Summary
Sumoylation is a reversible post-translational modification that plays roles in many processes, including transcriptional regulation, cell division, chromosome integrity and DNA damage response. Using a proteomics approach, we identified ~250 candidate targets of sumoylation in C. elegans. One such target is the cytoplasmic intermediate filament (cIF) protein named IFB-1, which is expressed in hemidesmosome-like structures in the worm epidermis and is essential for embryonic elongation and maintenance of muscle attachment to the cuticle. In the absence of SUMO, IFB-1 formed ectopic filaments and protein aggregates in the lateral epidermis. Moreover, depletion of SUMO or mutation of the SUMO acceptor site on IFB-1 resulted in a reduction of its cytoplasmic soluble pool, leading to a decrease in its exchange rate within epidermal attachment structures. These observations indicate that SUMO regulates cIF assembly by maintaining a cytoplasmic pool of non-polymerized IFB-1, and that this is necessary for normal IFB-1 function.
Introduction
SUMO, a small ubiquitin-like modifier, is attached covalently to target proteins via a series of enzymatic reactions carried out by SUMO-specific enzymes (Melchior, 2000). This posttranslational modification is essential for normal development and viability since it regulates a variety of essential biological processes, such as cell cycle, apoptosis, chromatin integrity and nucleocytoplasmic transport. Modification by SUMO serves as a signal for changing protein interactions that may modify substrate activity, localization and stability (Geiss-Friedlander and Melchior, 2007; Johnson, 2004; Melchior, 2000). Novel interactions between a SUMO-modified substrate and additional proteins can be mediated through a SUMO interaction motif (SIM) domain (Song et al., 2004).
The majority of the known targets of SUMO in yeast and mammalian cells are nuclear proteins including transcription factors, chromatin-associated proteins, DNA repair proteins and components of the PML (promyelocytic leukemia) nuclear bodies (Gill, 2005; Hay, 2005; Seeler and Dejean, 2003). However, SUMO also modifies substrates in the cytoplasm, mitochondria, plasma and ER membrane (Geiss-Friedlander and Melchior, 2007). One of the first identified substrates for SUMO were the S. cerevisiae septins (Johnson and Blobel, 1999; Takahashi et al., 1999). Septins are cytoskeletal proteins which form a belt of 10 nm filaments that encircles the bud neck. Mutagenesis analysis suggests that sumoylation of septins is required for the disassembly of the ring during cytokinesis (Johnson and Blobel, 1999).
SUMO proteins are highly conserved through evolution. Invertebrates have a single SUMO protein while mammalian cells ubiquitously express three SUMO paralogs (SUMO-1, SUMO-2 and SUMO-3) (Johnson, 2004). The C. elegans genome encodes for one SUMO gene, called smo-1, which is required for embryonic and postembryonic development (Broday et al., 2004; Jones et al., 2002). To further elucidate the role of SUMO in a multicellular organism, we have performed a proteomics screen designed to identify a wide range of proteins modified by sumoylation in C. elegans. Among the putative targets is a group of cytoplasmic and nuclear intermediate filament proteins. We focused on IFB-1, a protein that belongs to the cytoplasmic intermediate filament (cIF) family (Karabinos et al., 2001; Karabinos et al., 2003; Woo et al., 2004). cIFs assemble into 10 nm filament networks that provide cells with mechanical integrity (Herrmann and Aebi, 2004). In addition, cIFs were shown to play an important role in signaling platforms that sense stress, regulate organelle function and are involved in cell migration (Kim and Coulombe, 2007). The assembly of mature filaments was elucidated in vitro but the polymerization process and IF assembly in cells remains largely unknown. In vertebrates cIFs form a large multigene family of about 70 members, whereas the C. elegans genome contains 11 cIF genes (Karabinos et al., 2001). A subset of worm cIF proteins function in epidermal attachment structures that are structurally related to vertebrate hemidesmosomes and are required for C. elegans epidermal mechanical strength (Chisholm and Hardin, 2005; Fridkin et al., 2004). IFB-1 and additional cIFs are also essential for embryonic epidermal morphogenesis at the stage of embryonic elongation (Chisholm and Hardin, 2005). Here we demonstrate that IFB-1 is sumoylated at the C terminus and that SUMO regulates its assembly into the epidermal filaments, likely by serving as an IFB-1-sequestering protein.
Results
Identification of SUMO targets in C. elegans
To identify new substrates of the SUMO pathway in a multicellular organism we constructed a C. elegans strain that expresses His6- and Flag-tagged SMO-1/SUMO as its only copy of the SUMO gene. SUMO-conjugated proteins were isolated from this strain by a double-affinity purification procedure and components of the isolated protein mixture were then identified by subsequent LC-MS/MS analysis (Figure S1) (Denison et al., 2005a; Denison et al., 2005b). Using a mixed population of worms, we expected to identify targets from all developmental stages and tissues since SUMO is widely expressed in C. elegans (Broday et al., 2004). Candidate proteins were grouped according to molecular function and biological process (Figure 1, Table S1). The large variety of targets demonstrated the global role of SUMO modification in the regulation of many cellular processes. In addition to the expected high fraction of nuclear proteins we identified a large group of cytosolic, membrane, and other subcellular organelle proteins as has been found in similar proteomics studies in yeast, mammalian cells and Drosophila (Ganesan et al., 2007; Makhnevych et al., 2009; Nie et al., 2009; Panse et al., 2004; Rosas-Acosta et al., 2005; Wohlschlegel et al., 2004). Putative targets of note from the newly identified non-nuclear proteins are involved in post- translational modifications such as phosphorylation, glycosylation and myristoylation (in addition to known targets such as enzymes of the SUMO and ubiquitin pathways and proteosomal subunits). This highlights possible cross-talk between sumoylation and various post-translational modification pathways as has already been shown for ubiquitin (reviewed in (Perry et al., 2008)). Additional identified targets in this screen are cytoskeleton components that include actin-binding proteins, myosins, α- and β-tubulin and intermediate filament proteins (Table S1). IFB-1 is a cIF protein that is required for embryonic elongation and for maintenance of the mechanical linkage between the muscle and cuticle. The ifb-1 locus encodes two isoforms, IFB-1A and IFB-1B (Woo et al., 2004). We focus here on IFB-1A (hereafter referred to as IFB-1) and show that SUMO modification is required for its regulated assembly and function in the epidermal attachment structures.
Figure 1. SUMO putative targets in C. elegans.
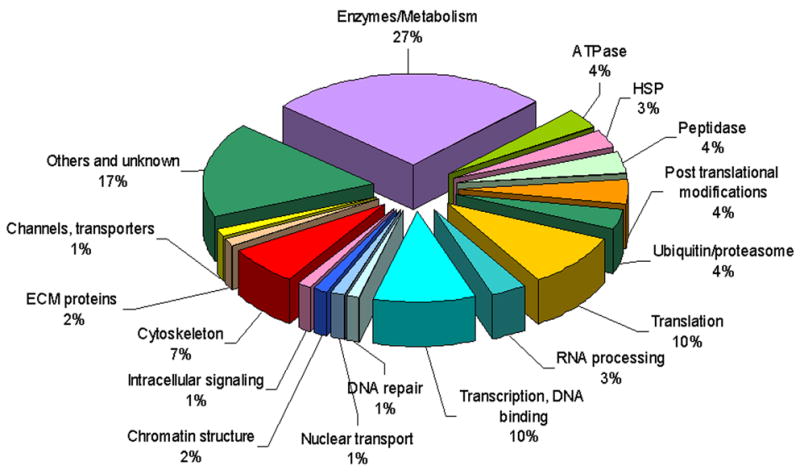
The identified SUMO putative targets are arranged in functional groups according to their relative abundance. Nuclear targets are labeled with blue colors. Cytoskeleton targets are labeled in red. The entire list of candidates is presented in Table S1. See also Figure S1.
Deletion of the smo-1 gene causes collapse of the normal IFB-1 pattern and formation of ectopic filaments and cytoplasmic inclusions
To elucidate the function of IFB-1 sumoylation, we analyzed its localization pattern in wild-type and smo-1 null worms. Immunostaining with anti-IFB-1 antibody and analysis of IFB-1::GFP reporter in wild-type animals showed that IFB-1 is localized at the basal and apical membrane of the epidermis in a pattern of circumferential stripes (Figure 2A,C,E and (Woo et al., 2004)), consistent with its localization in hemidesmosome-like structures (reviewed in (Cox and Hardin, 2004)). Homozygous smo-1(ok359) progeny (ok359 is a deletion allele of the smo-1 gene) of heterozygous mothers are viable due to maternally supplied SUMO product, but develop into sterile adults with aberrant somatic gonad, germ line and vulva, probably as a result of dilution/degradation of the maternal smo-1 gene product during larval development (Broday et al., 2004). In such smo-1 homozygous mutants derived from heterozygous parents, the normal pattern of IFB-1 was disrupted and ectopic cytoplasmic filamentous structures, mainly circular and long filaments were observed in the lateral epidermis (Figure 2B,D,F,G, short and long arrows). In addition, IFB-1 accumulated in two types of cytoplasmic inclusions. The first type of inclusion appeared as an enlarged ‘nucleation sites’ of the polymerizing filaments in the ventral and dorsal epidermis. These inclusions were arranged in linear arrays and were restricted to the region of the normal attachments (Figure 2B,D,F, bracket). The second type of inclusion was scattered throughout the cytoplasm in the lateral epidermis (Figure 2B,D,H,I, arrowhead). Analysis of the IFB-1::GFP reporter showed increasing accumulation of these inclusions during incremental developmental stages in smo-1(ok359) worms (Figure 2F–I). At the early L4 stage, there were mainly circular structures (Figure 2F, short arrow), whereas at the mid-L4 stage when the maternal product is thought to be depleted (Broday et al., 2004) additional long filaments (Figure 2G, long arrow) appeared in the lateral epidermis. By the late L4 stage large inclusions became visible (Figure 2H, arrowhead) and the apical and basal stripes were disorganized (Figure 2H, inset). In adults there were mainly GFP inclusions and abnormal weakly fluorescent apical and basal stripes (Figure 2I). To characterize the dynamics of the IFB-1::GFP molecules within the inclusions we performed fluorescence recovery after photobleaching (FRAP) analysis. Two main groups of inclusions were identified, inclusions that exhibit no recovery of fluorescence (10 min post-photobleaching), suggesting that they contain non-diffusing insoluble aggregates, and inclusions that recover slowly, especially by the mobility of molecules from the non-bleached region of the inclusion, and probably contain a larger fraction of soluble IFB-1::GFP (Figure S2). Accordingly, using a complementary fluorescence loss in photobleaching (FLIP) analysis, loss of fluorescence was detected in some of the inclusions and in all the intermediate circular structures (Figure S2). Indeed, the inclusions that contained soluble IFB-1::GFP species showed mainly internal recovery in FRAP, but FLIP analysis showed loss of fluorescence, indicating that these inclusions are not membrane-enclosed. This limited recovery provide evidence of a lack of cytosolic pool of IFB-1::GFP subunits in the smo-1(ok359) epidermis (see below). We next compared the localization of IFB-1 in smo-1(ok359) worms to its overexpression in a wild-type background using transgenic worms expressing hsp-16p::IFB-1::His6 (Figure S2). The exogenously expressed protein co-localized with the endogenous IFB-1, and when high levels of transgene were expressed, we observed formation of IFB-1 inclusions that were restricted to the basal and apical side of the hemidesmosome-like plaques and were therefore suggested to be enlarged ‘nucleation sites’. A few small aggregates were observed in the lateral epidermis similarly to aggregates previously reported with GFP-tagged cIFs and suggested to be a result of overexpression (Hapiak et al., 2003; Woo et al., 2004). Thus, overexpression of IFB-1 in the presence of SUMO in wild-type worms did not mimic the formation of ectopic inclusions and cytoplasmic filamentous structures in the lateral epidermis which had been detected in the smo-1 mutant worms (Figure 2).
Figure 2. Abnormal IFB-1 filaments and formation of inclusions in smo-1 deleted worms.
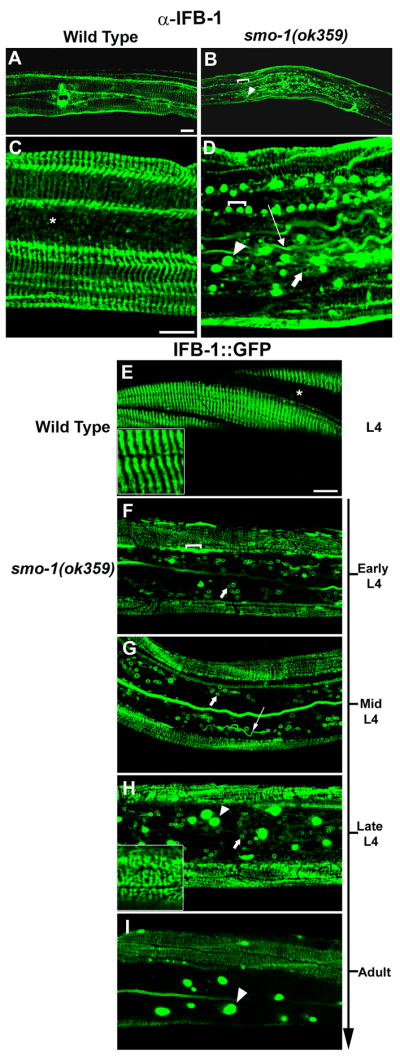
Localization of IFB-1 in wild-type and smo-1(ok359) mutant. (A–D) Immunostaining with anti-IFB-1 antibody at the late L4 stage; (A,C) IFB-1 localization in the epidermal circumferential bands in wild-type. There is no staining in the lateral epidermis (indicated by asterisk). (B,D) IFB-1 accumulates in the smo-1(ok359) worms in a linear array of inclusions (bracket), lateral inclusions (arrowhead), long filaments (long arrow) and abnormal circular filaments (short arrow). (E–I) Localization of IFB-1::GFP in wild-type and smo-1(ok359) mutants. (E) At the L4 wild-type larva expression is restricted to circumferential bands in the dorsal and ventral epidermis. The arranged parallel bands are shown in the inset. (F–I) IFB-1::GFP expression in smo-1(ok359) mutant. (F) early L4, circular filaments in the lateral epidermis (short arrow) (G) mid L4, long filamentous (long arrow) and circular structures (short arrow). (H) late L4, GFP inclusions (arrowhead), dorsal and ventral epidermal attachments (inset).(I) adult, GFP inclusions (arrowhead). Bar, 20 μm in A–B, 10 μm in C–I. See also Figure S2.
Other hemidesmosome-like components are disrupted in smo-1 mutant worms
To determine whether additional cIFs are affected by SUMO we used the MH4 antibody (Francis and Waterston, 1991) which recognizes IFA-1,2,3. IFA-2 (also known as MUA-6 (Hapiak et al., 2003) (Figure 3A), was identified among the putative targets in our screen (Table S1) and has been shown to hetero-polymerize with IFB-1 in vitro (Karabinos et al., 2003). In addition to disorganized apical and basal stripes, we observed abnormal circular filaments similar to the IFB-1 circular structures (Figure 3B). It is possible that these circular filaments resulted from non-sumoylated IFAs or that the association between IFB-1 and IFAs is not dependent on SUMO and also occurs in the smo-1 mutant worms that accumulate abnormal IFB-1.
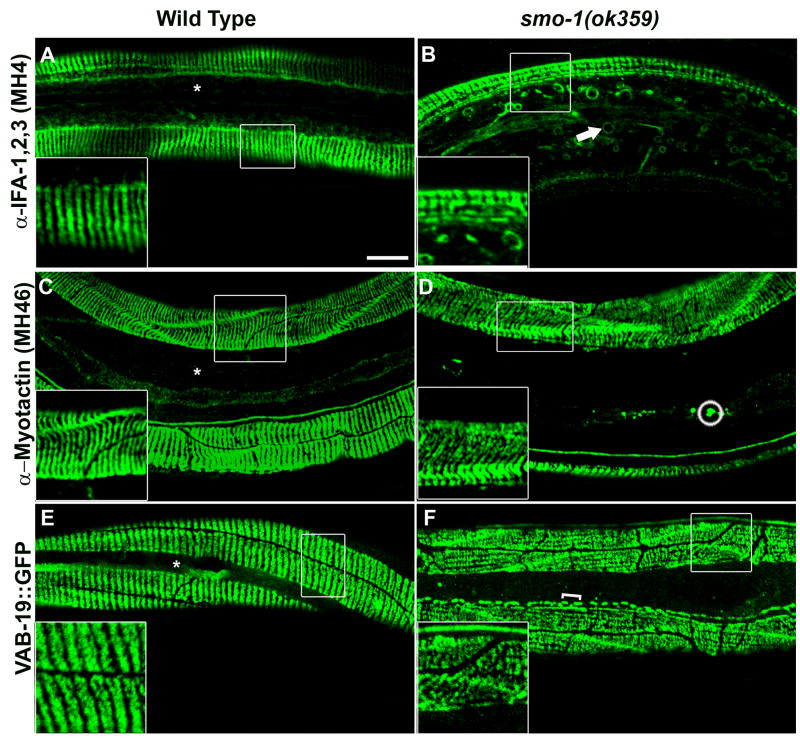
Figure 3. Additional components of the hemidesmosome-like attachments do not accumulate into large inclusions.
(A–B) Staining of wild-type and smo-1(ok359) mutant with the MH4 antibody. (A) the wild-type pattern of the MH4 antibody is similar to anti-IFB-1. (B) attachment structures in the smo-1 mutant (inset). Circular filaments are labeled with a short arrow. (C–D) Staining of wild-type and smo-1(ok359) mutant with the MH46 antibody. Small inclusions in the lateral epidermis are circled. (E–F) VAB-19::GFP expression in wild-type and smo-1(ok359) mutants. Brackets indicate elongated inclusions at the ventral epidermis. Asterisks indicate restriction of the wild-type staining to the dorsal and ventral epidermis. Bar, 10 μm.
To determine whether other components of the trans-epidermal attachments exhibit similar abnormalities we analyzed the expression pattern of myotactin and VAB-19/Kank (Figure 3C–F). Myotactin is a transmembrane adhesion molecule at the basal side of the epidermis and is predicted to link VAB-10A/plectin and cIFs (Bosher et al., 2003; Hresko et al., 1999; Plenefisch et al., 2000). VAB-19/Kank is a cytoplasmic adaptor protein that is localized to both the apical and basal attachments (Ding et al., 2003). Both components are required for maintenance of the hemidesomosome-like attachments. The staining pattern of both proteins, but especially of VAB-19, in smo-1(ok359) worms exhibited disorganization of the annular arrangement (Figure 3D,F). Short linear arrays of VAB-19::GFP inclusions were observed in the apical or basal hemidesmosome-like structures (Figure 3F, bracket). A few small aggregates of myotactin visualized with the MH46 antibody (Francis and Waterston, 1991) were observed in the lateral epidermis (Figure 3D, circle). The disorganized pattern of the circumferential stripes of VAB-19::GFP in the smo-1 deletion mutant resembled the IFB-1::GFP arrangement in smo-1 worms (Figure 3F compared to Figure 2H, insets) suggesting that VAB-19 localization in the hemidesmosome-like attachments depends on normal sumoylation of cIFs. We failed to observe circular filaments or large inclusions for either myotactin or VAB-19. In addition, we did not detect a muscle detachment (Mua) phenotype in smo-1(ok359) worms which has been observed in mytoactin and VAB-19 mutants, suggesting that although the attachment structures are disorganized they are still functional.
SUMO is required for cytoplasmic localization of IFB-1 and for embryonic elongation
During embryonic elongation, formation of the nascent hemidesmosome-like structures enabled us to observe de novo formation of the IF network. To examine the effect of SUMO on IFB-1 localization and function during this process, we used the feeding-RNAi approach to deplete SUMO. We selected for embryos that had enough SUMO to perform embryonic cell divisions until the comma stage (early elongation). IFB-1 expression during embryonic elongation was followed using the IFB-1::GFP reporter. Expression of IFB-1::GFP was initially detected at the comma stage in the cytoplasm. At the 1.5-fold stage an increased fluorescence signal was detected in the cytoplasm and was localized to the dorsal and ventral epidermis (detected in 34/35 embryos analyzed) (Figure 4A–D, inset). This suggested that a cytoplasmic pool of soluble IFB-1 is maintained during the formation of the epidermal attachment plaques and supplies building blocks that then incorporate into the nascent structures during the process of embryonic elongation. At the 2-fold stage, the protein localized to the newly formed hemidesmosome-like structures and the complete pattern of circumferential bands is observed before hatching (Figure 4E–H, inset). Accumulation of small aggregates (Figure 4D,F circle) could be a result of overexpression of the GFP reporter (Woo et al., 2004) and storage of IFB-1 prior to its polymerization as has been shown for type II keratins in mouse embryos (Lu et al., 2005)). Analysis of smo-1(RNAi) viable embryos revealed an abnormal pattern of IFB-1 expression (Figure 4I–P). As soon as GFP fluorescence could be detected at the comma stage and later, at 1.5-fold stage, IFB-1 was localized to abnormal thick and short filamentous structures in all regions of the epidermis (detected in 34/38 embryos analyzed) while cytoplasmic staining was very weak or undetectable (32/38 embryos) (Figure 4I–L, inset). We did not detect any normal embryos beyond the 1.5-fold stage. Embryos were arrested with severe elongation defects although they did display muscle twitching (Figure 4M–P). These results suggest that during embryonic elongation, SUMO is required to keep IFB-1 in a soluble fraction that serves as storage for free subunits. These subunits are then available for incorporation into the newly assembled filaments. When SUMO is depleted, this pool is eliminated, leading to aberrant polymerization of IFB-1. Such altered IFB-1 assembly could in turn result in the defective embryonic elongation seen in smo-1(RNAi) embryos.
Figure 4. SUMO is required for a cytoplasmic mobile pool of IFB-1.
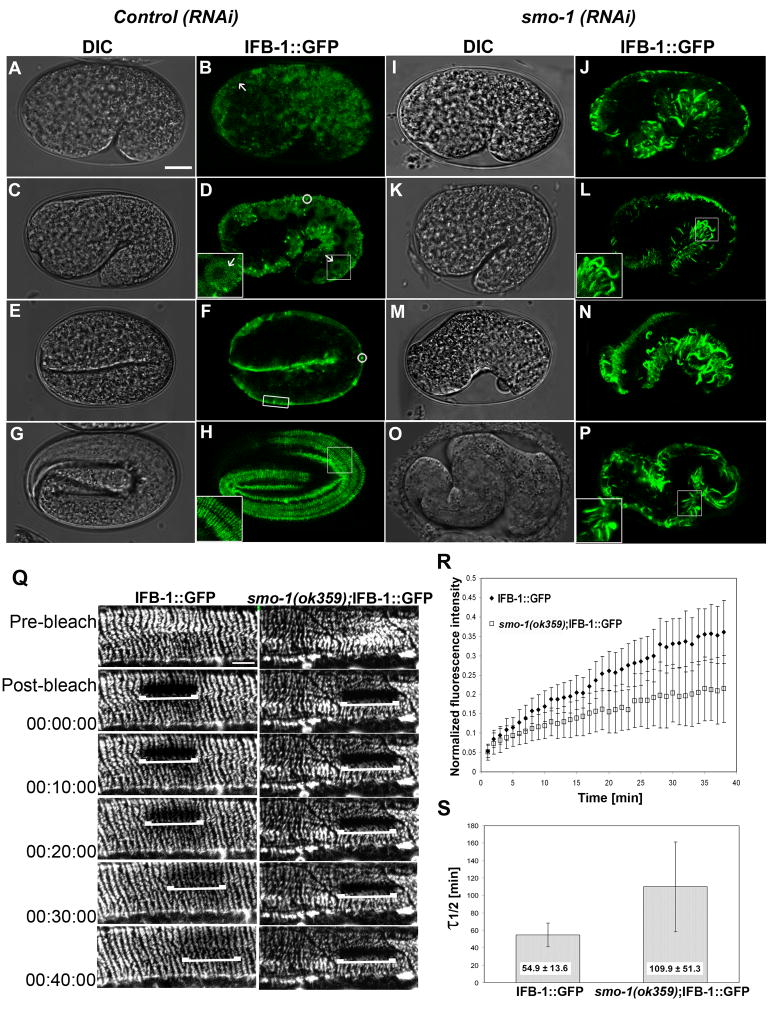
(A–H) DIC and expression pattern of IFB-1::GFP during elongation in wild-type embryos. (A–B) Cytoplasmic expression of IFB-1::GFP at the comma stage (arrow). (C–D) Cytoplasmic expression at 1.5-fold stage (arrow, inset). Small aggregates are circled. (E–F) The transition from cytoplasmic to localized expression in the nascent epidermal attachment sites at the 2-fold stage (box). (G–H) Fluorescence pattern of the circumferential bands of the mature epidermal attachment structures at the 3.5-fold stage (inset).
(I–P) DIC and expression pattern of IFB-1::GFP of embryos from hermaphrodites treated with smo-1(RNAi). (I–L) Formation of abnormal filaments and no cytoplasmic staining at the comma stage and 1.5-fold embryos (inset). (M–P) Embryos with severe elongation defects at estimated parallel stages exhibit abnormal pattern of filaments. Bar, 10 μm.
(Q–S) Depletion of SUMO decreases IFB-1 mobility measured by FRAP. (Q) Time-lapse micrographs of the bleached area before photobleaching (pre-bleach), immediately after photobleaching (post-bleach) and at intervals during recovery (min) of IFB-1::GFP reporter in wild-type and smo-1(ok359) animals. Bar, 5 μm. (R) Fluorescence intensities in the photobleached area are plotted against time. Values at each time point are normalized to a nonbleached area and to the fluorescence ratios before the bleach. Analysis was performed to n=12 worms from each genotype. (S) The half life time of fluorescence recovery, τ1/ 2. Mean values and standard deviations are shown. The differences between the τ1/ 2 values of the wild-type (n=12) and the smo-1(ok359) (n=12) worms are statistically significant (two sided t-test, p<0.0001). See also movies 1 and 2.
The cytoplasmic-to-filament exchange rate for IFB-1 is two-fold slower in smo-1 mutant animals
To test whether SUMO indeed maintains a cytoplasmic pool of IFB-1 that is available for incorporation into the hemidesmosome-like structures, we measured the FRAP time course of the IFB-1::GFP reporter in both wild-type and smo-1(ok359) epithelial attachments. FRAP analysis of IFA-1::GFP and IFB-1::GFP has been previously performed in the marginal cells of the pharynx, showing slow recovery with kinetics on the order of hours (Karabinos et al., 2003). Similarly, the half-life recovery time (τ1/2) of keratins heteropolymers is ~100 min, in contrast to a τ1/2 of ~ 6 min for vimentin homopolymers in cultured epithelial cells (Yoon et al., 2001). As shown in typical images in Figure 4Q (and in Supporting Materials movies 1,2), FRAP at the epithelial attachment sites occurred more rapidly in the wild-type background than in the smo-1 mutant worms. The ratio of the average normalized fluorescence intensity (smo-1(ok359)/wt) approached 0.6 after about 20 min, indicating slower recovery of the smo-1 mutant (Figure 4R). The average τ1/2 was 54.9 min for IFB-1::GFP in wild-type filaments and 109.9 min in the smo-1(ok359) background (Figure 4S). These observations imply that the exchange rate between soluble subunits and incorporated IFB-1 is slower when SUMO is depleted probably due to a decrease in the soluble pool of IFB-1.
IFB-1 is sumoylated on K460 in the C-terminal immunoglobulin-fold domain
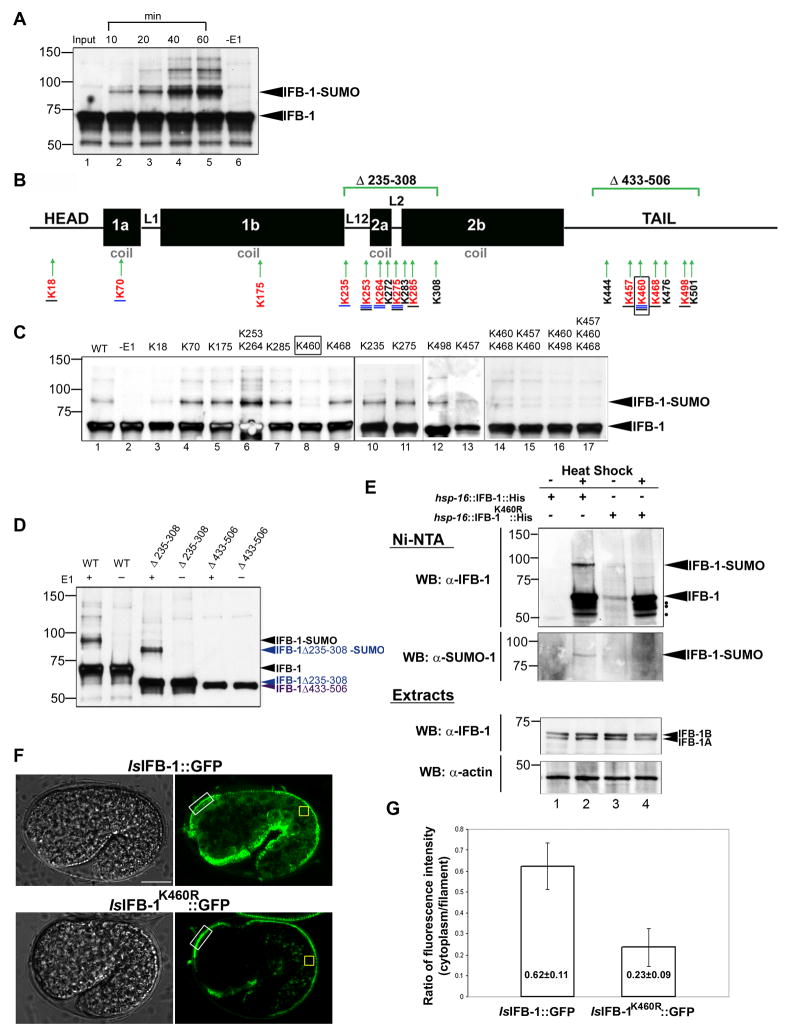
To determine whether IFB-1 is modified by SUMO and to map the SUMO acceptor site(s), we first performed in vitro sumoylation reactions with wild-type IFB-1. Bacterially expressed IFB-1 was incubated with SUMO-1, the E1- activating SUMO enzyme (SAE1 and SAE2) and Ubc9 in the presence of ATP. Western blot analysis with α-IFB-1 antibodies detected a time-dependent increase in the intensity of a slower migrating band with a molecular-weight shift of about 20 kDa (the molecular weight of IFB-1 is 63.7 kDa) (Figure 5A, lanes 2–5). Higher molecular weight bands that were observed following longer reaction periods may represent sumoylation at additional sites or formation of poly-SUMO chains on the primary site (Figure 5A, lanes 3–5). The slower migrating bands were not seen when the E1 enzyme was absent from the reaction (Figure 5A, lane 6). To identify the lysine(s) required for sumoylation of IFB-1, site-directed mutagenesis was carried out on each of the 12 lysine residues (mutated to arginine) that are predicted to be sumoylated (SUMOsp algorithm)(Xue et al., 2006) or/and are highly conserved (Figure 5B). In vitro sumoylation assays were performed with single mutants and/or mutant pairs and possible changes in the degree of sumoylation were monitored. This analysis identified K460, located in the C-terminal Ig-fold domain of IFB-1, as the primary sumoylation site. Instead of the clear sumoylation band at ~83 kDa (Figure 5C, upper arrow), two weaker bands above and below this band were observed following in vitro sumoylation reactions with the K460R mutant (Figure 5C, lane 8) as well as following reactions with double or triple mutants that included K460R (Figure 5C, lanes 14–17). Although we can not rule out completely the sumoylation, albeit at a very poor efficiency, on alternative residues, it is noteworthy that these bands do not correspond to the molecular change expected from the addition of one or two SUMO molecules, and are likely to represent background noise. To further verify these results, deletion constructs in two regions of IFB-1 that include several clustered predicted sumoylation sites were assayed in vitro (Figure 5B): IFB-1Δ235-308, which includes the 2a coil, linkers domains and part of the 2b coil (mid-helix) and IFB-1Δ433-506 in the tail domain (which includes K460). Sumoylation reactions revealed that the IFB-1Δ235-308 deletion construct is still sumoylated while sumoylation is abolished in IFB-1Δ433-506 (Figure 5D). These results establish that sumoylation of IFB-1 takes place primarily at its C-terminal tail.
Figure 5. IFB-1 is sumoylated in vivo and in vitro on K460.
(A) IFB-1 is modified by SUMO-1 in vitro. Recombinant IFB-1 was incubated for the indicated times (minutes) in modification reactions including E1 (SAE1 and SAE2), E2 (UBC9), SUMO-1 and ATP (lanes 1–5) or control reaction without E1 (lanes 6, 60 min incubation). (B) Schematic presentation of IFB-1 structure consisting of head, rod and tail domains. The four rod subdomains (coil 1a, 1b, 2a, 2b) are joined by three non-helical linkers (L1, L12, L2). Lysine residues predicted to be sumoylated (SUMOsp algorithm) are underlined in blue (the four lysines with the highest score are labeled with double line), conserved lysines in worm and/or human IFs are underlined in black, lysines included in the region of the deletions are in black and the lysines mutagenized in this work are in red. The primary sumoylation site, K460, is boxed. The IFB-1Δ235-308 and IFB-1Δ433-506 deletion constructs are marked. (C) In vitro sumoylation of wild-type and lysine-to-arginine mutant forms of IFB-1. Shown are single mutants (lanes 3–5, 7–13), double mutants (lanes 6, 14–16) or a triple mutant (lane 17). The sumoylated-IFB-1 band is marked by an arrowhead. (D) In vitro sumoylation of wild-type and IFB-1Δ235-308 (blue) and IFB-1Δ433-506 (purple) deletion constructs. Reactions were performed with (+) or without (−) E1 enzyme. Proteins were detected by immunoblotting with anti-IFB-1-specific antibody. (E) IFB-1 is sumoylated in C. elegans. Transgenic worms expressing His6-tagged IFB-1 (Ishsp-16::IFB-1::His) (lanes 1–2) or K460R mutant (Ishsp-16::IFB-1K460R::His) (lanes 3–4) were treated with heat shock (+) to induce IFB-1 expression, or non-treated controls (−). Lysates (~2 mg total protein) were subjected to affinity chromatography on Ni-NTA agarose followed by immunoblotting with anti-IFB-1 (upper panel) and anti-SUMO (lower panel) specific antibodies. The sumoylated-IFB-1 band is marked by an arrowhead. Possible proteolytic forms of the induced IFB-1 are labeled with black dots. The levels of actin and IFB-1 in total worm lysates (5% lysate) are shown in the lower panels (Extracts); the two IFB-1 isoforms (1A and 1B) are labeled. (F–G) Reduced cytoplasmic pool in IFB-1K460R::GFP embryos. Fluorescence intensity in the cytoplasm was measured in the dorsal or ventral epidermis of 1.5-fold embryos expressing the IFB-1::GFP (n=32) or IFB-1K460R::GFP (n=28) reporters. (F) Representative DIC and fluorescence images. (G) Fluorescence intensity ratios. Values in the cytoplasm (yellow box) are normalized to the intensity values in the nascent epidermal attachment structures (white box). Mean values and standard deviations are shown. The differences between the values of the IFB-1::GFP and the IFB-1K460R::GFP worms are statistically significant (two sided t-test, p<0.0001). See also Figures S3 and S4 and movie 3.
To test whether IFB-1 is modified by SUMO in the animals, we analyzed the functional fusion protein hsp-16::IFB-1::His6 and hsp-16::IFB-1K460R::His6 (see below, Figure 6) in which the sumo acceptor site K460 was mutated to arginine. The two constructs were expressed in transgenic worms under regulation of the hsp-16 heat-shock promoter. Nickel-affinity purification (Figure S3) of both wild-type and mutant IFB-1K460R extracts from heat shock treated and control transgenic worms was followed by western blotting using α-IFB-1 and α-SUMO antibodies. A slower migrating form with a molecular weight shift of about 20 kDa consistent with SUMO-modified IFB-1, was detected in extracts expressing wild-type IFB-1 (Figure 5E, lane 2). This higher molecular weight band was abolished in the K460R mutant, while increased IFB-1 levels following heat shock was confirmed in both extracts using α-IFB-1 antibodies (Figure 5E, lane 4). In both the wild-type and K460R, at least three bands were detected below IFB-1 (upper blot, black dots) which are likely to be a result of proteolytic cleavage of IFB-1, as has been shown for several vertebrate IF proteins (Byun et al., 2001; Ku and Omary, 2001). The results of this analysis indicate that IFB-1 is a direct substrate for SUMO modification and that K460 is the primary in vivo SUMO acceptor site of IFB-1.
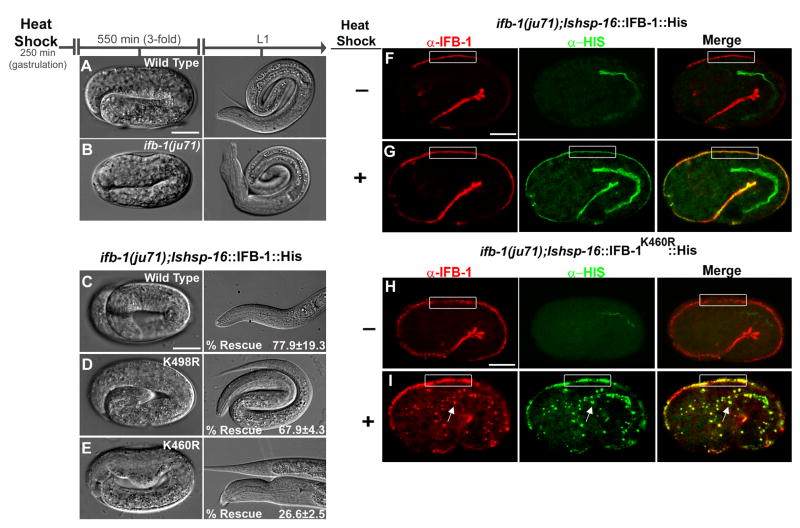
Figure 6. K460 is essential for IFB-1 function during embryonic elongation.
(A–B) Elongation of the epidermis in wild-type and ifb-1(ju71) mutant embryos following heat shock at gastrulation (control). (C–E) Rescue experiments of the ifb-1(ju71) embryos using heat-inducible transgenes expressing the wild-type IFB-1, IFB-1K498R and IFB-1K460R mutants (Table S2). (F–G) immunostaining of control (−) and heat shock treated ifb-1(ju71);hsp-16::IFB-1::His embryos (+). (HI) immunostaining of control (−) and heat shock treated ifb-1(ju71);hsp-16::IFB-1K460R::His embryos (+). Both endogenous and ectopically expressed IFB-1 are detected by the anti-IFB-1 antibody (red). Ectopically expressed IFB-1 is detected by the anti-His antibody (green). Box indicates the attachment structures in dorsal epidermal cells. Arrow indicates ectopic inclusions recognized by anti-IFB-1 and anti-His antibodies. Bar, 10 μm.
The sumoylation-deficient K460R protein exhibits a reduced cytoplasmic pool of IFB-1
To test the hypothesis that sumoylation of IFB-1 is required for maintenance of its cytoplasmic pool, we generated a fusion protein, IFB-1K460R::GFP, expressed under the control of the ifb-1 5′ regulatory sequences. Relative to the wild-type IFB-1::GFP reporter, we found a major reduction in cytoplasmic expression of the mutant transgene during embryonic elongation, when filament assembly occurs. The mean fluorescence-intensity ratio of cytoplasm to filament (cytoplasm values were normalized to the fluorescence intensity in the nascent filaments, see Experimental Procedures) was lower in the IFB-1K460R::GFP worms than in the wild-type IFB-1::GFP strain (0.23 ± 0.09 versus 0.62 ± 0.11, respectively, p<0.0001; Figure 5F–G). This finding indicates that sumoylation of IFB-1 on K460 is required to maintain its cytoplasmic pool during assembly of the epidermal attachment structures. To further characterize the sumoylation mutant form of IFB-1, we performed FRAP analysis on the IFB-1K460R::GFP filaments. The exchange rate between soluble and incorporated IFB-1 subunits was slower in IFB-1K460R::GFP than in the wild-type IFB-1::GFP. The average τ1/2 was 229 ± 138 min in the IFB-1K460R::GFP filaments (Figure S4) compared to 54.9 ± 13.6 min in the IFB-1::GFP filaments (Figure 4S), both measured in a wild-type genetic background (statistically significant difference; two sided t-test, p<0.0001). We conclude that the soluble pool of IFB-1K460R::GFP is smaller than that of the wild-type IFB-1::GFP. In agreement with these observations, the structure of the epidermal attachment sites in larvae expressing IFB-1K460R::GFP was impaired (Figure S4) and resembled the aberrant structures in the smo-1(ok359) background (Figure 2).
K460 is required for IFB-1 function during embryogenesis
The ifb-1(ju71) deletion allele causes abnormal embryonic elongation which results in incompletely penetrant embryonic arrest at the 3-fold stage, as well as an abnormal head epidermis (the Vab phenotype) (Woo et al., 2004) (Figure 6A–B). The His-tagged wild-type IFB-1 protein expressed from the hsp-16::IFB-1::His6 construct, complemented the ifb-1(ju71) mutant phenotype and was recognized by the α-IFB-1 antibody in the apical and basal epidermis of ifb-1(ju71) embryos (Figure 6C,F–G, Table S2). We assessed the functional activity of the IFB-1K460R mutant protein expressed from the hsp-16::IFB-1K460R::His6 construct by similar rescue experiments of the ifb-1(ju71) embryos. Strikingly, the degree of rescue achieved by the K460R mutant was limited to 26.6% whereas the wild-type IFB-1 or a control K498R mutant efficiently rescued (77.9% and 67.9%, respectively) the ifb-1(ju71) mutant embryos (Figure 6C–E, Table S2). Further analysis by immunostaining revealed accumulation of abnormal discontinuous attachments and ectopic inclusions in embryos expressing the K460R sumoylation mutant, but not in those expressing the wild-type IFB-1 protein (Figure 6F–I). We conclude that abnormal, probably nonfunctional attachments, combined with accumulation of inclusions in the lateral epidermis (which are probably toxic at this stage), prevent normal elongation.
Discussion
The role of SUMO in multicellular organisms is still unclear. Here we used a proteomics approach to identify diverse targets of SUMO in C. elegans. Among the identified targets are cytoplasmic intermediate filament proteins. We chose to study IFB-1, a member of the cIF family that links the apical and basal hemidesmosome-like structures of the epidermis. IFB-1 is sumoylated on lysine 460 that is located at the Ig-fold domain at the C-terminal tail of IFB-1. In vertebrates, this domain is found only in nuclear lamins (Dhe-Paganon et al., 2002; Krimm et al., 2002), while in C. elegnas it is found in both lamin and the cytoplasmic intermediate filament proteins (Karabinos et al., 2001). This motif is known to be involved in protein-protein interactions and is associated with ~26% of the known mutations in lamins causing laminopathies (Shumaker et al., 2008). It has been shown to negatively regulate lamin polymerization (Shumaker et al., 2005) which is in accordance to our suggested negative regulatory role of SUMO in IFB-1 polymerization (Figure S5). Sumoylation at the Ig-fold domain probably modulates the β-sheet structure and thus may transiently change protein interactions.
De novo assembly of cIFs takes place during embryonic elongation. Depletion of SUMO at this stage of embryogenesis by RNAi caused the formation of abnormal filaments and lack of a cytoplasmic pool. At late larval stages when only a maintenance process of the existing network takes place, a low rate of exchange of IFB-1 subunits within the existing filaments was detected in smo-1(ok359) mutant animals compared to the wild-type background in parallel to its accumulation into abnormal filaments and cytoplasmic inclusions. Similar phenotypes were observed when the sumoylation –deficient K460R protein was analyzed in the wild-type background. Notably, although the epidermal attachment sites of the IFB-1K460R::GFP larvae were severely disorganized, no ectopic filaments or inclusions were formed in their lateral epidermis, raising the possibility that additional proteins such as IFA-2 may also be sumoylated to prevent the formation of IFB-1 inclusions.
While abnormalities in the localization pattern were observed in smo-1 mutant animals also for the epidermal attachment components mytoactin and especially VAB-19, these proteins did not display ectopic filaments and cytoplasmic inclusions. This supports a specific role for SUMO in the regulation of cIFs polymerization and suggests that abnormal cIFs polymerization affects the recruitment of additional components of the hemidesmosome-like attachments such as VAB-19.
The amount of IFB-1 inclusions increases in smo-1 deleted adult animals, suggesting complete depletion of the maternal SUMO or overloading of the cellular degradation machinery. The pathology of many IF-related diseases involves accumulation of cytoplasmic inclusions, for example keratin-related liver disorders that result in formation of Mallory bodies, (Nam-On Ku, 2007) desmin-related myopathies (DRM) (Bar et al., 2005) and GFAP mutants that are associated with Alexander disease (Quinlan et al., 2007).
SUMO was found to accumulate in inclusions associated with several neurodegenerative diseases, including multiple system atrophy, Huntington’s disease and other polyglutamine disorders (Dorval and Fraser, 2007). On the other hand, our findings show that lack of SUMO causes the formation of cIF aggregates. Thus in different cellular environments, and in a target specific manner, SUMO can contribute to accumulation of protein aggregates or protect against their formation. Indeed, recently it was reported that the amyloid precursor protein (APP) is sumoylated and that its sumoylation is associated with decreased levels of Aβ peptides (Zhang and Sarge, 2008b).
The assembly properties of cIFs have been studied in depth in vitro (Herrmann and Aebi, 2004), but less is known about their in vivo assembly. Regulation of cIF polymerization in vivo is thought to be accomplished by post-translational modifications. cIFs are known to be heavily phosphorylated mainly on the N- and C- terminals (Omary et al., 2006). Reorganization of cIFs during mitosis was shown to be phosphorylation-dependent (Chou et al., 1989). More recently it was demonstrated that phosphorylation of keratins target them for degradation through the ubiquitin-proteasome pathway in response to shear stress (Jaitovich et al., 2008). Lower keratin ubiquitination and hyperphosphorylation are associated with Mallory body deposits in a variety of liver diseases (Ku and Omary, 2000). Sumoylation is an additional post-translational modification of cIFs that may be involved in disease. Recently, human Lamin A which is a nuclear IF protein (the C. elegans Lamin, LMN-1(Liu et al., 2000), was identified as a candidate in this screen) was shown to be sumoylated and its sumoylation associated with familial dilated cardiomyopathy (Zhang and Sarge, 2008a). Using a selective RNAi approach, we showed that the formation of abnormal filaments is accompanied by a major decrease in cytoplasmic accumulation of IFB-1. These data suggest that in vivo a soluble pool of cytoplasmic IFB-1 heterodimers (IFB-1 was shown to form heterotypic interaction with IFA proteins in vitro, (Karabinos et al., 2003)) or tetramers is maintained by sumoylation. A regulated desumoylation process may be responsible for the ability of the free subunits to polymerize into tonofibrils (Figure S5). Thus, SUMO modification may serve as an IFB-1-sequestering factor resembling the inhibitory mechanism of actin assembly by Thymosin β4 (Hannappel, 2007). Since the exchange rate between soluble subunits and polymerized IFB-1::GFP in the epidermal attachment sites depends on SUMO and on sumoylation of IFB-1, it supports such a role for SUMO, probably by regulating the interactions with its associated proteins. In summary, we demonstrated here that SUMO plays an important role in the process of cIF assembly by serving as a negative regulator of filament assembly in vivo.
Experimental Procedures
Strains
C. elegans was cultured as described (Brenner, 1974). The genes and alleles used in this study are: smo-1(ok359) (isolated and kindly provided by the C. elegans Gene Knockout Project team at Oklahoma Medical Research Foundation [OMRF]) (Broday et al., 2004) and ifb-1(ju71) (Woo et al., 2004). In addition, the following integrated transgenes or extrachromosomal arrays were used: juIs176[IFB-1A::GFP; rol-6] (Woo et al., 2004), juIs167[VAB-19::GFP; rol-6] (Ding et al., 2003), smo-1(ok359);tvEx25[psmo-1::His-FLAG-SMO-1; rol-6] (this study), tvIs32[hsp-16::IFB-1::His] (this study), tvIs37[hsp-16::IFB-1K460R::His] (this study), tvIs38[hsp-16::IFB-1K498R::His] (this study), tvIs41[IFB-1::GFP;rol-6] (this study), tvIs42[IFB-1K460R::GFP;rol-6] (this study).
Purification of SUMO conjugates for mass spectrometry analysis
For proteomics analysis the smo-1(ok359);tvEx25[psmo-1::His-FLAG-SMO-1; rol-6] strain was constructed. The psmo-1::His-FLAG-SMO-1; rol-6 construct contains the smo-1 genomic sequences and 5′ and 3′ regulatory sequences of the smo-1 genomic region and rescues the smo-1 genetic deletion. The rescued strain that expresses only the SUMO tagged protein was used for the proteomics analysis. Protein purification and mass spectrometry was performed according to (Denison et al., 2005b).
Antibodies
Antibodies used for western analysis are anti-SUMO (Zymed clone 21C7) (1:1000), anti-IFB-1 (1:6000) (Karabinos et al., 2001), anti-actin (Santa Cruz) (1:1000). Antibodies used for Immunofluorescence are Penta-His Alexa Flour 488 conjugate (Qiagen)(1:200), anti-IFB-1 (1:100), MH4 (1:10), MH46 (1:10).
In vitro sumoylation
Full length ifb-1 cDNA was cloned in the pET-20b(+) vector (Novagen), expressed in BL21(DE3) bacteria at 30°C and purified with 8M urea, 5 mM Tris (pH 7.6), 1 mM ETDA, 0.4 mM PMSF. Protein (0.2 mg/ml) was diluted 10 times in sumoylation buffer (10x buffer contains 200 mM Hepes pH 7.5, 50 mM MgCl2, and 20 mM ATP) to allow formation of soluble oligomers. Reactions were performed using in vitro SUMOylation kit purchased from LAE Biotech International (Rockville, MD). Reactions were incubated at 37°C at the indicated time points. Reactions with IFB-1 mutants and control reactions without E1 were incubated for 1 hr at 37°C.
In vivo sumoylation
IFB-1 expression was induced by heat shock in transgenic worms harboring the phsp16::IFB-1::His6 construct or phsp16::IFB-1K460R::His6. The heat shock treatment was as following: 1 hr at 32°C, 2 hr at 20 °C, 1 hr at 32 °C, collection of worms and immediate freezing in liquid nitrogen. Protease inhibitor cocktail (sigma P1860) was added (1:500) after the first heat shock treatment. Transgenic worms were lysed in 6M guanidinium-HCl, 0.1M Na2HPO4/NaH2PO4 (pH 8.0), 5 mM imidazole. Following sonication, lysates were incubated for 4 hr with Ni-NTA beads, and then washed and eluted according to (Treier et al., 1994) with an additional wash with 0.5% NP-40 before elution. The eluted proteins were analyzed by anti-SUMO antibody and anti-IFB-1 antibody. Total worm lysate was TCA precipitated before gel electrophoresis and analyzed by anti-actin and anti-IFB-1 antibodies.
RNAi treatment
smo-1(RNAi) plasmid (clone 1013 from J. Ahringer RNAi library) or empty L4440 vector were transformed into HT115 (DE3). RNAi by feeding was carried out as described (Fraser et al., 2000). An overnight culture was diluted 1:100, and at OD=0.4 IPTG (Sigma Inc., cat. I5502) was added to a final concentration of 1 mM for 4 h at 37°C. The induced bacteria expressing smo-1 dsRNA was then seeded directly onto NMG-RNAi plates containing 1mM IPTG and 50 μg/ml carbenicillin. Seeded plates were dried at room temperature overnight. Embryos expressing IsIFB-1::GFP (collected by bleaching of gravid adults) were grown on RNAi plates at 20°C for 4 days. For analysis of embryos, adults were cut and the embryos were transferred to an agar pad for fluorescent and DIC examination.
Rescue experiments of the ifb-1(ju71) mutant and immunostaining of embryos and L1 larvae
The integrated strain Is[hsp-16::IFB-1::His], Is[hsp-16::IFB-1K460R::His] and Is[hsp-16::IFB-1K498R::His] were crossed to the ifb-1(ju71) strain. Gravid homozygous adults laid eggs for 2 hr and then removed from plates. Induction of IFB-1 was performed using the following conditions: 30 min at 32°C, 2 hr at 20 °C x 3. One heat shock for 30 min at 32°C at late gastrulation was sufficient to rescue embryos that harbor the wild-type IFB-1 construct. For immunostaining, single induction was done by subjecting embryos (1.5hr after bleaching of gravid adults) to 30 min heat shock at 32°C. Five inductions of IFB-1 (Figure S2) were performed by subjecting embryos to 30 min heat shock, 1.5 hr at 20°C x2, 30 min heat shock, 18 hr at 20°C, 30 min heat shock, 1.5 hr at 20°C x2 and fixation of L1 larvae for immunostaining. Fixation and staining of embryos and L1 larvae were performed according to Finney (Finney and Ruvkun, 1990).
Statistical methods
Multiple repeats were performed in all experiments as indicated and mean and standard deviation values were calculated. Hypothesis testing was performed using two sided t-test after rejecting the non-equal variance H1 assumption. Error bars and error estimates (±) are the pertinent standard deviation values.
Microscopy and quantification
Quantification of cytoplasmic GFP in embryos
To quantify the difference in cytoplasmic fluorescence between embryos, the fluorescence intensity in the cytoplasm was normalized to the fluorescence intensity of the nascent epidermal attachment sites in each embryo. Mean values and standard deviation of normalized fluorescence intensity were plotted for each strain. Analysis was performed to three independent strains of each transgene in at least three independent experiments. Shown are results from one representative strain.
FRAP analysis of filaments
Fluorescence recovery after photobleaching (FRAP) studies were performed using a Zeiss LSM 5 EXCITER confocal scanning microscope. Worms at mid L4 stage were mounted on 4% agarose pads and anesthetized with 0.1 % tricaine, 0.01 % tetramisole, 30 min before FRAP. Images were acquired (1% laser power transmission) in a single Z-plane using open pinhole at 1 min intervals. Photobleaching was performed by 4× iterative scanning (100% transmission) focused on a region of 6.86 μm2. Mean fluorescence intensity at each time point within the bleached area and an adjacent unbleached region was quantified using the LSM 5 EXCITER software following manual corrections of the ROIs (due to animal movement). Data analysis. Normalized fluorescence intensity values were determined at each time point by the ratio (Tt/Ct)/(T0/C0). T0 is the intensity in the ROI before photobleaching, Tt is the intensity in the ROI at a given time point after bleaching. C0 and Ct are the intensities at time 0 and at time t, respectively, of a control unbleached area (Brignull et al., 2006).
The variation with time of the normalized fluorescence intensity values was averaged over n=12 animals for each strain. In addition, for each movie the normalized FRAP data was fitted to the equation 1−Ae−kt (Sprague and McNally, 2005), where k is the estimated rate of binding, i.e. the rate of dissociation-reassociation of the IFB-1::GFP molecules from and to the filament. Specifically, the logarithm of the fraction of the fluorescence recovered was plotted against time and the slope k was determined by linear regression for each movie. Mean value and standard deviation of k were calculated for n=12 animals from each strain. The half life time of fluorescence recovery, τ1/2, representing the dissociation-reassociation kinetics of IFB-1::GFP to the filaments, was calculated as τ1/ 2 = −ln 0.5/ k.
FRAP analysis of IFB-1 inclusions
Images were acquired using open pinhole at 10 sec intervals. Photobleaching was performed by 6× iterative scanning (100% transmission) focused on a region of 0.4 μm2.
FLIP analysis of IFB-1 inclusions
A region of 18 μm2 outside an inclusion was repeatedly bleached (1xscanning) at 10 sec intervals and images were acquired using open pinhole following each bleach. Measurements are shown for inclusions or circular-shaped filaments located at a distance of 10 μm from the bleached area.
Supplementary Material
Acknowledgments
We thank Rudolf Leube for the anti-IFB-1 antibody, Koret Hirschberg and David Broday for consulting on FRAP analysis and Ronen Zaidel-Bar for comments on the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The MH4 and MH46 monoclonal antibodies developed by the Waterston laboratory were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA. A.D.C. acknowledges funding from the NIH (GM54657). This research was supported by grants from the Israel Science Foundation (ISF 980/06) and the Israel Cancer Research Fund (06-203-RCDA) to L.B. and National Institutes of Health (NIH) grant GM67945 to S.P.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bar H, Mucke N, Kostareva A, Sjoberg G, Aebi U, Herrmann H. Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc Natl Acad Sci U S A. 2005;102:15099–15104. doi: 10.1073/pnas.0504568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher JM, Hahn BS, Legouis R, Sookhareea S, Weimer RM, Gansmuller A, Chisholm AD, Rose AM, Bessereau JL, Labouesse M. The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J Cell Biol. 2003;161:757–768. doi: 10.1083/jcb.200302151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull HR, Morley JF, Garcia SM, Morimoto RI, Kheterpal Indu, Ronald W. Methods in Enzymology. Academic Press; 2006. Modeling Polyglutamine Pathogenesis in C. elegans; pp. 256–282. [DOI] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Gupta BP, Sternberg PW, Podbilewicz B, Ronai Z. The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev. 2004;18:2380–2391. doi: 10.1101/gad.1227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001;8:443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- Chisholm AD, Hardin J. Epidermal morphogenesis. WormBook. 2005:1–22. doi: 10.1895/wormbook.1.35.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YH, Rosevear E, Goldman RD. Phosphorylation and disassembly of intermediate filaments in mitotic cells. Proc Natl Acad Sci U S A. 1989;86:1885–1889. doi: 10.1073/pnas.86.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EA, Hardin J. Sticky worms: adhesion complexes in C. elegans. J Cell Sci. 2004;117:1885–1897. doi: 10.1242/jcs.01176. [DOI] [PubMed] [Google Scholar]
- Denison C, Kirkpatrick DS, Gygi SP. Proteomic insights into ubiquitin and ubiquitin-like proteins. Current Opinion in Chemical Biology. 2005a;9:69–75. doi: 10.1016/j.cbpa.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A Proteomic Strategy for Gaining Insights into Protein Sumoylation in Yeast. Mol Cell Proteomics. 2005b;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the Globular Tail of Nuclear Lamin. J Biol Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Ding M, Goncharov A, Jin Y, Chisholm AD. C. elegans ankyrin repeat protein VAB-19 is a component of epidermal attachment structures and is essential for epidermal morphogenesis. Development. 2003;130:5791–5801. doi: 10.1242/dev.00791. [DOI] [PubMed] [Google Scholar]
- Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773:694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fridkin A, Karabinos A, Gruenbaum Y. Intermediate filaments in Caenorhabditis elegans. Methods Cell Biol. 2004;78:703–718. doi: 10.1016/s0091-679x(04)78024-3. [DOI] [PubMed] [Google Scholar]
- Ganesan AK, Kho Y, Kim SC, Chen Y, Zhao Y, White MA. Broad spectrum identification of SUMO substrates in melanoma cells. Proteomics. 2007;7:2216–2221. doi: 10.1002/pmic.200600971. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Current Opinion in Genetics & Development. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Hannappel E. beta-Thymosins. Ann N Y Acad Sci. 2007;1112:21–37. doi: 10.1196/annals.1415.018. [DOI] [PubMed] [Google Scholar]
- Hapiak V, Hresko MC, Schriefer LA, Saiyasisongkhram K, Bercher M, Plenefisch J. mua-6, a gene required for tissue integrity in Caenorhabditis elegans, encodes a cytoplasmic intermediate filament. Dev Biol. 2003;263:330–342. doi: 10.1016/j.ydbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: A History of Modification. Molecular Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Hresko MC, Schriefer LA, Shrimankar P, Waterston RH. Myotactin, a novel hypodermal protein involved in muscle-cell adhesion in Caenorhabditis elegans. J Cell Biol. 1999;146:659–672. doi: 10.1083/jcb.146.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitovich A, Mehta S, Na N, Ciechanover A, Goldman RD, Ridge KM. Ubiquitin-proteasome-mediated degradation of keratin intermediate filaments in mechanically stimulated A549 cells. J Biol Chem. 2008;283:25348–25355. doi: 10.1074/jbc.M801635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annual Review of Biochemistry. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Crowe E, Stevens TA, Candido EP. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2002;3:RESEARCH0002. doi: 10.1186/gb-2001-3-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabinos A, Schmidt H, Harborth J, Schnabel R, Weber K. Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc Natl Acad Sci U S A. 2001;98:7863–7868. doi: 10.1073/pnas.121169998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabinos A, Schulze E, Schunemann J, Parry DA, Weber K. In vivo and in vitro evidence that the four essential intermediate filament (IF) proteins A1, A2, A3 and B1 of the nematode Caenorhabditis elegans form an obligate heteropolymeric IF system. J Mol Biol. 2003;333:307–319. doi: 10.1016/j.jmb.2003.08.041. [DOI] [PubMed] [Google Scholar]
- Kim S, Coulombe PA. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 2007;21:1581–1597. doi: 10.1101/gad.1552107. [DOI] [PubMed] [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, Bonne G, Courvalin JC, Worman HJ, Zinn-Justin S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Ku NO, Omary MB. Keratins Turn Over by Ubiquitination in a Phosphorylation-modulated Fashion. J Cell Biol. 2000;149:547–552. doi: 10.1083/jcb.149.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku NO, Omary MB. Effect of mutation and phosphorylation of type I keratins on their caspase-mediated degradation. J Biol Chem. 2001;276:26792–26798. doi: 10.1074/jbc.M103315200. [DOI] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Hesse M, Peters B, Magin TM. Type II keratins precede type I keratins during early embryonic development. Eur J Cell Biol. 2005;84:709–718. doi: 10.1016/j.ejcb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C, Raught B. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol Cell. 2009;33:124–135. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Melchior F. SUMO-NONCLASSICAL UBIQUITIN. Annual Review of Cell and Developmental Biology. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- Nam-On Ku PSBHZGZTMBO. Keratins let liver live: Mutations predispose to liver disease and crosslinking generates Mallory-Denk bodies. Hepatology. 2007;46:1639–1649. doi: 10.1002/hep.21976. [DOI] [PubMed] [Google Scholar]
- Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One. 2009;4:e5905. doi: 10.1371/journal.pone.0005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Tao GZ, Toivola DM, Liao J. Heads and tails’ of intermediate filament phosphorylation: multiple sites and functional insights. Trends in Biochemical Sciences. 2006;31:383–394. doi: 10.1016/j.tibs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A Proteome-wide Approach Identifies Sumoylated Substrate Proteins in Yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- Perry JJP, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends in Biochemical Sciences. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Plenefisch JD, Zhu X, Hedgecock EM. Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development. 2000;127:1197–1207. doi: 10.1242/dev.127.6.1197. [DOI] [PubMed] [Google Scholar]
- Quinlan RA, Brenner M, Goldman JE, Messing A. GFAP and its role in Alexander disease. Experimental Cell Research. 2007;313:2077–2087. doi: 10.1016/j.yexcr.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A Universal Strategy for Proteomic Studies of SUMO and Other Ubiquitin-like Modifiers. Mol Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Lopez-Soler RI, Adam SA, Herrmann H, Moir RD, Spann TP, Goldman RD. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc Natl Acad Sci U S A. 2005;102:15494–15499. doi: 10.1073/pnas.0507612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, Strelkov SV, Aebi U, Cardoso MC, Goldman RD. The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J Cell Biol. 2008;181:269–280. doi: 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends in Cell Biology. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Iwase M, Konishi M, Tanaka M, Toh-e A, Kikuchi Y. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem Biophys Res Commun. 1999;259:582–587. doi: 10.1006/bbrc.1999.0821. [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., III Global Analysis of Protein Sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Woo WM, Goncharov A, Jin Y, Chisholm AD. Intermediate filaments are required for C. elegans epidermal elongation. Dev Biol. 2004;267:216–229. doi: 10.1016/j.ydbio.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KH, Yoon M, Moir RD, Khuon S, Flitney FW, Goldman RD. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J Cell Biol. 2001;153:503–516. doi: 10.1083/jcb.153.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Sarge KD. Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. J Cell Biol. 2008a;182:35–39. doi: 10.1083/jcb.200712124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Sarge KD. Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem Biophys Res Commun. 2008b;374:673–678. doi: 10.1016/j.bbrc.2008.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.