted to the sample/Lycopersicon ratio using the average of the Pisum/Lycopersicon ratio of 3·9807 (see Results). Relative DNA content was then calculated for each sample as the average of two values (one measurement series with two standards) on a global scale and six values (three measurement series with two standards) on a landscape and local scale, respectively.
The following measurements strategy was chosen to ensure maximum comparability of the results measured on different days, and to avoid the potential differences originating from the use of different individuals of standards. Both sample sets (global and local + landscape) were measured separately, in each with the only individuals of the standards. In both sample sets, the samples were measured in randomly generated order (random number generator in a Microsoft Excel spreadsheet). Each set was measured over 2 weeks in autumn 2004.
The relationship between DAPI and propidium iodide (PI) staining in the studied plants and observed differences were confirmed based on measurements taken in spring 2006, 17 months later. Twenty diploids and 20 tetraploids across the whole previously observed range of the relative DNA content were collected at the same day from the cultivation field and were measured simultaneously with DAPI and PI staining in the same laboratory using a PA-I Partec ploidy analyser (HBO lamp, DAPI) and Partec CyFlow (green laser 532 nm, 100 mW, PI). Sample preparation was similar to that described above with the following differences. (1) In step 1, twice the amounts of material (sample+standard Lycopersicon esculentum ‘Stupické polní tyčkové rané’) and Otto I buffer were used. (2) In step 2, suspension was divided into the two sample tubes to which 1 mL of Otto II buffer with DAPI or with PI + RNase was added, respectively. The final concentration of PI and RNase was 50 µg mL−1. Sample series of diploids and tetraploids were measured once, within 1 d, with the same individual of the standard.
Statistical treatment and variables
In the statistical analyses, two independent data sets were used (one for global scale samples; the other for landscape + local scale samples). Common descriptive statistics and statistical tests were calculated in the Microsoft Office Excel 2000, Statistica 7.1 and SPSS 8.0 programs. The relative range of DNA content was calculated as the highest/lowest DNA content ratio. Boxplots (see Figs 5, 7 and 8) show the median, interquartile range, outliers (circles) and extreme data points (stars).
On the global scale (within the whole distribution area), the data set was divided into diploid and tetraploid subsets. In both, the DNA content was tested with three groups of factors scored for each locality (see Table 4 and supplementary information). Distribution of DNA content was tested for normality relative to each factor using the Shapiro–Wilk normality test (P = 0·05). Differences or correlations among variables were consequently tested using (1) a t-test and (2) one-way ANOVA (both for data with normal distribution) or non-parametrically using (3) Spearman's rho correlation, and (4) Mann–Whitney test (see Table 4). Samples with unclear variable status were omitted from partial analysis. Eventual correlations were tested on the smaller subsets representing phytogeographical regions or tetraploid types.
On the local scale, relationships between DNA content and some microhabitat characters were tested within the populations. Only dramatically contrasting characters in particular populations were studied (e.g. plants from the top plateau versus the steep rocky face). For the six characters, (1) slope, (2) invasion of Robinia pseudacacia, (3) deviation of exposition from south-west, (4) level of exposure to the north, (5) altitudinal position on the slope, and (6) vegetation cover, samples in each population were categorized into minimum, intermediate or maximum categories. Only samples from the minimum and maximum categories (binary scored) were analysed. For the following two characters, (7) bare rock without vegetation cover and (8) ruderal habitat, the samples were binary scored in each population. The mean DNA content was calculated for samples in both extreme categories within each population. For all eight characters, differences in DNA content between the two binary scored categories were tested using the paired Wilcoxon non-parametric test. The diploid and tetraploid populations were analysed together.
Intrapopulation variability was tested using one-way ANOVA on the core data of the flow cytometric measurements as the ratio between inter- and intrasample variability (six sample/standard ratios per sample). The existence of intrapopulation variability in DNA content was tested on the core data of the flow cytometric measurements (six sample/standard ratios per sample).
The differences between relative genome sizes (monoploid relative DNA content) of different ploidy levels were tested using a t-test. The spatial relation of the relative genome size of tetraploids and diploids was tested with Procrustes analysis using PROTEST software (Procrustean randomization test; Peres-Neto and Jackson, 2000) with significance test from Jackson (1995) based on 9999 replications. Two matrices were compared. The first represented a matrix of the geographical distance of all diploids (93 samples = rows) from all possible combinations of tetraploids (62 samples = columns). The second represented the difference in the relative genome size of diploids and tetraploids (halved values) arranged in the same order as the previous matrix. The significant matching of matrices indicates that there is a geographical relationship of the genome size of diploids and tetraploids.
The DAPI/PI relationship and the reproducibility of the DAPI measurements were tested via linear regression. Forty DAPI measurements from spring 2006 were compared with those of the same samples from autumn 2004 using a paired t-test.
RESULTS
Accuracy and reproducibility of measurements with DAPI
The relative DNA content of the standard Lycopersicon esculentum ‘Stupické polní tyčkové rané’ was approximately 0·64-fold of the average diploid. The relative DNA content of the second standard, Pisum sativum ‘Ctirad’ was about 1·32 fold of the average tetraploid. None of the peaks of any of the samples or standards overlapped in any combination, either of the G0/G1 or of the G2 phase cells. The average ratio between standards was 3·9807 (264 measurements, CV 0·622 %); the average of all peak CVs of the standards was 1·703 %.
Measurements on local and landscape scales (88 samples) were repeated three times with both standards. For each of 88 samples, the CV was calculated from six sample/standard ratios. The average of these CVs was 0·545 %. The average CV of all 792 [(Lycopersicon + sample + Pisum) × 88 samples × 3 series] peaks in all measurements was 1·677 %.
For estimation of the measurement error of a single measurement, 264 (three series of 88 samples), those from the previous set were used. In each sample, the difference from the average of all three measurements was calculated. All 264 differences were ordered by size. Exactly 95 % of these differences were less than 1·010-fold. Therefore, at the same measurement accuracy, it was assumed that the following measurement of the same sample should not differ from the ideal mean value by more than ±1·010-fold at P < 0·05.
The measurement accuracy on the global scale was similar to the local and landscape scales (the average of all 480 peak CVs was 1.679 %). The use of a single repeated measurement in global scale is sensitive enough, regarding the measurement error of a single measurement (±1.010-fold) and the range of relative DNA content (1.121-fold at maximum) found in the previous measurements on local and landscape scales. The relative DNA contents of samples measured with DAPI in spring 2006 did not significantly differ from those measured in autumn 2004 (paired t-test, P > 0·05). The strong correlation between the results of both measurements indicate high reproducibility (P < 0·001, see Fig. 2; see also supplementary information).
DAPI versus PI in the study of intraspecific DNA content variation
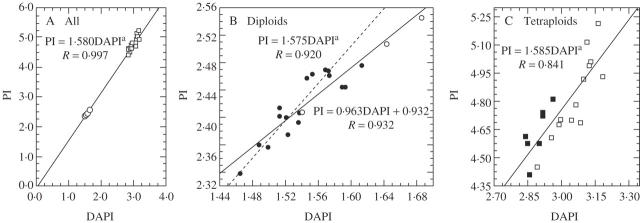
Simultaneous measurements with DAPI and PI staining carried out in spring 2006 and autumn 2004 were similar in measurement accuracy for DAPI (average CV of all peaks was 1·619 % in 2006, 1·679 % in 2004). The average CV of all peaks measured with PI was 3·100 %. A highly linear DAPI/PI correlation appeared both in diploids and in tetraploids (Fig. 1B, C—full lines; see also supplementary information). The relationship between DAPI and PI with Lycopersicon esculentum as a standard was the same for both ploidy levels, and also for all the tetraploids and most of the diploids of Festuca pallens analysed separately (PI = 1·58 DAPI, Fig. 1A, C—full lines; 1B—dashed line), indicating similar AT content for both ploidy levels. Only five diploid samples with the highest relative DNA content had a slightly higher AT content and decreased the slope of the regression line (Fig. 1B, full line). Nevertheless, including all diploids, the DAPI/PI relationship of all diploids remained highly significant as well as in tetraploids. This indicates that the variation as detected by DAPI correlates with absolute DNA content.
Fig. 1.
Comparison of the relative DNA content estimated with the AT-selective DAPI dye and intercalary PI dye in diploid (2x) and tetraploid (4x) Festuca pallens (Lycopersicon esculentum as the standard). Samples with known chromosome number (Šmarda and Kočí, 2003) are marked with closed symbols, those with unknown chromosome number with open ones. In ploidy levels together (A) and also in both ploidy levels separately (B, C), strong linear correlations appear between both dyes (full lines). The slope of the regression lines is around 1·58 in all three diagrams, if the five diploid samples with the highest relative DNA content are excluded (A, C - full lines, B - dashed line). These five samples appear to have a slightly higher AT content and decrease the slope of regression line of all diploids (B, full line). aIntercept non-significant (P > 0·05) and set to be zero.
Intra- and interpopulation variability of DNA content (local and landscape scales)
Intrapopulation variability in DNA content was shown in 16 of 24 tested populations (P < 0·001, Tables 1 and 2, Fig. 3). Intrapopulation variability was similar in both diploid and tetraploid populations (median difference, Table 2). The two most variable populations were tetraploid (Tables 1 and 2). A maximum intrapopulation difference of 1·121-fold was observed in the tetraploid population from Mikulov—Sv. Kopeček. All three plants there clearly differ in relative DNA content (standard sample ratios: 2·937, 3·066 and 3·291). This difference was independently confirmed in the simultaneous measurement of both extreme samples (1·119-fold difference, Fig. 4). Considerable variability was observed also in the tetraploid population from Praha—Prokopské údolí, where the DNA contents of samples differed 1·077-fold at maximum (Table 1).
Table 2.
Difference among plants in the populations
| Population (samples) no. | No. of variable populations (P < 0·01)* | Median difference (fold) | Maximum difference (fold) | |
|---|---|---|---|---|
| All populations (2x + 4x) | 24 (87) | 16 | 1·022 | 1·121 |
| All excluding Mikulov population | 23 (84) | 15 | 1·021 | 1·077 |
| Diploid (2x) populations | 10 (40) | 8 | 1·022 | 1·056 |
| Tetraploid (4x) populations | 14 (47) | 8 | 1·021 | 1·121 |
| 2x excluding Mikulov population | 13 (44) | 7 | 1·021 | 1·077 |
The only diploid sample from a mixed population was excluded from the analysis.
One-way ANOVA.
Fig. 2.
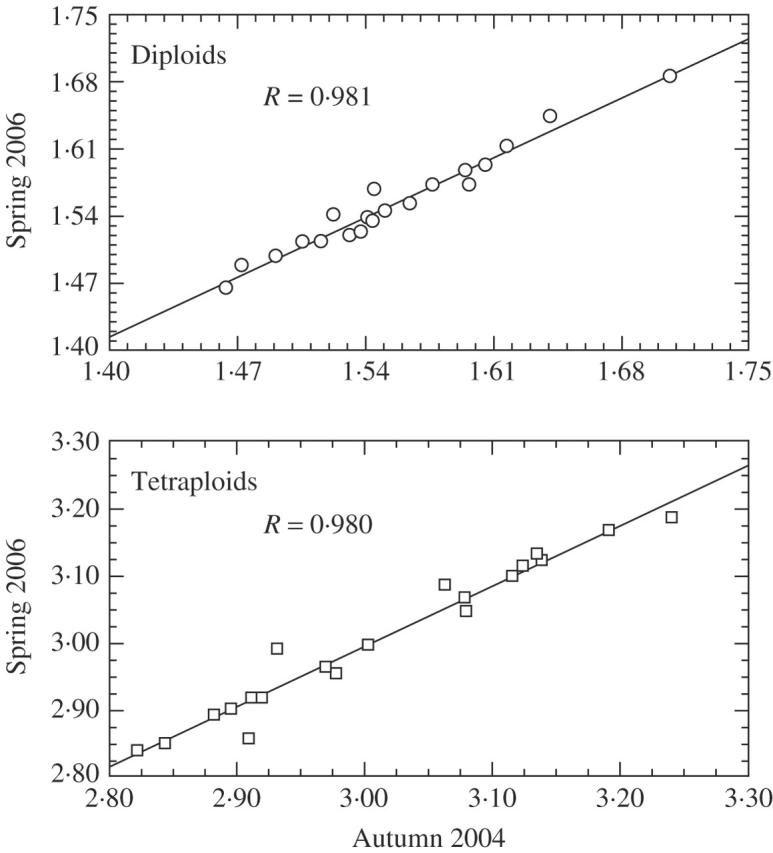
Reproducibility of measurements: comparison of DAPI measurements of diploid and tetraploid Festuca pallens carried out in autumn 2004 and spring 2006. Strong linear correlation of both indicates high reproducibility. No seasonal changes were detected (paired t-test, P > 0·05).
Fig. 3.
Observed intrapopulation and landscape pattern in relative DNA content on three investigated transects: circles represent diploids, squares tetraploids. Dark segment of symbols shows relative DNA content, and symbol size indicates position of the sample within the total observed range for the respective ploidy level (open: lower limit, closed: upper limit).
Fig. 4.
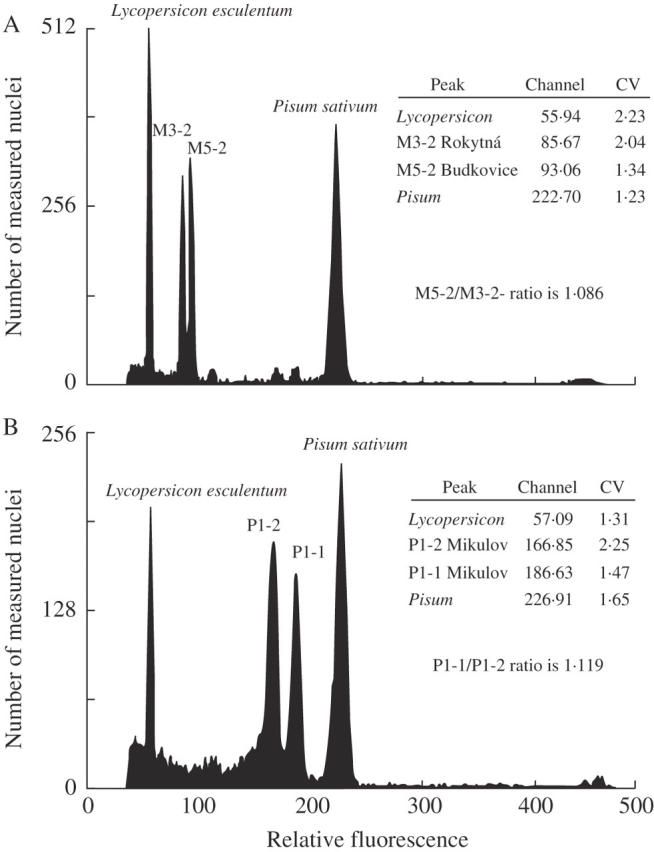
Difference in relative DNA content of two simultaneously measured diploid Festuca pallens samples: (A) maximal difference of diploid samples of two localities separated by 2 km from a transect along the Rokytná river; (B) maximal difference of tetraploid samples from the same population from Mikulov (Pálava hills transect); 10 000 cells were counted.
On the landscape scale within the transects, the mean relative DNA content was similar in almost all populations and no spatial relations were observed (Fig. 3, Table 3). However, two populations (Tables 1 and 2, Fig. 3) differed conspicuously from the others in mean or range of variability of relative DNA content. All three plants from the population near Budkovice (Rokytná transect) had apparently higher values (P < 0·001); the mean relative DNA content of this population was 1·088-fold higher than the population with the lowest mean DNA content from the same transect (Fig. 3, Table 3). The population of tetraploids from Mikulov—Sv. Kopeček showed the greatest variability, similar to the variability range within all the other tetraploids (Fig. 3, Table 2).
Table 3.
Landscape variability among populations within the transects
| Transect | Population (samples) no. | Max. difference among population means (fold) | Max. difference among all samples (fold) |
|---|---|---|---|
| Rokytná River (2x)* | 5 (16) | 1·088 | 1·106 |
| Pálava Hills (4x) | 5 (15) | 1·038 | 1·121 |
| Vltava River (rx) | 5 (15) | 1·029 | 1·065 |
| Rokytná River excluding Budkovice (2x)* | 4 (14) | 1·020 | 1·042 |
| Pálava Hills excluding Mikulov—Sv. Kopeček (4x) | 4 (12) | 1·020 | 1·027 |
The mixed population was excluded from the analysis.
No significant relationships between microhabitat factors and DNA content on the local and landscape scales were found. However, all four samples from sites invaded by Robinia pseudacacia were slightly smaller than those from non-invaded sites. Similarly, all three samples from ruderalized sites were larger than those from non-ruderalized sites from the same populations. The low significance in these cases may be due to the small number of samples available.
DNA content variation on a global scale
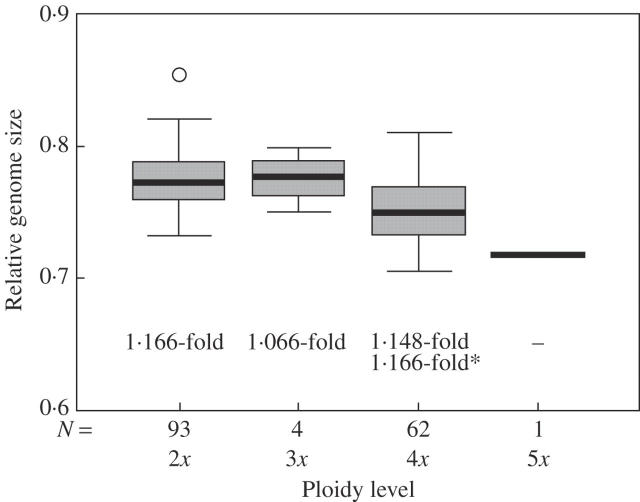
Within the whole distribution area, variability in all investigated ploidy levels was similar (Fig. 5). At maximum, an approximate 1·166-fold difference between samples was observed in diploids from Vadu Crişului (Romania) versus Děkovka (Czech Republic). On a global scale, a maximum 1·148-fold difference was found in tetraploids from Csór (Hungary) versus Friedrichsschwerz (Germany). Including those samples from the local scale, the difference increased up to the same level as in the diploids (1·166-fold), as a result of the largest sample from the Mikulov—Sv. Kopeček population. A similar range of variability (1·105-fold) was also observed in 29 karyologically homogeneous diploid samples (without B chromosomes): 1·065-fold in ten tetraploid samples.
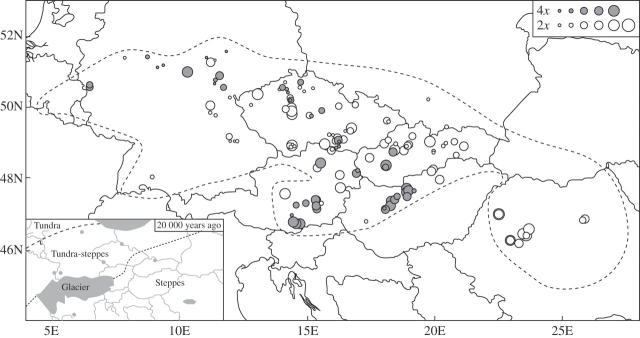
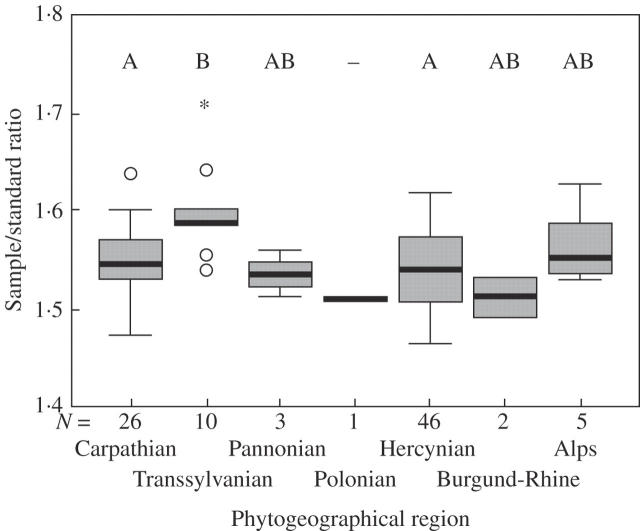
The relative DNA content in diploids correlated significantly with latitude and longitude (Table 4), increasing from the north-west to the south-east part of the distribution area (Fig. 6). This geographical gradient in DNA content also reflects the significant differences in DNA content among different phytogeographical regions (Fig. 7, Table 4). This observed pattern matches well with the character of palaeo-vegetation cover (Lang, 1994) at the end of the last ice age (20 000 years ago), with plants with larger genomes preferring areas of former steppes rather than steppe-tundra (P < 0·05, Fig. 6, Table 4). A strong correlations was also found between the high DNA content and the relict character of the locality (P < 0·01, Table 4). All these relationships were particularly strong for plants from Transylvania, which always occur in relict habitats and have a greater DNA content in comparison with the those from other phytogeographical regions (P < 0·001, Fig. 7). Only a minor correlation of DNA content with longitude remained significant (P < 0·05) in the remaining diploid samples when diploids from Transylvania were omitted from analysis.
Table 4.
Characters investigated on the global scale and their relationship to relative DNA content
| Location |
Palaeo-vegetation* |
Bedrock† |
Habitat |
Phytogeographical region‡ |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Latitude (Lat) | Longitude (Lon) | Altitude (Alt) | Steppetundra (Tun) | Steppe (Ste) | Siliceous (Si) | Calcareous (Ca) | Serpentine (Se) | River valley (Rv) | Relict habitat (Rh)§ | Castle or ruin (Cr) | Quarry (Qu) | Invaded by Robinia (Ro) | Carpathian (Car) | Transsilvanian (Tra) | Pannonian (Pan) | Polonian (Pol) | Hercynian (Her) | Burgund-Rhine (B-R) | Alps (Alp) | |
| Scoring: | °NL | °EL | m a.s.l. | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Statistics | Spearman's rho | M/M¶ | M/M | M/M | M/M | M/T | M/M | M/T | T/M | T/T | One-way ANOVA | |||||||||
| 2x | ||||||||||||||||||||
| Relationship | −0·345 | 0·324 | −0·001 | Yes | No | No | No | No | Yes | No | No | No | Yes | |||||||
| P | 0·001 | 0·002 | 0·992 | 0·018 | 0·931 | 0·672 | 0·390 | 0·939 | 0·006 | 0·225 | 0·280 | 0·754 | F = 4·469; P = 0·001 | |||||||
| 4x | ||||||||||||||||||||
| Relationship | −0·564 | 0·401 | 0·504 | Yes | Yes | Yes | No | No | No | No | No | No | Yes | |||||||
| P | <0·001 | <0·001 | 0·001 | <0·001 | <0·001 | 0·001 | 0·739 | 0·434 | 0·727 | 0·257 | 0·136 | 0·102 | F = 6·241; P = 0·001 | |||||||
Statistically significant relationships are indicated in bold.
At the end of the last ice age (20 000 years ago) according to Lang (1994); samples from glaciated areas are excluded.
Vulcanites and agglomerates excluded.
Meusel and Jäger (1992); samples from the border of two phytogeographical regions were included in both.
Deep canyon, mountain rock or ravine, karstic gorge; locality with relict species (see Introduction); unclear cases scored as non-relict.
M, Mann–Whitney test; T, t-test.
Fig. 5.
Relative genome size of separate ploidy levels of Festuca pallens. Relative genome size of higher polyploids (4x and 5x) is significantly smaller (t-test, P < 0·001). The numbers below the box plots give the total range of variation within the respective ploidy level. Relative genome size within all samples of all ploidy levels varied 1·201-fold. *Value calculated together with additional data from uncultivated population samples.
Fig. 6.
Observed difference in relative DNA content in the whole distribution area of Festuca pallens. Open symbols represent diploids, closed symbols tetraploids. The size of symbols reflects the relative DNA content divided into the five categories in a particular ploidy level with the 6th given to the extremely large diploid from Vadu Crişului. The natural range is marked by a dashed line. The generally higher DNA amounts in the south and east part of the natural range and in areas of steppes 20 000 years ago are of particular note.
Fig. 7.
Differences in relative DNA content of diploid plants from the different phytogeographical regions. Phytogeographical regions indicated with the same letter above the box-plots do not differ significantly (Bonferoni post-hoc comparison, P = 0·05).
Relative DNA content of tetraploids was also correlated significantly with the latitude and longitude (Table 4), with the same tendency of plants with larger genomes to concentrate in the south-east part of the distribution area (Fig. 6). In contrast to that for diploids, the DNA content of tetraploids was correlated with altitude (Table 4), with plants with larger genomes occurring at higher elevations. A clear difference of DNA content among tetraploid types was shown (one-way ANOVA, P < 0·001, Fig. 7), which may to some extent explain the observed correlations. When the dataset was divided into subsets representing these types, correlations with longitude disappeared. There remained only significant correlations (P < 0·05) of DNA content and altitude in the Scabrifolia (R = 0·431) and Pannonian (R = 0·507) types, and DNA content with latitude in the Pannonian type (R = −0·447). Differences in the DNA content of the tetraploid types can also explain differences among phytogeographical regions and type of bedrock. Indeed, plants with a lower DNA content of Scabrifolia type occur in the Hercynian region, namely on siliceous substrates, whereas those of the Pannonian and Alpine types occur on calcareous bedrock in Pannonia and the Alps. In separate subsets, the only significant difference remained between the DNA content of plants from different types of bedrock in plants of the Pannonian type. As for the case of correlation with latitude, the relationship with bedrock type in the Pannonian type was caused by a lower DNA content of plants from the siliceous hills of south-west Moravia.
As for diploids, the DNA content of tetraploids was correlated with the type of palaeo-vegetation (P < 0·001, Table 4). Other investigated parameters (Table 4) had no significant relationship to relative DNA content (α = 0·05).
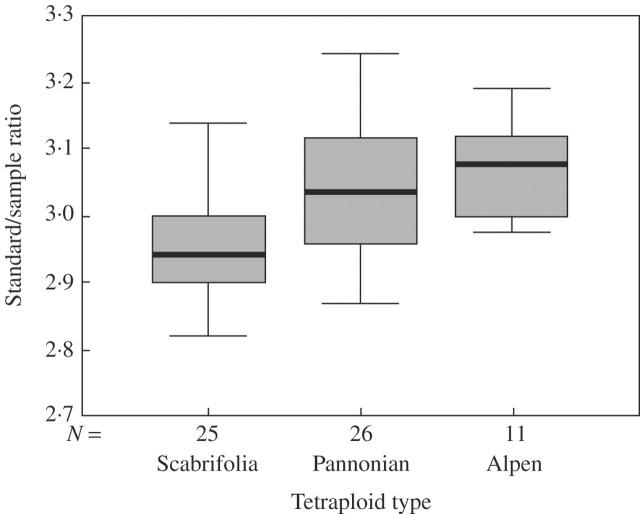
In general, the relative genome size of tetraploids was smaller than those of two-fold diploids (P < 0·001, Fig. 5). A strong spatial coincidence of the relative DNA content of tetraploids and diploids was revealed by Procrustean analysis (P < 0·001). Both tetraploids and diploids with a higher genome size tended to occur in the same regions. Within the tetraploids, the Scabrifolia type had a smaller genome size than the other two types (P < 0·001, Fig. 8).
Fig. 8.
Relative DNA content of separate tetraploid types. One-way ANOVA revealed significant differences among these types (P < 0·001). The relative DNA content of the Scabrifolia type is significantly smaller than in the two others (t-test, P < 0·001).
DISCUSSION
The results of the current study indicate the existence of a clear geographical gradient in DNA content within the whole distribution area of Festuca pallens, which may be explained and correlated with a number of macroecological, geographical and evolutionary factors. The observed variability in DNA content decreases generally in logical sequence with decreasing geographical scale studied; the highest variability appears within the whole distribution area, less on the landscape scale and the least on the intrapopulation level. However, although several variables explain well the observed pattern of DNA content, the present results do not provide an answer to the question of whether this pattern is a result of random genetic processes, environmental pressure or evolutionary events. The existence of some extreme populations, with very different DNA content (Budkovice) or high intrapopulation variability (Mikulov), indicates that interpretation of the observed pattern is more complex, and that the DNA content of an individual or a population is probably conditioned by multiple factors. Further study of such contrasting populations in combination with molecular methods provide the best opportunity for understanding some of these processes. The majority of recent genome size variability in angiosperms is related to the different mechanisms of retrotransposon proliferation and deletion (Bennetzen et al., 2005). The activity of retrotransposons may differ in contrasting microhabitats (Kalendar et al., 2000), of relevance for species with a high diversification of extreme habitats such as Festuca pallens. Changes in genome structure and selfish elements in grasses are summarized and discussed by Jones and Pasakinskiené (2005).
Intraspecific genome size variation on a global scale
An extremely high 2·5-fold variation in genome size (caused by differences in repetitive DNA content) has been documented on a global scale in the mosquito Aedes albopictus (Rai and Black, 1999). In plants, a survey of species with the highest reported intraspecific DNA content variation was given by Ohri (1998). Wide ranges in intraspecific DNA content variability were reported for grasses, e.g. 1·176-fold between the mean genome sizes of populations and 1·662-fold between individual genome sizes for Dasypyrum villosum (Caceres et al., 1998), 1·37-fold among maize cultivars (Rayburn et al., 1985), 1·36-fold between accessions from botanical gardens and plants from the field for Milium effusum (Bennett and Bennett, 1992), 1·28-fold among the mean population DNA content in Dactylis glomerata from altitudinal transects of the Mediterranean region (Reeves et al., 1998), and 1·125-fold among the African and Indian populations of Eleusine coracana (Hiremath and Salimath, 1991). The wide variation in Dasypyrum villosum (Greilhuber, 2005; Obermayer and Greilhuber, 2005) is considered to be a consequence of technical noise, although these authors also found differences in the DNA content in this species, 1·07-fold between populations and 1·12-fold between individuals, and confirmed this result with double peaks in flow cytometry (Obermayer and Greilhuber, 2005). This range of intrapopulation and interpopulation genome size variability is very similar to our presented results for Festuca pallens. Conversely, a low level of intraspecific DNA content variation was detected in Dactylis glomerata (1·021-fold among the mean DNA content of populations in the Slovenian Alps, Vilhar et al., 2002) in Lolium perenne (1·041-fold among the mean size of populations of various cultivars, Sugyiama et al., 2002) and Hordeum spontaneum (1·049-fold among wild populations in Israel, Turpeinen et al., 1999). A very low but statistically confirmed interpopulation difference of 1·6 % (=1·016-fold interpopulation range of variation) was reported for another grass, Sesleria albicans, by Lysák et al. (2000); the maximum difference among individuals of S. albicans reported by these authors was 6·1 % (cf. Lysák et al., 2002, table 3). They argued for intrapopulation genome stability based on a unimodal distribution within a flow-cytometry histogram of simultaneously measured plants with the greatest difference of genome size from the same population. The same test was used here for the most variable tetraploid and diploid populations (Fig. 4); the results confirm real intrapopulation variability in Festuca pallens.
Genome size and macro-ecological or historical factors
The correlation between the relative DNA content on a global scale and the type of vegetation at the end of the last ice age is interesting from the viewpoint of the ecological stability of most natural rocky habitats of Festuca pallens. These habitats were not covered by forest during either glacial or interglacial periods, nor covered by soil, due to which they did not change ecologically for a long period of time, and the occurrence of Festuca pallens in these habitats is considered to be of ancient origin given the co-occurrence of various types of relicts (see Introduction).
Correlations of genome size with latitude have been hitherto documented particularly on an interspecific level, for example among crop plant species (Bennett, 1976; see the review by Knight et al., 2005). Among grasses, a negative latitudinal correlation with intraspecific genome size variation was confirmed, for example, in Zea mays in North America (Laurie and Bennett, 1985; Rayburn et al., 1985) or in natural populations of Festuca arundinacea in Europe (Ceccarelli et al., 1992). An intraspecific genome size correlation with latitude for Hordeum marinum in western Europe and H. pubiflorum in South America was not confirmed (Jakob et al., 2004). By contrast, these authors found a correlation with climate (mean July temperature); a similar correlation (with mean January temperature) was reported by Turpeinen et al. (1999) in wild populations of Hordeum spontaneum in Israel. Within the framework of the relatively low Central European distribution of Festuca pallens, the SE–NW climatic gradient may play an important role in relation to DNA content. A negative correlation with continental climate expressed by a negative correlation with longitude was found in related European Cirsium species by Bureš et al. (2004b).
In F. pallens, it was not possible to confirm the relation between altitude and genome size repeatedly documented in other grasses, for example Dasypyrum villosum, Zea mays, Secale, Teosinte and Dactylis glomerata (Knight et al., 2005, and references therein). However, the potential role of macroclimate can be eliminated by the extreme contrast of such climates in the rocky habitats of Festuca pallens. In Sesleria albicans growing in similar habitats to F. pallens, altitudinal correlation with genome size was not confirmed (Lysák et al., 2000).
Microgeographical genome size differentiation
Among plant species, a well-documented and widely accepted case of ecologically induced intraspecific genome size variability is flax, Linum usitatissimum (Cullis, 2005, and references therein). Intrapopulation genome size variability in relation to extreme local ecological gradients was reported from the Evolution Canyon (Nahal Oren, Israel) population of Hordeum spontaneum and caused by variations in the BARE-1 copy number (Kalendar et al., 2000), and in the population of Ceratonia siliqua from the same locality (Bureš et al., 2004a) as a consequence of genome diversity induced by environmental stress (Nevo, 2001). This relationship to the most-contrasted tested microhabitat characters between samples within the investigated variable populations was not confirmed in the present study.
Intraplant genome size variation
Intraplant variation in genome size has been documented in Zea mays (Biradar and Rayburn, 1993) and in the roots of Vicia faba (caused by a decrease of the highly repetitive DNA in developed tissues; Bassi et al., 1984). Such variations can be dismissed here given the the same part of the tissue was used: the young, whitish, basal part of the just developing tiller leaf.
Seasonal variation in cytosolic compounds
Secondary cytosolic compounds can influence flow cytometry measurements (Noirot et al., 2000, 2003; Price et al., 2000). Some authors have found seasonal changes in the fluorescence intensity of nuclei in various plant species or genera, for example in Actinidia deliciosa (e.g. Hopping, 1994). Such changes can then be interpreted as genome size variability. In the present study, potential differences caused by different habitat conditions and date of collection were eliminated by the homogeneous cultivation of Festuca pallens samples representing the global distribution, collecting samples on the local and landscape scales over 4 d, minimizing measurements of both sample sets over a period of just 2 weeks as well as the measurement of the same part of the young yellowish parts of leaves containing a minimum of secondary compounds. For the same reasons, it is possible to exclude any analogies with other seasonal changes in genome size documented, for example in Helianthus (Cavallini et al., 1989; Michaelson et al., 1991). An excellent example of the combination of seasonally influenced fluorescence intensity of 1·04- to 1·08-fold within various Spanish and Canarian populations (probably caused by furanocoumarins) of Bituminaria bituminosa (Fabaceae), clearly separated from geographically influenced true intraspecific genome size variation of 1·121-fold among populations as confirmed by double peaks, was given by Walker et al. (2006). The double peak histograms (Fig. 4) of the simultaneous measurement of samples of different genome size in the present study, strong linear correlation and similarity of measurements from autumn 2004 and spring 2006 (Fig. 2) all argue that the DNA content variation in Festuca pallens is not a fluorescence artefact of seasonal changes (e.g. in cytosolic compounds).
B chromosomes
In addition to the more or less continual variation in relative DNA content of Festuca pallens reported here, larger differences were found between some individuals within the populations (up to 12·1 % at the local scale). These ‘jump’ differences can linked to the presence of B chromosomes (cf., for example, Jones and Rees, 1982; Delgado et al., 2004), which are commonly reported in grasses (Jones and Díez, 2004). In Festuca, B chromosomes were documented for several species (cf. Jones and Díez, 2004) and also for F. pallens (Šmarda and Kočí, 2003). The detected variation here seems not to be caused exclusively by the presence of B chromosomes, as a 1·105-fold range of DNA content was observed in diploids with no B chromosomes (see the supplementary information). The contribution of B chromosomes to the total DNA content is still unclear. First, there is a positive correlation between genome size and the number of B chromosomes (Ayonoadu and Rees, 1971; Jones and Rees, 1982; Rosato et al., 1998; Delgado et al., 2004); by contrast, the genome size of plants with B chromosomes may in some cases be even smaller that those with no B chromosomes (Porter and Rayburn, 1990; Poggio et al., 1998). B chromosomes are, moreover, widely accepted to be derived from A-chromosomes (Jones and Houben, 2003, and references therein; Leach et al., 2005) and interchanges with normal A-chromosomes have also been documented (Nokkala and Nokkala, 2004; Ribeiro et al., 2004).
DAPI applicability for DNA content study
DAPI is an AT-selective dye and the DNA content measured with DAPI may be influenced by base composition. Therefore, the initial genome size reports based upon DAPI should be verified using a stain that is independent of base ratio (Doležel et al., 1992; Johnston et al., 1999). Although among higher plants as a whole genome size and AT level are not correlated (Barow and Meister, 2002), in most known cases the base composition does not vary strongly among closely related species. This is supported by the close correlation between the real genome size (as estimated with PI) and the apparent genome size (as estimated with DAPI) documented in different Lolium individuals (Sugiyama et al., 2002, fig. 2; R = 0·883), Cirsium species (Bureš et al., 2004b, cf. table 1, R = 0·993) and Trifolium species (Vizintin et al., 2006, R = 0·99). The present analysis indicates a strong linear correlation of the results with AT-selective DAPI and intercalary PI staining (Fig. 1). This justifies the use of a base-specific dye in order to obtain maximal resolution, and enables the results for Festuca pallens to be interpreted in terms of absolute genome size. The rather low AT content variability found in Festuca pallens diploids seems not to affect dramatically the strong linear DAPI/PI correlation. Use of DAPI seems to be more suitable for the study of intraspecific genome size variability in those cases where high a DAPI/PI correlation has been verified, especially due to the lower CVs, and thus higher accuracy, resolution and reproducibility of DAPI measurements in relation to PI.
DNA content in diploid and tetraploid level
Plants of the two ploidy levels in F. pallens are very similar in morphology but otherwise clearly differ from the other Central European species. The lower relative genome size (Cx-value) found in tetraploids argues, among many others examples, for the downsizing of the genome in polyploids described by Leitch and Bennett (2004). Variability within both ploidy levels is very similar (1·170-fold in diploids, and 1·164-fold in tetraploids). The spatial relationship of genome size in diploids and tetraploids may indicate an evolutionary link between both ploidy levels, although a similar situation may also be the result of the adaptation of both ploidy levels to similar macroclimatic factors. A further explanation may be the advantage of plants with a smaller genome in colonization of new habitats in north-west Europe from their refuges in deep canyons and rocky steppes in south-east Europe during the last glatiation, as indicated by the relationship of higher DNA content to the relict habitats and palaeo-steppes in the present analysis. Differences in DNA content among tetraploid types may also indicate their probable polytopic origin and may serve as a taxonomic criterion for their delimitation.
SUPPLEMENTARY INFORMATION
Supplementary information are available online at http://www.aob.oupjournals.org giving details of sample localities, results of measurements, and character states at the local, landscape and global scales, together with results of simultaneous measurements with DAPI and PI staining.
Acknowledgments
We are indebted to all colleagues who collected plants (see supplementary information). This study was supported by the Grant Agency of the Czech Republic (GA ČR 206/03/0228 ‘Phylogeography of polyploid complexes in Europe’) and by the Ministry of Education of the Czech Republic (MSM0021622416 and LC06073).
LITERATURE CITED
- Auquier P. 1977. Taxonomie et nomenclature de quelques Festuca tétraploides du groupe de Festuca ovina L. s.l. en Europe moyenne. Bulletin du Jardin Botanique National de Belgique 47: 99–116. [Google Scholar]
- Ayonoadu U, Rees H. 1971. The effects of B-chromosomes on the nuclear phenotype in root meristems of maize. Heredity 27: 365–383. [Google Scholar]
- Barow M, Meister A. 2002. Lack of correlation between AT frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry 47: 1–7. [DOI] [PubMed] [Google Scholar]
- Bassi P, Cionini PG, Cremonini R, Seghizzi P. 1984. Under representation of nuclear DNA sequences in differentiating root cells of Vicia faba. Protoplasma 123: 70–77. [Google Scholar]
- Bennett MD. 1976. DNA amount, latitude and crop plant distribution. Environmental and Experimental Botany 16: 93–108. [Google Scholar]
- Bennett MD. 1985. Intraspecific variation in DNA amount and the nucleotypic dimension in plant genetics. In: Freeling M, ed. Plant genetics (UCLA Symposia on Molecular and Cellular Biology, New Ser. Vol. 35). New York: Alan Liss, 283–302.
- Bennett ST, Bennett MD. 1992. Variation in nuclear DNA amount between wild and cultivated populations of Milium effusum (2n = 28). Genome 35: 1050–1053. [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. 2005. Mechanisms of recent genome size variation in flowering plants. Annals of Botany 95: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biradar DP, Rayburn AL. 1993. Intraplant nuclear DNA content variation indiploid nuclei of maize (Zea mays L.). Journal of Experimental Botany 44: 1039–1044. [Google Scholar]
- Bureš P, Pavlíček T, Horová L, Nevo E. 2004a. Microgeographic genome size differentiation of the carob tree, Ceratonia siliqua, at “Evolution Canyon”, Israel. Annals of Botany 93: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureš P, Wang Y-F, Horová L, Suda J. 2004b. Genome size variation in Central European species of Cirsium (Compositae) and their natural hybrids. Annals of Botany 94: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres ME, Pace CD, Mugnozza GTS, Kotsonis P, Ceccarelli M, Cionini PG. 1998. Genome size variations within Dasypyrum villosum: correlations with chromosomal traits, environmental factors and plant phenotypic characteristics and behavior in reproduction. Theoretical and Applied Genetics 96: 559–567. [Google Scholar]
- Cavallini A, Zolfino C, Natali L, Cionini G, Cionini PG. 1989. Nuclear DNA changes within Helianthus annuus L.: origin and control mechanism. Theoretical and Applied Genetics 77: 12–16. [DOI] [PubMed] [Google Scholar]
- Ceccarelli M, Falistocco E, Cionini PG. 1992. Variation of genome size and organization within hexaploid Festuca arundinacea. Theoretical and Applied Genetics 83: 273–278. [DOI] [PubMed] [Google Scholar]
- Cullis CA. 2005. Mechanisms and control of rapid genomic changes in flax. Annals of Botany 95: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M, Caperta A, Ribeiro T, Viegas W, Jones RN, Morais-Cecílio L. 2004. Different numbers of rye B chromosomes induce identical compaction changes in distinct A chromosome domains. Cytogenetic and Genome Research 106: 320–324. [DOI] [PubMed] [Google Scholar]
- Doležel J, Bartoš J. 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany 95: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Göhde W. 1995. Sex determination in dioecious plants Melandrium album and Melandrium rubrum using high-resolution flow cytometry. Cytometry 19: 103–106. [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. 1992. Comparison of three fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum 85: 625–631. [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R. 1998. Plant genome size estimation by flow cytometry: interlaboratory comparison. Annals of Botany 82 (Suppl. A): 17–26. [Google Scholar]
- Evans GM, Durrant A, Rees H. 1966. Associated nuclear changes in the induction of flax genotrophs. Nature 212: 697–699. [Google Scholar]
- Greilhuber J. 1988. Critical reassessment of DNA content variation in plants. In: Brandham PE, ed. Kew Chromosome Conference III. London: HMSO, 39–50.
- Greilhuber J. 1997. The problem of variable genome size in plants (with special reference to woody plants). In: Borzan Z, Schlarbaum SE, eds. Cytogenetic studies of forest trees and shrub species. Zagreb: Croatian Forests, Inc. and Faculty of Forestry, University of Zagreb, 13–34.
- Greilhuber J. 1998. Intraspecific variation in genome size: a critical reassessment. Annals of Botany 82 (Suppl. A): 27–35. [Google Scholar]
- Greilhuber J. 2005. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany 95: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Ebert I. 1994. Genome size variation in Pisum sativum. Genome 37: 646–655. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Obermayer R. 1997. Genome size and maturity group in Glycine max (soybean). Heredity 78: 547–551. [Google Scholar]
- Greilhuber J, Obermayer R. 1998. Genome size variation in Cajanus cajan (Fabaceae): a reconsideration. Plant Systematics and Evolution 212: 135–141. [Google Scholar]
- Hiremath SC, Salimath SS. 1991. Quantitative nuclear DNA changes in Eleusine (Gramineae). Plant Systematics and Evolution 178: 225–233. [Google Scholar]
- Hopping ME. 1994. Seasonal changes in fluorescence intensity of kiwifruit nuclei stained with propidium iodide and measured by flow cytometry. New Zealand Journal of Botany 32: 237–245. [Google Scholar]
- Jackson DA. 1995. PROTEST: a PROcrustean randomization TEST of community environment concordance. Ecoscience 2: 297–303. [Google Scholar]
- Jakob SS, Meister A, Blattner FR. 2004. The considerable size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology and speciation rates. Molecular Biology and Evolution 21: 860–869. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ. 1999. Reference standards for determination of DNA content of plant nuclei. American Journal of Botany 86: 609–613. [PubMed] [Google Scholar]
- Jones N, Houben A. 2003. B chromosomes in plants: escapees from the A chromosome genome? Trends in Plant Sciences 8: 417–423. [DOI] [PubMed] [Google Scholar]
- Jones N, Pasakinskiené I. 2005. Genome conflict in the gramineae. New Phytologist 165: 391–409. [DOI] [PubMed] [Google Scholar]
- Jones RN, Rees H. 1982. B chromosomes. New York: Academic Press.
- Jones RN, Díez M. 2004. The B chromosome database. Cytogenetic and Genome Research 106: 149–150 (http://www.bchromosomes.org/bdb/). [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. 2000. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences of the USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany 95: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. 1994. Quartäre Vegetationsgeschichte Europas. Jena: Gustav Fischer.
- Laurie DA, Bennett MD. 1985. Nuclear content in the genera Zea and Sorghum. Intergeneric, interspecific and intraspecific variation. Heredity 55: 307–313. [Google Scholar]
- Leach CR, Houben A, Field B, Pistrick K, Demidov D, Timmis JN. 2005. Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris. Genetics 171: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. 2004. Genome downsizing in polyploidy plants. Biological Journal of the Linnean Society 82: 651–663. [Google Scholar]
- Lysák MA, Rostková A, Dixon JM, Rossi G, Doležel J. 2000. Limited genome size variation in Sesleria albicans. Annals of Botany 86: 399–403. [Google Scholar]
- Markgraf-Dannenberg I. 1980. Festuca. In: Tutin TG, Heywood VH, Burges NA, et al., eds. Flora Europaea, Vol. 5. Cambridge: Cambridge University Press, 125–153.
- Meusel H, Jäger EJ. 1992. Vergleichende Chorologie der Zentraleuropäischen Flora. Karten, Literatur, Register. Band III. Jena: Gustav Fischer.
- Michaelson MJ, Price HJ, Johnston JS, Ellison JR. 1991. Variation of nuclear DNA content in Helianthus annuus (Asteraceae). American Journal of Botany 78: 1238–1243. [Google Scholar]
- Murray BG. 2005. When does intraspecific C-value variation become taxonomically significant? Annals of Botany 95: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E. 2001. Evolution of genome–phenome diversity under environmental stress. Proceedings of the National Academy of Sciences of the USA 99: 6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot M, Barre P, Louarn C, Duperray C, Hamon S. 2000. Nucleus–cytosol interactions—a source of stochimetric error in flow cytometric estimation of nuclear DNA content in plant. Annals of Botany 86: 309–316. [Google Scholar]
- Noirot M, Barre P, Louarn C, Duperray C, Hamon S. 2003. Effects of caffeine and chlorogenic acid on propidium iodide accessibility to DNA. Consequence on genome size evaluation in coffee tree. Annals of Botany 92: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot M, Barre P, Duperray C, Hamon S, De Kochko A. 2005. Investigation on the causes of stochiometric error in genome size estimation using heat experiments: consequences on data interpretation. Annals of Botany 95: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokkala S, Nokkala C. 2004. Interaction of B chromosomes with A or B chromosomes in segregation in insects. Cytogenetic and Genome Research 106: 394–397. [DOI] [PubMed] [Google Scholar]
- Obermayer R, Greilhuber J. 1999. Genome size in Chinese soybean accessions—stable or variable? Annals of Botany 84: 259–262. [Google Scholar]
- Obermayer R, Greilhuber J. 2005. Does genome size in Dasypyrum villosum vary with fruit colour? Heredity 95: 91–95. [DOI] [PubMed] [Google Scholar]
- Ohri D. 1998. Genome size variation and plant systematics. Annals of Botany 82 (Suppl. A): 75–83. [Google Scholar]
- Otto F. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z, eds. Methods in cell biology: flow cytometry, Vol. 33. San Diego, CA: Academic Press, 105–110. [DOI] [PubMed]
- Peres-Neto PR, Jackson DA. 2000. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129: 169–178. [DOI] [PubMed] [Google Scholar]
- Pils G. 1981. Karyologie und Verbreitung von Festuca pallens Host in Österreich. Linzer Biologische Beiträge 13: 231–241. [Google Scholar]
- Poggio L, Rosato M, Chiavarino AM, Naranjo CA. 1998. Genome size and enviromental correlations in maize (Zea mays ssp. mays, Poaceae). Annals of Botany 82 (Suppl. A): 107–115. [Google Scholar]
- Porter HL, Rayburn AL. 1990. B-chromosome and C-band heterochromatin variation in Arizona maize population adapted no different altitude. Genome 33: 659–662. [Google Scholar]
- Price HJ. 1988. Nuclear DNA content variation within angiosperm species. Evolutionary Trends in Plants 2: 53–60. [Google Scholar]
- Price HJ, Hodnett G, Johnston JS. 2000. Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propridium iodide fluorescence. Annals of Botany 86: 929–934. [Google Scholar]
- Rai KS, Black WC. 1999. Mosquito genomes: structure, organization and evolution. In: Hall JC, Dunlap JC, Friedmann T, Gianelli F, eds. Advances in genetics, Vol. 41. San Diego: Academic Press, 1–33. [DOI] [PubMed]
- Rayburn AL, Price HJ, Smith JD, Gold JR. 1985. C-band heterochromatin and DNA content in Zea mays. American Journal of Botany 72: 1610–1617. [Google Scholar]
- Reeves G, Francis D, Davies MS, Rogers HJ, Hodkinson TR. 1998. Genome size is negatively correlated with altitude in natural populations of Dactylis glomerata. Annals of Botany 82 (Suppl. A.): 99–105. [Google Scholar]
- Ribeiro T, Pires B, Delgado M, Viegas W, Jones N, Morais-Cecilio L. 2004. Evidence for ‘cross-talk’ between A and B chromosomes of rye. Proceedings of the Royal Society of London, Series B-Biological sciences 271 (Suppl. 6): 482–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato M, Chiavarino AM, Naranjo CA, Camara Hernandez J, Poggio L. 1998. Genome size and numerical polymorphism for the B chromosome in races of Maize (Zea mays ssp. mays, Poaceae). American Journal of Botany 85: 168–174. [PubMed] [Google Scholar]
- Sugiyama S, Yamaguchi K, Yamada T. 2002. Intraspecific phenotypic variation associated with nuclear DNA content in Lolium perenne L. Euphytica 128: 145–151. [Google Scholar]
- Šmarda P. 2001. Středoevropské taxony Festuca ser. Psammophilae M. Pawlus. Master thesis, Masaryk University, Brno.
- Šmarda P. 2006. DNA ploidy levels and intraspecific DNA content variability in Romanian fescues (Festuca L., Poaceae) measured in fresh and herbarium material. Folia Geoboanica 41 (in press).
- Šmarda P, Kočí K. 2003. Chromosome number variability in Central European members of the Festuca ovina and F. pallens groups (sect. Festuca). Folia Geobotanica 38: 65–95. [Google Scholar]
- Šmarda P, Müller J, Vrána J, Kočí K. 2005. Ploidy level variability of some Central European fescues (Festuca L. subg. Festuca, Poaceae). Biologia (Bratislava) 60: 25–36. [Google Scholar]
- Temsch EM, Greilhuber J. 2000. Genome size variation in Arachis hypogaea and A. monticola re-evaluated. Genome 43: 449–451. [PubMed] [Google Scholar]
- Tracey R. 1980. Beiträge zur Karyologie, Verbreitung und Systematik des Festuca ovina Formenkreises im Osten Österreichs. PhD thesis, University of Vienna, Austria.
- Turpeinen T, Kulmala J, Nevo E. 1999. Genome size variation in Hordeum spontaneum populations. Genome 42: 1094–1099. [DOI] [PubMed] [Google Scholar]
- Vilhar B, Vidic T, Jogan N, Dermastia M. 2002. Genome size and nucleolar number as estimators of ploidy level in Dactylis glomerata in Slovenian Alps. Plant Systematics and Evolution 234: 1–13. [Google Scholar]
- Vizintin L, Javornik B, Bohanec B. 2006. Genetic characterization of selected Trifolium species as revealed by nuclear DNA content and ITS rDNA region analysis. Plant Science 170: 859–866. [Google Scholar]
- Walker DJ, Monino I, Correal E. 2006. Genome size in Bituminaria bituminosa (L.) C. H. Stirton (Fabaceae) populations: separation of “true” differences from environmental effects on DNA determination. Environmental and Experimental Botany 55: 258–265. [Google Scholar]