Abstract
In mammals, glucose-dependent insulinotropic polypeptide (GIP) is synthesized predominately in the small intestine and functions in conjunction with insulin to promote nutrient deposition. However, little is known regarding GIP expression and function in early vertebrates like the zebrafish, a model organism representing an early stage in the evolutionary development of the compound vertebrate pancreas. Analysis of GIP and insulin (insa) expression in zebrafish larvae by RT-PCR demonstrated that although insa was detected as early as 24 h postfertilization (hpf), GIP expression was not demonstrated until 72 hpf, shortly after the completion of endocrine pancreatic development but prior to the commencement of independent feeding. Furthermore, whole mount in situ hybridization of zebrafish larvae showed expression of GIP and insa in the same tissues, and in adult zebrafish, RT-PCR and immunohistochemistry demonstrated GIP expression in both the intestine and the pancreas. Receptor activation studies showed that zebrafish GIP was capable of activating the rat GIP receptor. Although previous studies have identified four receptors with glucagon receptor-like sequences in the zebrafish, one of which possesses the capacity to bind GIP, a functional analysis of these receptors has not been performed. This study demonstrates interactions between the latter receptor and zebrafish GIP, identifying it as a potential in vivo target for the ligand. Finally, food deprivation studies in larvae demonstrated an increase in GIP and proglucagon II mRNA levels in response to fasting. In conclusion, the results of these studies suggest that although the zebrafish appears to be a model of an early stage of evolutionary development of GIP expression, the peptide may not possess incretin properties in this species.
Keywords: incretin hormones, enteroinsular axis, endocrine pancreas
glucose-dependent insulinotropic polypeptide (GIP) was identified as a 42-amino acid mammalian gastrointestinal regulatory peptide synthesized in endocrine K-cells located in the mucosa of the small intestine (1, 2). GIP has been designated as an incretin hormone by virtue of its ability to stimulate insulin release from pancreatic islet β-cells (9, 10, 16, 30). GIP is released into the circulation principally following the ingestion of carbohydrates and fats (6, 19) and, to a lesser extent, after the administration of a protein hydrolysate or specific amino acids (5, 27, 32). During the fasting state, circulating levels of GIP are low, and GIP release occurs within minutes of nutrient ingestion, immediately prior to insulin secretion, reaching a peak in 15–30 min before returning to basal levels 3 h after stimulation (8). Previous studies demonstrated that GIP accounts for ∼70% of nutrient-induced insulin release, thereby accounting for its designation as a major physiological incretin (11, 29). However, whereas the biological effects of GIP have been well characterized in mammals, little is known regarding its role in early vertebrates.
It has been postulated that during evolution islet cells translocated from the gastrointestinal tract, leading to the development of a distinct pancreas to protect islet cells from potentially harmful contaminants in food such as bacterial pathogens, which could severely damage the intestinal mucosa. Any ensuing destruction of insulin-producing β-cells could thereby lead to serious metabolic consequences and ultimately to the organism's death. As a result, communication between the gastrointestinal tract and the translocated endocrine pancreas, designated the enteroinsular axis (29), became necessary and was accomplished by the development of GIP and other incretins (25).
The zebrafish represents an appropriate model organism representing an early stage in the evolutionary development of the compound pancreas of vertebrates, and we accordingly selected this organism to characterize the expression pattern and function of GIP and to elucidate when during evolution incretins may have developed. We have analyzed the expression patterns of GIP and insulin in both larval and adult zebrafish and have indirectly assessed zebrafish GIP (zfGIP) functional activity by examining receptor activation and the effect of food deprivation on mRNA levels. Our studies indicate that, in contrast to mammals, GIP is expressed principally in the pancreas of zebrafish. Moreover, although zfGIP activates the rat GIP receptor (GIPR) in vitro, the native target of GIP binding appears to be a putative glucagon receptor. Finally, our results are consistent with the hypothesis that although a primitive form of GIP is expressed in the zebrafish, its location in the pancreas, along with its pattern of expression and binding, indicates that the peptide has not assumed its mammalian role as an incretin hormone.
METHODS
Animals.
Adult male and female zebrafish (Danio rerio, AB wild-type strain) were housed in 14:19-h light-dark cycle at a temperature of (26.5°C) and a pH of (7.0–7.4) in a controlled multitank recirculating water system (Aquaneering, San Diego, CA). Fish were fed twice daily with live brine shrimp (Brine Shrimp Direct, Ogden, UT) and flake food (TetraMin; Tetra, Blacksburg, VA). The experimental protocol was approved by the Boston University School of Medicine animal welfare committee.
RNA isolation and RT-PCR.
After homogenization with a PowerGen 125 rotor-stator homogenizer (Fisher Scientific, Pittsburgh, PA), total RNA was extracted from zebrafish larvae and adult tissues using RNeasy as recommended by the manufacturer (Qiagen, Valencia, CA). cDNA was synthesized from 5 μg of mRNA and 50 pmol of oligo(dT) primer using the Thermoscript RT-PCR system (Invitrogen, Carlsbad, CA), and one-tenth of this reaction volume was then used for analysis in subsequent PCR reactions. The reactions were carried out in a 50-μl volume containing 25 units Taq DNA polymerase (Roche, Mannheim, Germany). Primers used for the amplification of GIP and insulin are listed in Table 1.
Table 1.
Primer list for PCR reactions
| Gene | Forward Primer | Reverse Primer | Ta | Cycle No. |
|---|---|---|---|---|
| zfGIP | GTTTGATTTGCCTTGGAAGTGTCTGG | CATTCTAGATACATAACAAGACACTTGAC | 65° | 30 |
| zf insa | CCACCATTCCTCGCCTCTGC | GAGCATTAAGGCCTGTGTGCAA | 60° | 30 |
| zfGIP, qPCR | TTAAGAGCAGACCCAGGTCC | CTGAACCATCGAGTCCACAA | 60° | 30 |
| zf insa, qPCR | ACCATGGCAGTGTGGCTTCA | CAAAGTCAGCCACCTCAGTT | 60° | 30 |
| zf proglucagon II, qPCR | TTTGCGTCCCACTGCAAGGCGA | GTGTAGGTGCCCTATGCATCA | 60° | 30 |
| zfβ-actin, qPCR | GGCACGAGAGATCTTCACTC | CTGAGCCTCATCTCCCACAT | 60° | 30 |
| zfGIP, in situ probe | AGCTATTTAGGTGACACTATAGGTTTTGATTTGCCTTGGAAGTGTCTGG | TAATACGACTCACTATAGGGCATTCTAGATACATAACAAGACACTTGAC | 65° | 30 |
| zfinsa, in situ probe | AGCTATTTAGGTGACACTATAGCCACCATTCCTCGCCTCTGC | TATTACGACTCACTATAGGGGAGCATTAAGGCCTGTGTGCAA | 65° | 30 |
| zfGR, putative glucagon R | ATAGGTACCATGACTGGCCCCTGGTACCTGT | ATAGAATTCTCAGCAGTAGCTCTCGGAATA | 60° | 30 |
| zfGLP-2R | GGGGATCCTCCCAGCAGCAGCAGTACAG | CCGAATTCCTAGAACTCGCTCTCCTCGAT | 60° | 30 |
| zf proglucagon II, cloning | AAAAGCTTGGAAGACTCAGCTGTTAAAATG | AAGAATTCCTACTCTTGCTTGGGCTGTC | 60° | 30 |
| zfGIP, cloning | GGAAGCTTATACACCAGTTTAAGAGCAGACCC | AAAGAATTCCATGCATTATTGATTGAAAGTG | 60° | 30 |
Ta, annealing temperature; zf, zebrafish; GIP, glucose-dependent insulinotropic polypeptide; insa, GIP and insulin; qPCR, quantitative RT-PCR; GR, glucagon receptor; GLP-2R, GLP-2 receptor.
Collection of fed and fasted larvae.
Adult zebrafish were subjected to timed matings, and embryos were collected the morning after mating and divided into groups of 100. Starting at 6 days postfertilization (dpf), larvae were either fed paramecia four times per day or were fasted for 24–60 h. Larvae were harvested 1 h after feeding time (age ranging from 7 dpf to 8.5 dpf) and subsequently at 4-h intervals at which time they were flash frozen in liquid nitrogen and stored at −80°C until RNA isolation. Samples were analyzed using quantitative RT-PCR. The first group of fed fish (7.2 dpf, 28 h postfeeding) collected was used to calculate baseline expression, and was designated as the control group. GIP, glucagon, and insulin transcript levels were corrected for β-actin expression to control for variations in RNA preparations and cDNA synthesis reactions, and they were then expressed relative to the control group (28 h postfeeding).
Real-time RT-PCR.
RNA was analyzed using real-time RT-PCR (quantitative PCR) for the expression of GIP, insa, proglucagon II, and β-actin. First-strand synthesis was performed as described above, and PCR reactions were performed following the manufacturer's directions using primer (Invitrogen) and PCR conditions listed in Table 1. Reactions were carried out in a DNA Engine Opticon 2 cycler (Bio-Rad, Hercules, CA), and the expression level of each target gene was measured using β-actin as a control for loading.
Whole mount in situ hybridization.
cDNA for probe generation was isolated from zebrafish internal organs as described above. Primers used to generate the GIP and insulin probes were synthesized by Invitrogen with either a T7 or Sp6 consensus binding site at the 5′ end. Primers sequences are shown in Table 1. PCR products were amplified, isolated, and purified using the QIAQuick Gel Extraction Kit following the manufacturer's instructions (Qiagen). Digoxigenin-labeled sense and antisense RNA probes were synthesized using a Sp6/T7 labeling kit (Roche). Whole mount in situ hybridization of zebrafish larvae was performed using a modification of a previously described protocol (28). Larvae were depigmented by treatment with a 3% H2O2/0.5% KOH solution until they became transparent (1 to 2 h), and proteinase K digestion was performed for 30 min.
Immunohistochemistry.
Adult zebrafish were anesthetized with MS222 and killed for subsequent immunohistochemistry. Isolated tissue was fixed overnight in 4% paraformaldehyde and cryoprotected in 30% sucrose/PBS overnight. Samples were then embedded in Tissue-Tek OCT Compound (Sakura, Torrance, CA) and frozen in 2-methylbutane. Sections were cut at 7 microns using a cryostat (Spencer Scientific, Derry, NH) and stored at −80°C until incubation with antibodies to GIP, insulin, and amylase; antibodies and corresponding secondary antibodies are listed in Table 2.
Table 2.
Antibodies for immunohistochemistry
| Antigen | Species | Concentration | Company |
|---|---|---|---|
| GIP | Rabbit anti-zebrafish | 1:200 | New England Peptide |
| Insulin | Goat anti-human | 1:200 | Santa Cruz Biotechnology |
| Amylase | Rabbit anti-human | 1:400 | Santa Cruz Biotechnology |
Before staining, sections were allowed to equilibrate to room temperature and then fixed in 4% PFA, followed by several washes. The fixation reaction was quenched using 50 mM glycine pH 7.4. Sections were permeabilized with 0.25% Triton X-100/PBS, blocked in 5% BSA for 2 h, and then incubated with primary antibody overnight at 4°C at the indicated concentrations (Table 3). On the following day, sections were washed in PBS and then incubated with the appropriate secondary antibodies listed in Table 1 for 1 h. Sections were then washed and mounted under a coverslip with UltraCruz Mounting Media containing 1.5 μg/ml DAPI (Santa Cruz Biotechnology, Santa Cruz, CA) and examined under a fluorescent microscope. Rabbit anti-zfGIP (1:200) antiserum was custom made by New England Peptide, and goat anti-human insulin (1:200) and rabbit anti-human amylase (1:400) antisera were purchased from Santa Cruz Biotechnology. Photomicrographs were obtained using SPOT analysis software (Diagnostic Instruments).
Table 3.
Antibody concentrations
| Species | Conjugate | Concentration | Company |
|---|---|---|---|
| Donkey anti-rabbit | Cy3 | 1:500 | Jackson ImmunoResearch |
| Donkey anti-goat | Cy3 | 1:500 | Jackson ImmunoResearch |
| Donkey anti-rabbit | FITC | 1:250 | Jackson ImmunoResearch |
| Donkey anti-goat | FITC | 1:250 | Jackson ImmunoResearch |
Putative zebrafish glucagon receptor, zebrafish GLP-2 receptor, zebrafish proglucagon II, and zfGIP.
Sequences for one of the putative zebrafish glucagon receptors (zfGR; XM_685886), predicted zebrafish glucagon-like peptide-2 receptor (zhGLP-2R; XM_001332426), zebrafish proglucagon II (AJ133697), and zfGIP (EF010535) were obtained from the NCBI website. To allow cloning of full-length cDNA sequences, primers were designed with restriction sites on the 5′ ends; zfGR (KpnI and EcoRI), GLP-2R (BamHI and EcoRI), and proglucagon II and GIP (HindIII and EcoRI) (Table 1). cDNA sequences were amplified from zebrafish pooled internal organ RNA by RT-PCR and cloned into the pcDNA3 expression vector, and inserts were sequenced to confirm identity.
zfGR and zfGLP-2R bioassay.
The putative zfGR and zfGLP-2R expression plasmids were transfected into LVIP cells, and clonal cell lines were created by selecting cells 24 h later with G418 sulfate and hygromycin at concentrations of 450 μg/ml and 250 μg/ml, respectively. Bioassays were performed as previously described (15), with the exception that bioassay cells were plated on 96-well plates. Absorbance was read at a wavelength of 570 nm with an EXL 800 plate reader (BioTek, Winooski, VT).
Cell culture.
Cell lines (LVIP and LGIPR2, Dr. Ted Usdin, Bethesda, MD; GH3, American Type Tissue Collection, Bethesda, MD; zfGR; and zfGLP-2R) were grown in DMEM (MediaTech, Manassas, VA) containing 10% FBS (Hyclone, Logan, UT) at 37°C in an atmosphere of 5% CO2 in the presence of 100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (MediaTech). Bioassay cell lines were grown in the presence of 450 μg/ml of G418 sulfate (MediaTech) and 250 μg/ml of hygromycin B (Invitrogen). LVIP cells are a mouse fibroblast line possessing a VIP/β-galactosidase expression plasmid stably integrated into its genome.
Transient transfections.
One day prior to transfection, cells were plated in 6-well plates at a density of ∼1–2 × 105 cells per well. A mixture containing 1 μg of expression plasmid, 3 μl of Fugene6 (Roche), and 100 μl serum-free medium was incubated at room temperature. After 15 min, cells were washed with 2.0 ml of DMEM containing 10% FBS, and 100 μl of the DNA/Fugene mixture were added to each well. After 24 h, the medium was replaced with DMEM containing 10% FBS.
Preparation of conditioned medium.
GH3 cells plated in 6-well plates were transfected with the appropriate expression plasmids. After 24 h, medium was replaced with 2 ml serum-free DMEM. The following day, medium containing secreted peptides was collected and assessed using various receptor bioassays.
Receptor bioassay.
Bioassay cells were plated on to 96-well plates at ∼80% confluency per well and allowed to grow overnight in DMEM containing 10% FBS. The following day, the cells were washed and then incubated with either standard peptides diluted in serum-free DMEM or 100 μl of conditioned media. All samples were analyzed in triplicate. After a 6 h incubation, the cells were washed twice with PBS and placed at −80°C overnight. The following day, plates were removed from the freezer and incubated for 10 min at 37°C in 0.01 M sodium phosphate pH 8.0, 0.2 mM MgSO4 and 0.01 mM MnCl2. After the addition of 200 μl PM2 buffer (0.4% Triton X-100, 0.1 M sodium phosphate pH 8.0, 2 mM MgSO4, 0.1 M MnCl2), cell lysates were mixed by tapping the plate, and 50 μl of a 5 mg/ml solution of chloramphenicol red-β-d-galactopyranoside (Roche Molecular Bioanalyticals, Indianapolis, IN) were added. Plates were incubated at 37°C until the color was sufficiently developed, and absorbance was read at a wavelength of 570 nm with an EXL 800 plate reader (BioTek).
Competitive cAMP ELISA.
GH3 cells were plated on 48-well plates, allowed to attach overnight, and then serum starved for 24 h. Cells were then pretreated for 1 h with 1× IBMX, and incubated with 10−5 to 10−12 M of either zebrafish or porcine GIP for 10 min, in the presence or absence of the GIPR antagonist ANTGIP (10−5 M). Assays were performed in triplicate following the manufacturer's instructions (Millipore, Billerica, MA). Plates were read for 1 s using a POLARstar OPTIMA luminometer (BMG Labtech, Durham, NC).
Statistical analysis.
Data were analyzed using Student's t-test for unpaired samples, and statistical significance was assigned for P < 0.05.
RESULTS
GIP is expressed in the developing zebrafish larvae.
Before analysis was started, zebrafish and mammalian GIP amino acid sequences were evaluated to determine the level of identity present. The peptide sequences of human, mouse, and rat GIP are 90% identical. To determine the amino acid sequence similarity between zebrafish and mammalian GIP, sequences were aligned using ClustalW. Because mammalian GIP contains 42-amino acids, the first 42-amino acids of each posttranslational product were compared. Therefore, amino acids predicted or known to be cleaved during processing of the mature peptide are delineated in gray lettering (Fig. 1). As demonstrated in Fig. 1, alignment of mature peptides showed an overall identity between mammalian GIP and zfGIP of 36% at the amino acid level. However, if only the first 31 amino acids are examined, the length of predicted zfGIP, the degree of identity increases to 48%. Whether this degree of homology is sufficient to conclude that the identified zebrafish peptide, in fact, represents GIP cannot be established with certainty, and this was examined in the remainder of this manuscript.
Fig. 1.
Alignment of vertebrate incretin hormone sequences. A: human (NM_004123), rat (NM_019630), mouse (NM_008119), chicken (EF010535), frog (EF010533), and zebrafish (EF010535) glucose-dependent insulinotropic polypeptide (GIP) protein sequences were aligned using the ClustalW alignment program.
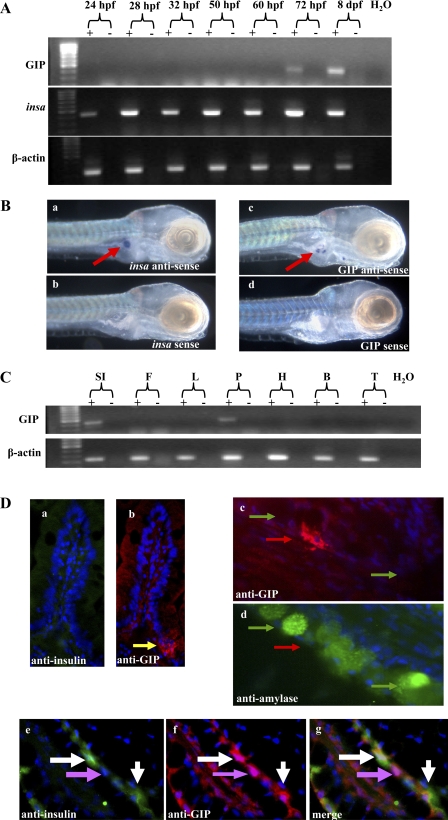
Under the temperature conditions employed in conventional developmental studies (28°C), the majority of zebrafish hatch at ∼72 h postfertilization (hpf), commence independent feeding at ∼5 dpf, and reach sexual maturity between 10 and 12 wk (24, 31). Zebrafish were analyzed using RT-PCR at developmental time points ranging from 24 hpf to 8 dpf, and in situ hybridization was performed to determine the relationship between GIP and insulin (insa) gene expression. Although the zebrafish possesses coding sequences for two distinct insulin genes, we elected to focus on insa, which has previously been shown to be pancreatic-specific, whereas expression of the second transcript, insb, has been demonstrated in both the brain and the pancreas (18). GIP expression was first evident at 72 hpf and was shown to increase at 8 dpf (Fig. 2A, top), confirming that GIP expression begins shortly after the completion of endocrine pancreatic development (∼60 hpf), but prior to the commencement of independent feeding (∼5 dpf). In contrast, insa expression (Fig. 2A, middle) was detected at all time points analyzed, consistent with previous studies in which zebrafish insa transcripts were detected as early as 1 hpf, most likely representing maternal transcripts important for proliferation. Figure 2A, bottom demonstrates β-actin transcripts in all larval samples analyzed, confirming cDNA integrity.
Fig. 2.
Analysis of GIP and insulin expression in zebrafish. A: zebrafish eggs/larvae were collected after timed matings. Total RNA was extracted, and cDNA was used as a template for PCR reactions to examine GIP (top), insa (middle), and β-actin (bottom) expression. B: zebrafish larvae were examined using whole mount in situ hybridization. Larvae were hybridized with insa antisense probe (a), insa sense probe (b), GIP antisense probe (c), or GIP sense probe (d). Red arrows indicate insa or GIP RNA expression. C: zebrafish adult tissues were assayed for GIP and β-actin expression. SI, small intestine; F, fat; L, liver; P, pancreas; H, heart; B, brain; T, tail. D: immunohistochemical analysis of adult zebrafish tissue. Intestinal sections were probed with goat anti-human insulin antiserum (a, e), rabbit anti-zebrafish GIP antiserum (b, c, f), or rabbit anti-human amylase antiserum (d). A merged image showing coexpression of GIP and insulin is shown in g. Yellow arrow in b indicates the location of GIP-positive cells. In c and d, green arrows in indicate amylase-positive cells and red arrows indicate GIP-positive cells. Purple arrows (e, f, g) indicate GIP-positive, insulin-negative cells. White arrows (e, f, g) indicate cells positive for both GIP and insulin. Nuclei (blue) were counterstained with DAPI. zfGIP, zebrafish GIP.
After the examination of temporal expression patterns, whole mount in situ hybridization was performed to compare the relative localization of GIP and insa transcripts in zebrafish larvae. Insa RNA was detected in a region consistent with pancreatic expression (Fig. 2B-a), and hybridization with a zfGIP α-sense probe (Fig. 2B-c) detected multiple small areas of expression in the same region. In contrast, in situ hybridization with zebrafish insa (2B-b) or GIP (Fig. 2B-d) sense probes demonstrated no expression, confirming the specificity of our probes for their respective targets.
Spatial relationship between GIP and insulin expression in the adult zebrafish.
Next we analyzed the expression pattern of GIP in adult zebrafish by RT-PCR and immunohistochemistry. Several tissues, including the intestine, pancreas, adipocytes, liver, heart, brain, and tail, were analyzed by RT-PCR for the presence of GIP transcripts. Fig. 2C demonstrates the detection of GIP transcripts in only the intestine and pancreas.
Adult zebrafish frozen tissue sections were subjected to immunohistochemistry using GIP, insulin, and amylase antisera. GIP expression was detected in the zebrafish intestine (Fig. 2D-a), confirming RT-PCR results. However, whereas GIP is expressed in K-cells throughout villi in the mammalian small intestine, GIP expression in the adult zebrafish was limited to isolated cells at the villous base. In addition, insulin expression was not detected in the zebrafish intestine (Fig. 2D-b).
We next examined GIP and insulin expression in the zebrafish pancreas. To establish whether GIP expression in the pancreas is confined to endocrine cells, we compared expression patterns of amylase, a known pancreatic-specific gene expressed in pancreatic exocrine cells, and GIP with serial sections. Although both GIP (red; Fig. 2D-c) and amylase (green; Fig. 2D-d) were expressed in the pancreas, expression was detected in different cellular subsets, suggesting that GIP expression, like insulin, is confined to the endocrine cells of the adult zebrafish pancreas.
Finally, adult zebrafish internal organ sections were double labeled with antisera to insulin and GIP to determine colocalization. Both insulin (Fig. 2D-e) and GIP (Fig. 2D-f) were expressed in the pancreas, and the merged image depicted in Fig. 2D-g demonstrated colocalization of GIP and insulin expression. These results indicate that the two peptides are both expressed individually and coexpressed in a subset of pancreatic endocrine cells.
zfGIP binds and activates the rat GIPR but not the rat GLP-1R.
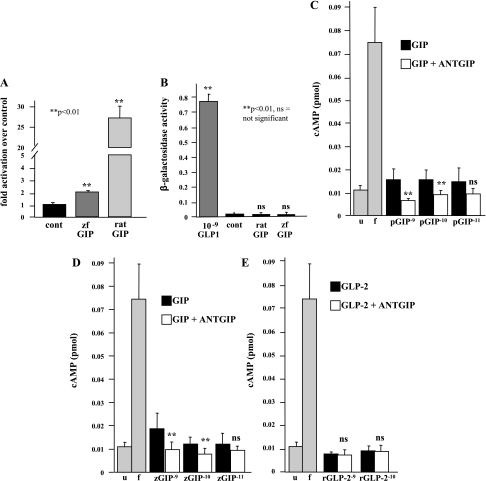
Preliminary searches conducted in our laboratory of the zebrafish genome failed to identify a significant homolog to mammalian GIPR. Therefore, we examined whether zfGIP could activate the mammalian GIPR. Expression plasmids for rat and zfGIP were transfected into GH3 cells. Conditioned media were then collected, and the ability of secreted GIP to activate the rat GIPR was assessed via bioassay. As expected, rat GIP (positive control) activated the rat GIPR. In addition, incubation with zfGIP resulted in a small, but statistically significant, activation of the rat GIPR (Fig. 3A).
Fig. 3.
Analysis of rat GIP receptor (GIPR) and GLP-1R activation by zfGIP. A and B: GH3 cells were transfected with empty vector, zfGIP expression plasmid, or rat GIP expression plasmid. After 48 h, media were collected, and the ability of secreted GIP to activate GIPR (A) or GLP-1R (B) was assessed using specific bioassays. Activation of GIPR was expressed as fold activation over control (A) or raw activity (B). P values were calculated using Student's t-test. C–E: GH3 cells were plated on 48-well plates, incubated in DMEM without FBS overnight and pretreated with 1× IBMX to inhibit endogenous phosphodiesterase activity. Cells were then treated with 10–11 M to 10–9 M porcine GIP (C), 10–11 M to 10–9 M zfGIP (D), or 10–10 M to 10–9 M rat GLP-2 purified peptides (E) in the presence or absence of the GIPR antagonist ANTGIP. In addition, cells were treated with 10 μM forskolin (f) as a positive control, and untreated cells (u) were used as a negative control. Cell lysates were generated, and intracellular cAMP levels were measured using a competitive ELISA. P values were calculated using Student's t-test. **P < 0.05, ns = not significant.
We next examined the specificity of the interaction between zfGIP and the rat GIPR by determining whether zfGIP could activate the closely related rat GLP-1R. It has previously been demonstrated that mammalian GLP-1R is specific for GLP-1 and that mammalian GIP peptides do not activate mammalian GLP-1R. We thus used a second bioassay to examine the ability of zfGIP to activate the rat GLP-1R. As shown in Fig. 3B, neither zebrafish nor rat GIP activated the GLP-1R. In contrast, a synthetic GLP-1 peptide did activate the GLP-1R in a concentration-dependent manner, thereby confirming the integrity of the GLP-1R bioassay.
Previous studies have reported that mammalian GIP signals through cAMP-dependent pathways in numerous cell types. Specifically, activation of GIPRs by mammalian GIP in GH3 cells, a rat pituitary adenoma cell line, has been reported to lead to an increase in intracellular cAMP. Consequently, we evaluated whether activation of the rat GIPR on GH3 cells by zfGIP could increase the accumulation of cAMP. Increasing concentrations of both zebrafish (Fig. 3D) and porcine GIP (Fig. 3C) significantly increase intracellular cAMP. The observed increase in intracellular cAMP levels stimulated by porcine (Fig. 3C) or zebrafish (Fig. 3D) GIP was abolished by the concomitant treatment of GH3 cells with 10−5 M ANTGIP. In contrast, ANTGIP had no effect on GLP-2-generated cAMP levels in GH3 cells (Fig. 3E). These findings indicate that zfGIP possesses sufficient identity with mammalian GIP to specifically activate the mammalian GIPR.
zfGIP binds and activates a zfGR, but not zfGLP-2R.
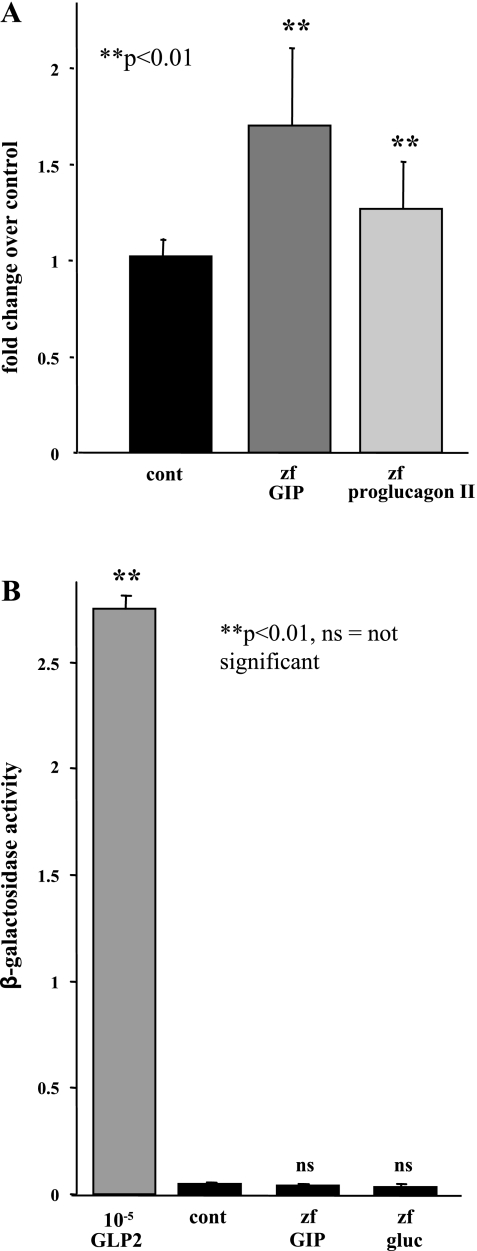
We next performed studies aimed at identifying the native receptor for zfGIP. A receptor previously identified as a putative zfGIPR, but classified by NCBI as similar to glucagon receptor, was identified as having the highest degree of identity to mammalian GIPR. Therefore, we cloned this putative glucagon receptor from zebrafish internal organ cDNA to create a zfGR bioassay cell. To enable us to compare the pattern of receptor activation, we also cloned the cDNA for zebrafish proglucagon II (translated products are glucagon and GLP-1) into the pcDNA3 expression plasmid. Proglucagon II and GIP expression plasmids were transfected into GH3 cells, and the media were assayed for the ability of the secreted peptide to bind to the putative zebrafish glucagon receptor. As demonstrated in Fig. 4A, conditioned media from transfected cells with the zebrafish proglucagon II expression plasmid activated the putative glucagon receptor to a small, but significant, degree. The level of activation of the putative glucagon receptor may have been less than predicted because the expression plasmid encompassed the cDNA for the entire glucagon polyprotein, and the posttranslational processing of zebrafish glucagon may have been incomplete in these cells. zfGIP similarly activated the putative glucagon receptor by ∼70% over control. These data thus suggest that the putative zebrafish glucagon receptor may be a potential target for zfGIP.
Fig. 4.
Analysis of putative zebrafish glucagon (zh gluc) receptor and GLP-2R activation by zfGIP. GH3 cells were transfected with empty vector, zfGIP expression plasmid, or zf proglucagon II expression plasmid. After 48 h, media were collected, and the ability of secreted GIP and glucagon to activate zfGR (A) or zfGLP-2R (B) was assessed using specific bioassays. Activation of GIPR was expressed as fold activation over control (A) or raw activity (B). cont, Control. P values were calculated using Student's t-test.
The specificity of zfGIP for the putative zebrafish glucagon receptor was assessed by determining whether it could bind to another related glucagon receptor family member. As demonstrated in Fig. 4B, neither zfGIP nor zebrafish proglucagon II was capable of activating the zebrafish GLP-2 receptor, as predicted. Commercially available rat GLP-2 peptide was used as a positive control and activated the zebrafish GLP-2R in a concentration-dependent manner, confirming the integrity of the GLP-2R bioassay. Therefore, GLP-2R does not appear to be an in vivo target for zfGIP.
Food deprivation in larval zebrafish alters mRNA expression of GIP, proglucagon II, and insa.
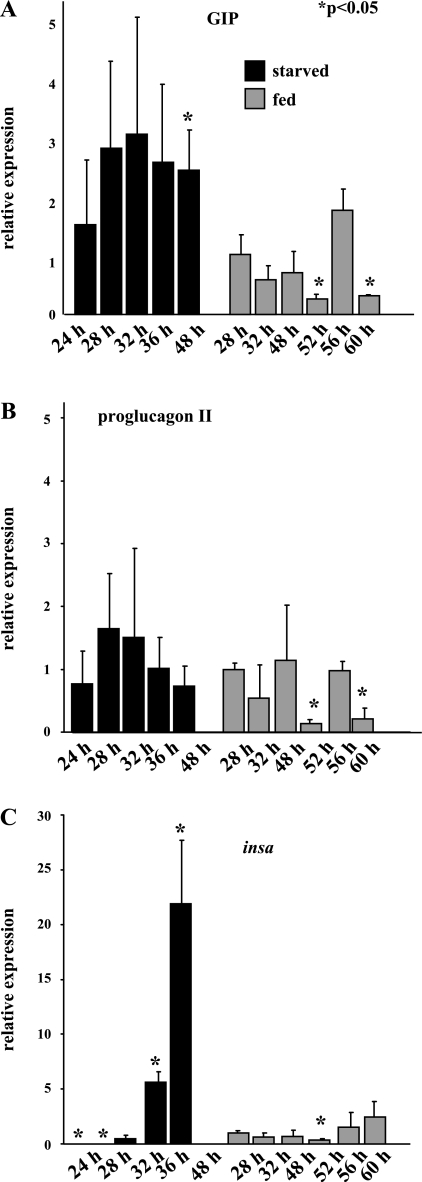
Food deprivation studies were used to indirectly determine whether GIP might function as an incretin hormone in the zebrafish. Prior studies in adult rats have demonstrated that GIP mRNA levels in the small intestine decrease significantly after a 2-day period of starvation (12). If this incretin pattern were to be maintained in zebrafish, it would be expected that food deprivation would likewise decrease GIP expression.
Small changes in GIP (Fig. 5A) and proglucagon II (Fig. 5B) levels in fed larvae were detected over the time course that larvae were collected, but were likely attributable to daily fluctuations in peptide expression. In addition, although these fluctuations were similar for GIP and proglucagon II, insa expression did not vary to any significant degree during the time when fed larvae were collected. In zebrafish larvae, food deprivation increased GIP mRNA levels from 24 to 32 h after fasting, reaching a level approximately threefold greater than expression in the control group (Fig. 5A). Although GIP expression began to decrease with prolonged starvation, transcript levels were still significantly increased over baseline values measured in fed larvae.
Fig. 5.
Examination of food deprivation on GIP, proglucagon II, and insa expression levels. Larvae were collected from timed adult matings, and starting at 6 days postfertilization were fed a standard paramecium diet or had food withheld. Total RNA was isolated and analyzed by quantitative PCR for the expression of GIP (A), proglucagon II (B), and insa (C). mRNA levels were corrected for β-actin expression, and data were expressed as means ± SE or expression relative to expression at 28 h after feeding. P values were calculated using Student's t-test. *P < 0.05.
In zebrafish larvae, food deprivation similarly increased proglucagon II expression from 24 to 32 h after fasting, reaching a peak of 1.6-fold greater expression than the control group and then appeared to decrease (Fig. 5B). Proglucagon II expression appeared to parallel GIP expression, although the absolute increase was smaller. In contrast to the effects of food deprivation on GIP and proglucagon II expression, insa expression (Fig. 5C) decreased from baseline values after 24 to 32 h of starvation. However, insa mRNA levels increased by 5.7-fold and 22-fold over baseline after 36 and 48 h of starvation, respectively (Fig. 5C). Thus, GIP expression in response to food deprivation and feeding in the zebrafish was the reverse of the mammalian pattern, but was remarkably similar to the proglucagon II pattern of expression. Moreover, in contrast to their parallel patterns of expression in mammals, no correlation between GIP and insa expression in the zebrafish was detected.
DISCUSSION
Zebrafish, a vertebrate that possesses a distinct pancreas due to fusion of the exocrine and endocrine anlagen, appears to represent an important evolutionary transition point in the development of incretin hormones. We have provided the first evidence for the differential expression patterns of GIP in mammals and zebrafish. Examination of the adult zebrafish intestine demonstrated that GIP expression was restricted to cells located near the base of the villi, differing from K-cell GIP expression throughout the villi in mammals. Although crypts, per se, are absent in the zebrafish intestine, the base of the villi possesses a population of differentiated proliferating cells (17). Early in the development of teleost fish like the zebrafish, most intestinal cells appear to possess the capacity to proliferate. As maturation proceeds, however, these proliferating cells diminish until they become restricted to a small population of cells located within the basal folds (22). Immunohistochemical analysis of GIP expression in the zebrafish intestine suggested that this population of proliferating cells expresses GIP. The detection of GIP in these cells and the absence of GIP-positive cells throughout other regions of the villi, supports the theory that the K-cell, as defined in mammals, may not yet exist in zebrafish.
Additionally, in contrast to mammals, GIP expression was demonstrated in zebrafish pancreatic endocrine cells, with colocalization with insulin detected in a subset of these cells. These observations represent the first instance in which GIP expression has been localized to the pancreas and the initial demonstration of GIP and insulin coexpression. Previous studies conducted in our laboratory and by others (14) have shown that of the five fully sequenced members of the Actinopterygii class (zebrafish, medaka, fugu, tetraodon, and stickleback), the zebrafish is the only species to possess a GIP-like sequence. This expression pattern suggests that the Actinopterygii might be the first evolutionary lineage in which a GIP-like protein exists and supports the theory that the enteroinsular axis may have evolved in the zebrafish by virtue of being the earliest vertebrate identified to express GIP (14).
Because of variations in the expression patterns of GIP between mammals and zebrafish, we conducted an analysis of food deprivation to study the function of zfGIP. Food deprivation studies in zebrafish larvae demonstrated that both GIP and proglucagon II levels increased during 32 h of starvation, after which expression remained constant or slowly declined. In contrast, insa levels remained low until increasing after 36 h of starvation and lagged 12 h behind the increase in GIP and proglucagon II release. The absence of a parallel temporal relationship between GIP and insa expression in the larvae is inconsistent with an incretin role for GIP in this species. This observation confirms our previous studies suggesting that GIP does not function as an incretin hormone in zebrafish, but rather may be glucagon mimetic. The results are also consistent with studies that demonstrated the divergence of peptide function in zebrafish compared with mammals. For example, in contrast to its role as an incretin in mammals, GLP-1 is not insulinotropic in teleost fish (35). In addition, although in-depth studies of GLP-1 expression have not been conducted in the zebrafish, in other teleost fish, GLP-1 is produced by pancreatic islet cells (35). Furthermore, unlike the glucose-lowering effects of GLP-1 seen in mammals, the effects of GLP-1 on glucose metabolism in fish hepatocytes are similar to those of glucagon. In fact, in most teleost fish examined, GLP-1 was more potent than glucagon in its ability to stimulate gluconeogenesis and glycogenolysis (21). Finally, in the fish endocrine pancreas, insulin secretion is not regulated by glucose levels (33), and consequently, teleost fish are considered to be glucose intolerant, releasing insulin in response to amino acid secretogogues. These observations suggest that in fish, insulin functions primarily as a growth-regulating hormone (33). They also provide further evidence against a role for either GIP or GLP-1 as an incretin hormone in the zebrafish, and indicate that the exquisite balance between insulin and glucagon expression required for glucose homeostasis in mammals may not be necessary in fish.
Our receptor activation studies demonstrated that zfGIP possessed the capacity to activate the rat GIPR. This observation suggests that while GIP may not be acting as an incretin in the zebrafish, sufficient identity exists between zebrafish and mammalian GIP peptides to enable receptor activation. This observation is consistent with previous studies showing that human glucagon can bind and activate both goldfish and frog glucagon receptors, suggesting that this family of G-protein coupled receptors is highly conserved across species (4, 23, 34). Interestingly, fish GLP-1 also possesses the capacity to stimulate insulin release from perfused dog and rat pancreata, as well as from pancreatic cell cultures, suggesting that when introduced to mammals, it functions as an incretin hormone (7, 20, 26). These observations also suggest that despite the capacity of ligands that share areas of conserved amino acid residues sufficient to activate corresponding receptors in higher-order vertebrates, evolution of the receptor appears to be equally important. Notwithstanding its insulinotropic properties in mammals, the absence of a native fish GLP-1R precludes a similar capacity for GLP-1 in fish. Moreover, in spite its ability to activate mammalian GIPR, GIP does not appear to be insulinotropic in the zebrafish.
We were also interested in identifying potential in vivo targets of zfGIP. Comparative studies by other groups, although identifying putative GIPR and GLP-1Rs in teleost fish, have not examined the function of these receptors (3, 13). Our studies indicated that the closest GIPR homolog in the zebrafish was a putative glucagon receptor. Although previously designated as a putative GIPR (13), it has been classified by NCBI as “similar to glucagon receptor.” This study demonstrated that both zfGIP and translational products from zebrafish proglucagon II were able to bind and activate this putative glucagon receptor. While this observation is inconsistent with previous mammalian studies reporting that each ligand activates a specific receptor, it has also been demonstrated that ligands in earlier vertebrates can interact with multiple receptors. For example, studies in goldfish have shown that both goldfish GLP-1 and goldfish glucagon can activate the goldfish GLP-1R to a similar degree (34). Based on these studies and the fact that GLP-1 and GIP appear to have acquired incretin properties after the divergence of fish and mammals (23), it has been hypothesized that the ancestral function of glucagon, GLP-1, and GIP may have been similar to the recognized mammalian functions of glucagon. This theory is supported by studies of the goldfish GLP-1R demonstrating that binding and activation of this receptor could be achieved by five different ligands: human glucagon, human GLP-1, human GIP, goldfish GLP-1, and goldfish glucagon (34).
Perspectives and Significance
The results of our studies are consistent with the following hypothesis. During evolution, insulin biosynthesis translocated to the pancreas, possibly to protect β-islet cells from enteric organisms, which then necessitated a messenger from the intestine to complete an enteroinsular axis. The eventual development of GIP and other incretins fulfilled this requirement. The additional survival benefit offered by GIP was its capacity to not only stimulate the release of insulin but to possess insulin mimetic properties, including effects on glucose homeostasis and lipid deposition in adipocytes. This physiological redundancy ensured the survival of organisms during times when food was scarce. Future studies will be necessary to more fully define the evolutionary development of incretins and their receptors and will serve to elucidate their precise role in the pathogenesis of obesity and related conditions.
GRANT
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant RO1-DK-53158 (to M. M. Wolfe).
DISCLOSURE
M. C. Musson, L. I. Jepeal, P. D. Mabray, I. V. Zhdanova and W. V. Cardoso have nothing to declare. M. M. Wolfe is an inventor on U.S. Patent 7-091-183.
REFERENCES
- 1.Brown JC, Dryburgh JR. A gastric inhibitory polypeptide. II. The complete amino acid sequence. Can J Biochem 49: 867– 872, 1971 [DOI] [PubMed] [Google Scholar]
- 2.Buchan AM, Polak JM, Capella C, Solcia E, Pearse AG. Electronimmunocytochemical evidence for the K cell localization of gastric inhibitory polypeptide (GIP) in man. Histochemistry 56: 37– 44, 1978 [DOI] [PubMed] [Google Scholar]
- 3.Cardoso J, Power D, Clark M. Comparative study of family 2 GPCRs in Fugu rubripes. Ann NY Acad Sci 1040: 257– 260, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Cardoso JC, Pinto VC, Vieira FA, Clark MS, Power DM. Evolution of secretin family GPCR members in the metazoa. BMC Evol Biol 6: 108, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr RD, Larsen MO, Winzell MS, Jelic K, Lindgren O, Deacon CF, Ahren B. Incretin and islet hormonal responses to fat and protein ingestion in healthy men. Am J Physiol Endocrinol Metab 295: E779– E784, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Cataland S, Crockett SE, Brown JC, Mazzaferri EL. Gastric inhibitory polypeptide (GIP) stimulation by oral glucose in man. J Clin Endocrinol Metab 39: 223– 228, 1974 [DOI] [PubMed] [Google Scholar]
- 7.D'Alessio DA, Fujimoto WY, Ensinck JW. Effects of glucagonlike peptide I-(7–36) on release of insulin, glucagon, and somatostatin by rat pancreatic islet cell monolayer cultures. Diabetes 38: 1534– 1538, 1989 [DOI] [PubMed] [Google Scholar]
- 8.Drucker DJ. The biology of incretin hormones. Cell Metab 3: 153– 165, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Dupre J, Ross SA, Watson D, Brown JC. Stimulation of insulin secretion by gastric inhibitory polypeptide in man. J Clin Endocrinol Metab 37: 826– 828, 1973 [DOI] [PubMed] [Google Scholar]
- 10.Ebert R, Illmer K, Creutzfeldt W. Release of gastric inhibitory polypeptide (GIP) by intraduodenal acidification in rats and humans and abolishment of the incretin effect of acid by GIP-antiserum in rats. Gastroenterology 76: 515– 523, 1979 [PubMed] [Google Scholar]
- 11.Gault VA, O'Harte FP, Harriott P, Mooney MH, Green BD, Flatt PR. Effects of the novel (Pro3)GIP antagonist and exendin(9–39)amide on GIP- and GLP-1-induced cyclic AMP generation, insulin secretion and postprandial insulin release in obese diabetic (ob/ob) mice: evidence that GIP is the major physiological incretin. Diabetologia 46: 222– 230, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Higashimoto Y, Opara EC, Liddle RA. Dietary regulation of glucose-dependent insulinotropic peptide (GIP) gene expression in rat small intestine. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 110: 207– 214, 1995 [DOI] [PubMed] [Google Scholar]
- 13.Irwin DM, Wong K. Evolution of new hormone function: loss and gain of a receptor. J Hered 96: 205– 211, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Irwin DM, Zhang T. Evolution of the vertebrate glucose-dependent insulinotropic polypeptide (GIP) gene. Comp Biochem Physiol Part D Genomics Proteomics 1: 385– 395, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Jepeal LI, Boylan MO, Wolfe MM. GATA-4 upregulates glucose-dependent insulinotropic polypeptide expression in cells of pancreatic and intestinal lineage. Mol Cell Endocrinol 287: 20– 29, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreymann B, Williams G, Ghatei MA, Bloom SR. Glucagon-like peptide-1 7–36: a physiological incretin in man. Lancet 2: 1300– 1304, 1987 [DOI] [PubMed] [Google Scholar]
- 17.Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development 123: 321– 328, 1996 [DOI] [PubMed] [Google Scholar]
- 18.Papasani MR, Robison BD, Hardy RW, Hill RA. Early developmental expression of two insulins in zebrafish (Danio rerio). Physiol Genomics 27: 79– 85, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Pederson RA, Schubert HE, Brown JC. Gastric inhibitory polypeptide. Its physiologic release and insulinotropic action in the dog. Diabetes 24: 1050– 1056, 1975 [DOI] [PubMed] [Google Scholar]
- 20.Plisetskaya E, Duguay SJ. Pancreatic hormones and metabolism in ectotherm vertebrates: current views. In: The Endocrinology of Growth, Development, and Metabolism in Vertebrates, edited by Schreibman MP, Scanes CG, Pang PKT. San Diego, CA: Academic, 1993, p. 265– 287 [Google Scholar]
- 21.Plisetskaya EM, Mommsen TP. Glucagon and glucagon-like peptides in fishes. Int Rev Cytol 168: 187– 257, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Rombout JH, Stroband HW, Taverne-Thiele JJ. Proliferation and differentiation of intestinal epithelial cells during development of Barbus conchonius (Teleostei, Cyprinidae). Cell Tissue Res 236: 207– 216, 1984 [DOI] [PubMed] [Google Scholar]
- 23.Sivarajah P, Wheeler MB, Irwin DM. Evolution of receptors for proglucagon-derived peptides: isolation of frog glucagon receptors. Comp Biochem Physiol B Biochem Mol Biol 128: 517– 527, 2001 [DOI] [PubMed] [Google Scholar]
- 24.Skauli KS, Reitan JB, Walther BT. Hatching in zebrafish (Danio rerio) embryos exposed to a 50 Hz magnetic field. Bioelectromagnetics 21: 407– 410, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Song DH, Getty-Kaushik L, Tseng E, Simon J, Corkey BE, Wolfe MM. Glucose-dependent insulinotropic polypeptide enhances adipocyte development and glucose uptake in part through Akt activation. Gastroenterology 133: 1796– 1805, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki S, Kawai K, Ohashi S, Mukai H, Murayama Y, Yamashita K. Reduced insulinotropic effects of glucagonlike peptide I-(7–36)-amide and gastric inhibitory polypeptide in isolated perfused diabetic rat pancreas. Diabetes 39: 1320– 1325, 1990 [DOI] [PubMed] [Google Scholar]
- 27.Thomas FB, Mazzaferri EL, Crockett SE, Mekhjian HS, Gruemer HD, Cataland S. Stimulation of secretion of gastric inhibitory polypeptide and insulin by intraduodenal amino acid perfusion. Gastroenterology 70: 523– 527, 1976 [PubMed] [Google Scholar]
- 28.Tsao PN, Chen F, Izvolsky KI, Walker J, Kukuruzinska MA, Lu J, Cardoso WV. Gamma-secretase activation of notch signaling regulates the balance of proximal and distal fates in progenitor cells of the developing lung. J Biol Chem 283: 29532– 29544, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng CC, Kieffer TJ, Jarboe LA, Usdin TB, Wolfe MM. Postprandial stimulation of insulin release by glucose-dependent insulinotropic polypeptide (GIP). Effect of a specific glucose-dependent insulinotropic polypeptide receptor antagonist in the rat. J Clin Invest 98: 2440– 2445, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unger RH, Eisentraut AM. Entero-insular axis. Arch Intern Med 123: 261– 266, 1969 [PubMed] [Google Scholar]
- 31.Westerfield M. The Zebrafish Book. A Guide For The Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press, 2000 [Google Scholar]
- 32.Wolfe MM, McGuigan JE. Release of gastric inhibitory peptide following a peptone meal in the dog. Gastroenterology 83: 864– 872, 1982 [PubMed] [Google Scholar]
- 33.Wright JR, Jr, O'Hali W, Yang H, Han XX, Bonen A. GLUT-4 deficiency and severe peripheral resistance to insulin in the teleost fish tilapia. Gen Comp Endocrinol 111: 20– 27, 1998 [DOI] [PubMed] [Google Scholar]
- 34.Yeung CM, Mojsov S, Mok PY, Chow BK. Isolation and structure-function studies of a glucagon-like peptide 1 receptor from goldfish Carassius auratus: identification of three charged residues in extracellular domains critical for receptor function. Endocrinology 143: 4646– 4654, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, Irwin DM. Fish proglucagon genes have differing coding potential. Comp Biochem Physiol B Biochem Mol Biol 137: 255– 264, 2004 [DOI] [PubMed] [Google Scholar]