Abstract
Proteins are transported into and out of the cell nucleus via specific signals. The two best-studied nuclear transport processes are mediated either by classical nuclear localization signals or nuclear export signals. There also are shuttling sequences that direct the bidirectional transport of RNA-binding proteins. Two examples are the M9 sequence in heterogeneous nuclear ribonucleoprotein A1 and the heterogeneous nuclear ribonucleoprotein K shuttling domain (KNS) sequence in heterogeneous nuclear ribonucleoprotein K, both of which appear to contribute importantly to the export of mRNA to the cytoplasm. HuR is an RNA-binding protein that can stabilize labile mRNAs containing AU-rich elements in their 3′ untranslated regions and has been shown to shuttle between the nucleus and cytoplasm (18, 19). We have identified in HuR a shuttling sequence that also possess transcription-dependent nuclear localization signal activity. We propose that HuR first may bind AU-rich element-containing mRNAs in the nucleus and then escort them through the nuclear pore, providing protection during and after export to the cytoplasmic compartment.
Keywords: nuclear localization, RNA degradation, nuclear export
The mRNAs of many protooncogenes, cytokines, and lymphokines are targeted for rapid degradation by AU-rich elements (AREs) located in their 3′ untranslated regions (1, 2). This is an important mechanism of posttranscriptional regulation of gene expression (for reviews, see refs. 3 and 4). To decipher the nature of the degradation machinery, a number of ARE-specific RNA-binding proteins have been identified and studied (5–15). We have focused on HuR, a ubiquitously expressed member of the Hu family of RNA-binding proteins (16). HuR exhibits specific affinity for ARE-containing RNA sequences in vitro (16, 17). Recently, it has been shown (18–20) that HuR overexpression enhances the stability of ARE-containing reporter mRNAs, establishing an in vivo role for HuR in mRNA decay.
There are four proteins belonging to the Hu family. HuR (also called HuA) is expressed ubiquitously in all tissues (21), whereas Hel-N1 (or HuB), HuC, and HuD are neuron-specific (22–25). The latter three proteins have been identified as target antigens in paraneoplastic encephalomyelitis sensory neuronopathy (the Hu syndrome) associated with small cell lung cancer (22–26). HuR was first cloned by using a degenerate oligonucleotide-directed PCR strategy designed to capture ubiquitously expressed Hu proteins (21). All Hu proteins contain three RNA binding domains of the RRM (RNA recognition motif; also called RBD) superfamily (27) and are related to the Drosophila RNA-binding protein ELAV (21–25). The RRMs are conserved among all four Hu family proteins, whereas the hinge region between RRMs 2 and 3 differs (28). Each individual Hu protein, however, is highly conserved among vertebrates. In the case of HuR, the human protein is 99.7% identical to that of mouse, 98.2% to chicken, and >90% to Xenopus (28–30).
HuR is localized predominantly in the nucleus (18). Yet, ARE-mediated mRNA decay is generally believed to occur in the cytoplasm. Through an interspecies heterokaryon fusion experiment, we have shown that HuR actually shuttles between the nucleus and cytoplasm (18). Therefore, it is possible that HuR may initially bind to ARE-containing mRNAs in the nucleus, accompany them into the cytoplasm, and then rapidly return to the nucleus after release from the mRNA, which is then subject to rapid decay. The shuttling of HuR depends on the transcriptional activity of RNA polymerase II (pol II) (19).
Specific sequences have been reported to be responsible for intracellular protein trafficking (for reviews, see refs. 31–33). The well characterized classical nuclear localization signal(NLS) either contains a cluster of basic amino acids [as in the simian virus 40 (SV40) large T antigen] or is bipartite, with two basic amino acids in the first cluster separated by a 10-aa spacer from a second cluster of three or more basic amino acids (as in nucleoplasmin or HIV-1 Rev). Translocation of proteins containing an NLS from the cytoplasm to the nucleus is independent of temperature and ongoing transcription. The classical nuclear export sequence (NES) is comprised of a stretch of 10 amino acids rich in leucine [as in Rev or the protein kinase inhibitor PKI; for reviews, see refs. 33 and 34].
In addition to the NLS and NES motifs, two shuttling sequences, namely the 38-aa M9 sequence of heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and A2 (35, 36) and the 24-aa KNS (hnRNP K shuttling domain) of hnRNP K (37), have been identified. These shuttling sequences possess both NLS and NES activities, with their NLS activities depending on the transcriptional activity of pol II (36–41). In the case of hnRNP K, it has been shown that the transcription-independent NLS overrides the transcriptional requirement of KNS and allows retention of the protein in the nucleus when cells are treated with the transcription inhibitor actinomycin D (37).
The HuR hinge region (amino acids 190–244) has a basic sequence (205-RRFGGPVHHQAQRFRF-220) similar but not identical to the consensus bipartite NLS in that it has only two basic amino acids in the second cluster. HuR does not, however, have any M9-, KNS-, or NES-like sequence, suggesting that it may contain a unique shuttling/NES motif. We set out to identify the NLS/NES/shuttling sequence(s) in HuR. Through extensive deletion analyses using two protein reporter systems, we have identified a sequence containing both NLS and NES (i.e., shuttling) activity in the HuR hinge region. These results broaden our understanding of nucleocytoplasmic protein-transport pathways.
MATERIALS AND METHODS
Plasmid Constructions.
C-terminal Flag-tagged HuR-ΔhRRM3 (HuR-ΔhRRM3-C-Flag) and HuR-ΔRRM3 (HuR-ΔRRM3-C-Flag) were generated by using PCR from the previously described expression constructs pCDNA3-HuR-M1 and -M2, respectively (18), and cloned back into the EcoRI and XhoI sites in pCDNA3 (Invitrogen). The N-terminal myc-tagged pCDNA3 expression vectors, including the pyruvate kinase (PK) construct, the PK-hnRNP A1 M9 (PK-M9) construct, and the nucleoplasmin-nuclear localization signal construct (NPc-NLS), were all gifts from G. Dreyfuss’ laboratory at the University of Pennsylvania and have been described (35, 36). To generate the myc-tagged PK-HuR-mutants 1–12 (PK-M 1–12, as shown in Fig. 3B), DNA fragments encoding the HuR hinge region were amplified by using PCR and inserted at the C terminus of PK at KpnI and NotI sites in the same orientation as in PK-M9 (35). The myc-tagged NPc-HuR mutant constructs (NPc-M1, -M7, and -M10, as shown in Fig. 4B) were generated by using PCR amplification of each HuR fragment followed by insertion into the EcoRI and XhoI sites C-terminal to NPc to replace the NLS sequence in NPc-NLS.
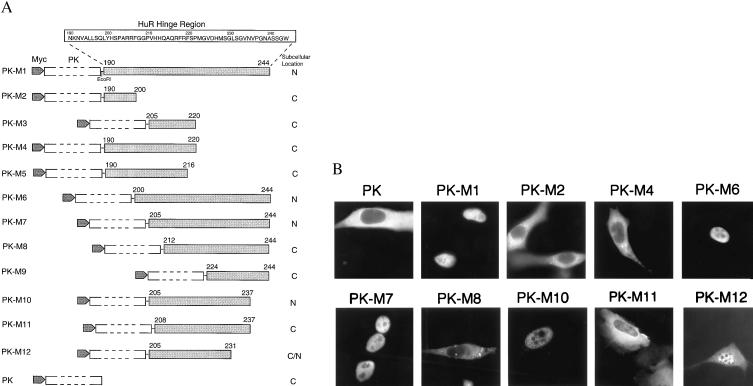
Figure 3.
Delineation of the HuR nuclear localization signal. (A) Schematic diagram and summary of the intracellular localization of PK fusion proteins containing HuR fragments M1 to M12. (B) NIH 3T3 cells were transiently transfected with myc-tagged PK expression vectors containing HuR fragments and processed as described in Fig. 1B, except for use of anti-myc antibody 9E10. myc-tagged PK was transfected as a control.
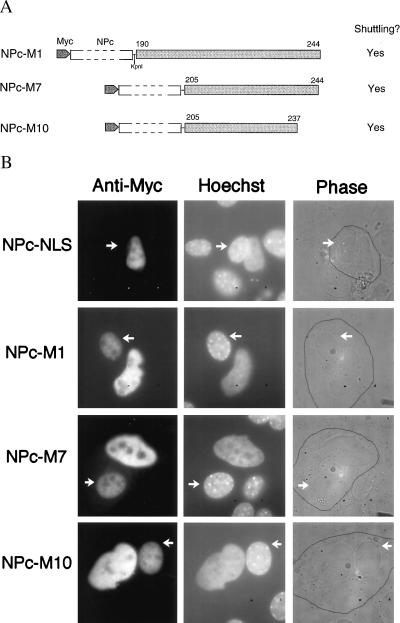
Figure 4.
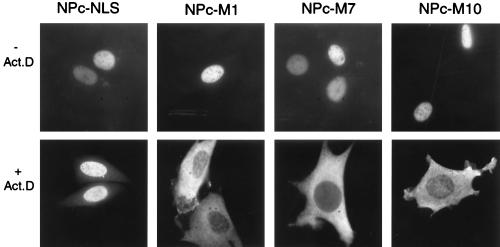
The HuR NLS confers shuttling activity. (A). Schematic representation of the NPc fusion proteins assayed in Fig. 4B. (B) Heterokaryon fusion assays of NPc-M1, -M7, and -M10. HeLa cells were transiently transfected with NPc fusion proteins containing HuR sequences M1, M7, or M10. Heterokaryon fusion and staining assays were performed as described in Fig. 2, except for use of anti-myc mAb 9E10. Mouse nuclei in the heterokaryons are indicated with arrows. The NPc-NLS was analyzed as a negative control.
Cell Culture and Transient Transfections.
HeLa and NIH 3T3 cells were passed in DMEM (GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO). Cells were grown on 60-mm plates overnight to 50% confluency and transfected by using the FuGene system (Boehringer Mannheim): plasmid DNA (2 μg) was incubated with 6 μl of FuGene solution in 200 μl of DMEM for 15 min and overlaid onto the cells. After overnight incubation, the culture medium was changed to fresh 10% FBS/DMEM. Cells were fixed or harvested 48 hr after transfection. For each transfection, 50–100 stained cells were examined; similar staining patterns were observed throughout. To inhibit pol II activity, cells were incubated with actinomycin D at 5 μg/ml for 3 hr before fixation and staining.
Immunofluorescence Microscopy.
Transfected HeLa or NIH 3T3 cells grown on coverslips were fixed with 3% paraformaldehyde in PBS for 20 min at room temperature, washed with PBS twice for 5 min, permeabilized with 0.5% Triton X-100/1% normal goat serum (Jackson ImmunoResearch) in PBS for 15 min, and washed with 1% normal goat serum/PBS three times for 10 min each. The coverslips were then incubated with primary antibodies in 1% normal goat serum/PBS at room temperature for 1 hr. The anti-Flag tag mAb (Sigma) was diluted to 10 μg/ml; culture supernatant from the monoclonal cell line producing anti-myc tag antibody 9E10 (gift of G. Hu, University of Michigan, Ann Arbor, MI) was used at a dilution of 1:20. The coverslips were washed twice for 10 min and incubated with goat anti-mouse IgG secondary antibody coupled to fluorescein isothiocyanate (Jackson ImmunoResearch) at 5.6 μg/ml in 1% normal goat serum/PBS at room temperature for 1 hr. For analyses of HeLa-3T3 heterokaryons, Hoechst dye 33258 (Sigma) was included at 1 μg/ml with the secondary antibody incubation. Hoechst dye produces punctate staining of mouse (but not HeLa) nuclei (42). The samples were then washed three times for 10 min each and mounted. Figures were photographed by using a charge-coupled device camera through an Axioskop microscope (Zeiss) with a ×100 Plan-NeoFluor oil objective.
Heterokaryon Formation.
Heterokaryons of HeLa and mouse L929 were formed as described (36, 18) with the following modifications. HeLa cells were seeded onto glass coverslips at 2 × 105 cells per coverslip 24 hr posttransfection. After overnight incubation, 3T3 cells that had been incubated for 30 min in the presence of 75 μg/ml cycloheximide (Sigma) were seeded onto the same coverslips at 3 × 105 cells per coverslip. The cocultures were then incubated for 3 hr in 75 μg/ml cycloheximide, fused with 50% polyethylene glycol (PEG 3350; Sigma) for 2 min, washed in PBS, and returned to medium containing 100 μg/ml cycloheximide for another 3-hr incubation, followed by paraformaldehyde fixation for immunostaining as described above.
RESULTS
The HuR Hinge Region Contains a Shuttling Signal.
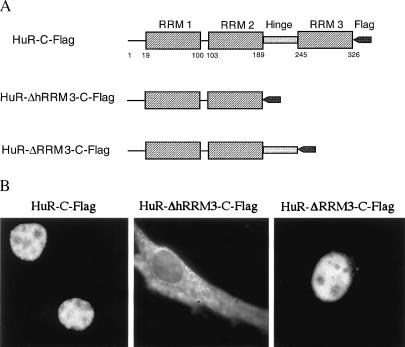
It has been shown that the nuclear protein HuR is able to shuttle between the nucleus and cytoplasm (18, 19) and that Flag-tagged HuR is located identically to the endogenous HuR protein (18). To investigate the HuR shuttling signal, we first compared the subcellular localization of two Flag epitope-tagged HuR deletion mutants—HuR-ΔhRRM3-C-Flag and HuR-ΔRRM3-C-Flag (Fig. 1A)—lacking either the hinge + RRM 3 (ΔhRRM3) or RRM 3 alone (ΔRRM3). Briefly, NIH 3T3 cells were transiently transfected with a DNA plasmid encoding the protein of interest; 36 hr later, the cells were fixed and stained for immunofluorescence microscopy to examine the intracellular distribution of the tagged protein. As shown in Fig. 1B, HuR-ΔRRM3 displays a nuclear staining pattern, the same as that of wild-type Flag-tagged HuR (HuR-C-Flag). A further deletion including the hinge region (HuR-ΔhRRM3) results in the fusion protein being distributed in both the nucleus and the cytoplasm, suggesting that the hinge region is critical for the nuclear localization of HuR.
Figure 1.
The HuR hinge region is essential for nuclear localization. (A) Schematic representation of wild-type and mutant HuR proteins assayed in B. (B) Subcellular localization of wild-type and truncated HuRs. NIH 3T3 cells were transiently transfected with expression vectors containing HuR-C-Flag, HuR-ΔhRRM3-C-Flag, or HuR-ΔRRM3-C-Flag, fixed with 3% paraformaldehyde for 20 min, permeabilized with 0.5% Triton X-100, and stained with monoclonal anti-Flag antibody (Sigma).
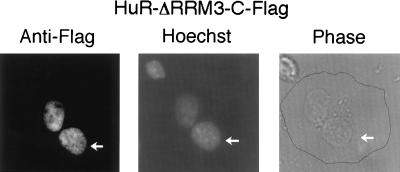
We next analyzed the nucleocytoplasmic shuttling ability of HuR-ΔRRM3-C-Flag by using the transient transfection interspecies heterokaryon assay previously described (18). HeLa cells transiently transfected with HuR-ΔRRM3-C-Flag were fused with mouse NIH 3T3 cells to form heterokaryons in the presence of the protein synthesis inhibitor cycloheximide. As shown in Fig. 2, 3 hr after fusion, HuR-ΔRRM3-C-Flag is present in both the mouse and human nuclei of heterokaryons as was previously observed for intact HuR-C-Flag (18). Because the only difference between HuR-ΔhRRM3 and -ΔRRM3 is the hinge region between RRMs 2 and 3, we conclude that this 55-aa region is essential for shuttling as well as for nuclear localization.
Figure 2.
HuR-ΔRRM3 shuttles between the nucleus and the cytoplasm. HeLa cells were transiently transfected with the HuR-ΔRRM3-C-Flag expression vector, fused with mouse NIH 3T3 cells to form heterokaryons by using 50% polyethylene glycol, and incubated for another 3 hr in the presence of the protein synthesis inhibitor cycloheximide. The coculture was then fixed and stained by using the anti-Flag mAb with costaining by Hoechst dye 33258, which distinguishes the human from the mouse nuclei (indicated by arrows).
The HuR Hinge Region Contains a Nuclear Localization Signal.
Because HuR-ΔRRM3 still contains two RRMs that can bind RNA and may therefore lead to passive shuttling of HuR, we used a non-RNA-binding reporter protein to study the shuttling activity conferred by the HuR hinge region. We first investigated its nuclear localization activity by using a myc-tagged cytoplasmic protein PK as the reporter. This system had been used previously to study the M9 NLS activity of hnRNP A1 (35). The 55-aa full-length HuR hinge region was cloned in-frame into the C terminus of myc-tagged PK to produce expression vector PK-M1 (see Fig. 3A). When transiently transfected into mouse NIH 3T3 cells, this sequence was able to direct the otherwise cytoplasmic PK reporter protein into the nucleus (Fig. 3B), confirming that HuR’s hinge region possesses NLS activity independent of RNA binding.
To further delineate the nuclear localization sequence within this 55-aa region, we made a series of N- and C-terminal deletions (Fig. 3A). First, we focused on the two clusters of basic amino acids near the N terminus of the hinge (amino acids 205–206 and 217–219) and assayed PK-M3, -M4, and -M5. To our surprise, each of these constructs was cytoplasmic (Fig. 3B, and data not shown for -M3 and -M5), indicating that these basic amino acids are not sufficient to target a fusion protein to the nucleus. Fusion proteins containing N-terminal deletions of the hinge region, PK-M6 and -M7, on the other hand, were nuclear, suggesting that the NLS resides downstream of amino acid 205. We then made further N- and C-terminal deletions PK-M8 to -M12. Among these fusions, only PK-M10, containing residues 205–237, RRFGGPVHHQAQRFRFSPMGVDHMSGLSGVNVP, was exclusively nuclear. Deletions from the N or C terminus of PK-M10, as in -M8, -M9, and -M11, result in fusion-protein localization mostly in the cytoplasm (Fig. 3B, and data not shown for PK-M9) or in both nucleus and cytoplasm (for PK-M12, see Fig. 3B). These results suggest that M10 comprises the minimal nuclear localization sequence in HuR.
The HuR Nuclear Localization Signal Also Contains Nuclear Export Activity.
Whereas heterokaryon experiments indicated that the HuR hinge region contains nucleocytoplasmic shuttling activity (Fig. 2), deletion analysis using the PK reporter system revealed an NLS signal within this region (Fig. 3). To define the shuttling signal within the hinge region, we asked whether any of the HuR sequences sufficient for nuclear localization—M1, M7, or M10—also possess NES activity. We utilized the NPc reporter system, which has been used previously to study the NES activity of shuttling signals M9 and KNS in hnRNP A1 and K, respectively (36, 37). We fused HuR sequences M1, M7, and M10 to the C terminus of myc-tagged NPc (Fig. 4A), and performed HeLa-3T3 cell heterokaryon experiments. As shown in Fig. 4B, all of these sequences, including the shortest (M10), are sufficient to direct the fusion protein across the HeLa nuclear membrane into the cytoplasm and subsequently into the 3T3 cell nucleus. The negative control, NPc-NLS, which contains the classical NLS of SV40 T antigen, remains confined to the HeLa nucleus. We therefore conclude that M10 (amino acids 205–237) is the minimal shuttling sequence for HuR and designate it as HNS (HuR Nucleocytoplasmic Shuttling sequence).
The NLS Activity of HNS Depends on the Transcriptional Activity of pol II.
HNS does not exhibit sequence homology to M9 or KNS (35, 37) but does contain two clusters of basic amino acids near its N terminus (amino acids 205–220). Deletion of the first three amino acids (RRF in PK-M11, Fig. 3) abolished the NLS activity of HNS, suggesting that the first cluster of basic amino acids is important for nuclear targeting.
To compare the nuclear localization activity of HNS with that of the classical NLS, M9, and KNS sequences, we investigated the subcellular localization of myc-tagged NPc fusion proteins containing HuR sequences M1, M7, and M10 (HNS) in cells in which pol II transcription had been inhibited with actinomycin D. Cells were transfected with DNA constructs encoding the fusion proteins and were treated with actinomycin D at 5 μg/ml. As shown in Fig. 5, each of these NPc fusion proteins containing portions of the HuR shuttling sequence HNS appears in the cytoplasm in the presence of actinomycin D, whereas NPc-NLS is nuclear. Thus, the two clusters of HuR basic amino acids within HNS do not retain the fusion proteins in the nucleus, indicating that the NLS activity of HNS is different from the classical NLS (31). Instead, HNS is similar to M9 and KNS in that its NLS activity is pol II transcription-dependent (35, 37).
Figure 5.
The NLS activity of HuR depends on pol II transcription. The HuR-nucleoplasmin fusion proteins NPc-M1, -M7, and -M10, as well as the negative control NPc-NLS, were transiently transfected into mouse NIH 3T3 cells, incubated with actinomycin D at 5 μg/ml for 3 hr, and fixed and subjected to immunofluorescence staining by using the anti-myc 9E10 antibody.
DISCUSSION
The RNA binding protein HuR has been shown to stabilize ARE-containing mRNAs (18–20) and shuttle between nucleus and cytoplasm (18, 19). To identify the HuR shuttling domain, we tested two HuR mutants, HuR-ΔhRRM3 and HuR-ΔRRM3, and found that shuttling activity resides in the hinge region between RRMs 2 and 3. We then assayed a series of PK fusion proteins containing sequences from the hinge region and determined that the minimal nuclear localization sequence resides in amino acids 205–237 of HuR. Finally, we investigated the NES activity of the HuR nuclear localization sequence by using the NPc reporter system and established that the minimal NLS also is able to direct fusion-protein shuttling. We designate this shuttling sequence HNS (for HuR Nucleocytoplasmic Shuttling).
The HuR shuttling signal HNS lies within the amino acid sequence that is the least conserved in the four Hu family proteins (refs. 28–30; Fig. 6A). The other three neuron-specific Hu proteins, Hel-N1 (HuB), HuC, and HuD all have insertions in the hinge region between RRMs 2 and 3 (Fig. 6A). This provides a good explanation for why the Hu family proteins appear to have different subcellular localization: HuR is nuclear and shuttles between the nucleus and cytoplasm (18, 19), whereas Hel-N1 (HuB), HuC, and HuD have been detected in both the nuclei and the cytoplasm of cells in the central and peripheral nervous system (22–26). Furthermore, Hel-N1 RNPs have been reported to be associated with tubulin, decorating the microtubules of medulloblastoma cells (43).
Figure 6.
Sequence comparisons of HNS. (A) Amino acid alignment of the hinge region of Hu family proteins. The hinge-region amino acid sequences of human Hu proteins Hel-N1 (HuB), HuC, and HuD and the Xenopus HuR homolog ElrA are compared with human HuR (28, 29). Residues identical to the HuR consensus are shown in dark gray, whereas conservative substitutions are shaded light gray. (B) Amino acid alignment of human HuR HNS and human hnRNP A1 M9 (36). Identical residues are shown in dark gray. The Gly-274 residue that is essential for shuttling in M9 has been indicated by an arrow.
It is not clear at present whether the HuR HNS shares a transport factor(s) with any of the previously investigated NLS, NES, or shuttling sequences. Studies of the Xenopus HuR homolog, ElrA, suggested the involvement of trans-acting factors that are regulated through development (44). ElrA is >90% identical to HuR at the amino acid level; in its hinge region it contains a sequence almost identical to HNS, except for two Gly-to-Ser changes at residues 230 and 233 and a substitution of Ala for Pro at position 237 (Fig. 6A). Yet, unlike HuR, ElrA is present predominantly in the oocyte cytoplasm, where it contributes to the polyadenylation of certain maternal mRNAs containing the embryonic-type cytoplasmic adenylation element (44). We therefore propose that one or more of the trans-acting factors essential for HuR shuttling in mature, differentiated mammalian cell lines may be absent during Xenopus early development. Alternatively, it is possible that the two Gly residues at positions 230 and 233 of HuR are absolutely essential for its shuttling activity, which remains to be investigated.
Different shuttling/NLS/NES sequences are recognized by different trans-acting protein factors. The classical NLS is bound by the importin α/β complex, docked to the nuclear pore complex, and translocated to the nucleoplasm through the Ran GTPase cycle in a temperature- and transcription-independent manner (for review, see refs. 32–34). The CRM 1 (Chromosomal Region Maintenance) protein, which exhibits homology to importin β, has been shown recently to be responsible for intracellular transport mediated by the NES; Ran GTPase also is involved (45–47). In the case of M9, Ran GTPase and transportin (which is related to importin β) are required for protein translocation through the nuclear pore complex (40, 41, 48–50). These factors appear to be non-tissue-specific and are conserved among vertebrates, with homologs existing even in yeasts (50). Some of them, such as Ran GTPase, are shared by several different types of protein localization signals.
Although there is no extensive sequence homology between HNS and M9, HNS does have a high glycine and serine content (like M9), and the sequence at its C terminus exhibits several amino acid similarities to the N-terminal half of M9 (Fig. 6B). Mutational analysis has shown that Gly-274 of M9, which is absolutely conserved, is essential for shuttling; its replacement by alanine abolishes both NLS and NES activities (36). However, at the comparable position in HNS (amino acid 221), there is a serine residue instead of glycine (Fig. 6B), suggesting that HNS is a unique shuttling sequence with a transport mechanism potentially different from that of M9.
Proteins containing the two types of previously well characterized shuttling sequences (M9 and KNS) are all hnRNPs, whose nucleocytoplasmic shuttling is important for general mRNA export. HuR appears to be the first shuttling/RNA-binding protein that is involved in the metabolism of a specific group of mRNAs. Overexpression of HuR has been shown to retard ARE-conferred mRNA decay (18–20), which is believed to occur in the cytoplasm. However, both endogenous and overexpressed HuR were found to be predominantly localized in the nucleus (18), raising the question of where HuR functions as a stabilizer. Our finding that HuR shuttles between nucleus and cytoplasm suggests a solution for this apparent paradox: HuR may first bind to ARE-containing mRNAs in the nucleus and translocate with them to the cytoplasm, providing continued protection. The identification of HNS provides supporting evidence for our hypothesis. Future introduction of mutations specifically within the HNS as opposed to the three RRMs should establish the importance of shuttling versus RNA binding to HUR’s functioning in cellular mRNA metabolism.
Acknowledgments
We are grateful to Dr. T. Gelehrter and members of his laboratory for providing reagents, sharing equipment, and offering helpful suggestions. We thank Drs. N. Maizels, G. Dreyfuss, J. Keene, and I. Gallouzi for comments on the manuscript; Dr. G. Dreyfuss, H. Siomi, and S. Nakielny for providing the anti-myc antibody 9E10; Dr. T. Glover for sharing a microscope; and P. Dillon for help preparing the manuscript. This work was supported by Grant CA 16038 from the National Institutes of Health. J. Steitz is an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- NLS
nuclear localization signal
- NES
nuclear export signal
- ARE
AU-rich element
- RRM
RNA recognition motif
- pol II
RNA polymerase II
- PK
pyruvate kinase
- NPc
nucleoplasmin
- HNS
HuR nucleocytoplasmic shuttling sequence
- hnRNP
heterogeneous nuclear ribonucleoprotein
References
- 1.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 2.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belasco J, Brawerman G. Control of Messenger RNA Stability. San Diego: Academic; 1993. [Google Scholar]
- 4.Chen C-Y A, Shyu A-B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 5.Malter J S. Science. 1989;246:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 6.Malter J S, Hong Y. J Biol Chem. 1991;266:3167–3171. [PubMed] [Google Scholar]
- 7.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. J Biol Chem. 1992;267:6302–6309. [PubMed] [Google Scholar]
- 9.Brewer G. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton B J, Nagy E, Malter J S, Arrick B A, Rigby W F C. J Biol Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- 12.Hamilton B J, Burns C M, Nichols R C, Rigby W F C. J Biol Chem. 1997;272:28732–28741. doi: 10.1074/jbc.272.45.28732. [DOI] [PubMed] [Google Scholar]
- 13.Katz D A, Theodorakis N G, Cleveland D W, Lindsten T, Thompson C B. Nucleic Acids Res. 1994;22:238–246. doi: 10.1093/nar/22.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vakalopoulou E, Schaack J, Shenk T. Mol Cell Biol. 1991;11:3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myer V E, Lee S I, Steitz J A. Proc Natl Acad Sci USA. 1992;89:1296–1300. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myer V E, Fan X C, Steitz J A. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan X C, Myer V E, Steitz J A. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan X C, Steitz J A. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng S S-Y, Chen C-Y A, Xu N, Shyu A-B. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy N S, Chung S, Furneaux H, Levy A P. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 21.Ma W-J, Cheng S, Campbell C, Wright A, Furneaux H. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 22.Dalmau J, Graus F, Rosenblum M K, Posner J B. Medicine. 1991;71:59–72. doi: 10.1097/00005792-199203000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Posner J B. Perspect Biol Med. 1995;38:167–181. doi: 10.1353/pbm.1995.0043. [DOI] [PubMed] [Google Scholar]
- 24.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner J B, Furneaux H M. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 25.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell R B. Proc Natl Acad Sci USA. 1996;93:4529–4536. doi: 10.1073/pnas.93.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 28.Good P. Proc Natl Acad Sci USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano H J, Darnell R B. J Neurol. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wakamatsu Y, Weston J A. Development (Cambridge, UK) 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- 31.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 32.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 33.Nakielny S, Dreyfuss G. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 34.Izaurralde E, Mattaj I W. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 35.Siomi H, Dreyfuss G. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 37.Michael W M, Eder P S, Dreyfuss G. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinol-Roma S, Dreyfuss G. Science. 1992;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- 39.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 40.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izaurralde E, Kutay U, Von Kobbe C, Mattaj I W, Gorlich D. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moser F G, Dorman B P, Ruddle F H. J Cell Biol. 1975;66:676–680. doi: 10.1083/jcb.66.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antic D, Keene J D. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, Good P J, Richter J D. Mol Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 46.Fornerod M, Ohno M, Yoshida M, Mattaj I. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 47.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 48.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 49.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 50.Siomi M C, Fromont M, Rain J-C, Wan L, Wang F, Legrain P, Dreyfuss G. Mol Cell Biol. 1998;18:4141–4148. doi: 10.1128/mcb.18.7.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]