Abstract
• Background and Aims Vella pseudocytisus subsp. paui (Cruciferae) is a narrow endemic plant to the Teruel province (eastern Spain), which is listed in the National Catalogue of Endangered Species. Two distinct ploidy levels (diploid, 2n = 34, and tetraploid, 2n = 68) have been reported for this taxon that belongs to the core subtribe Vellinae, a western Mediterranean group of shrubby taxa with a chromosome base number of x = 17. Allozyme and AFLP analyses were conducted (a) to test for the ploidy and putative palaeo-allopolyploid origin of this taxon, (b) to explore levels of genetic diversity and spatial structure of its populations, and (c) to address in-situ and ex-situ strategies for its conservation.
• Methods Six populations that covered the entire geographical range of this taxon were sampled and examined for 19 allozyme loci and three AFLP primer pair combinations. In addition, the gametic progenies of five individuals were analysed for two allozyme loci that showed fixed heterozygosity.
• Key Results Multiple banded allozyme profiles for most of the surveyed loci indicated the polyploidy of this taxon. Co-inherited fixed heterozygous patterns were exhibited by the gametophytic tissues of the mother plants. Both allozyme and AFLP markers detected high levels of genetic diversity, and a strong micro-spatial genetic structure was recovered from AFLP phenetic analyses and Mantel correlograms.
• Conclusions Allozyme data support the hypothesis of an allotetraploid origin of Vella pseudocytisus subsp. paui that could be representative of other taxa of the core Vellinae group. AFLP data distinguished three geographically distinct groups with no genetic interaction among them. Allotetraploidy and outcrossing reproduction have probably contributed to maintenance of high levels of genetic variability of the populations, whereas habitat fragmentation may have enhanced the high genetic isolation observed among groups. In-situ microgenetic reserves and a selective sampling of germplasm stocks for ex-situ conservation of this taxon are proposed.
Keywords: AFLP, allotetraploidy, allozymes, conservation, endangered species, genetic diversity, Iberian endemics, Mantel correlograms, palaeopolyploidy, spatial structure, Vella pseudocytisus subsp. paui, Vellinae
INTRODUCTION
Vella pseudocytisus L. (Vellinae, Cruciferae) includes three allopatric subspecies of the western Mediterranean area restricted to the cold steppes of eastern, centre and southern Iberian Peninsula and of north-western Africa (Gómez-Campo, 1981, 1993; Crespo et al., 2000; Charco, 2001).
Vella pseudocytisus and other congeners, together with the monotypic endemic genera Boleum Desv. and Euzomodendron Coss., form the core of subtribe Vellinae (Crespo et al., 2000). Vellinae is one of the seven recognized subtribes of tribe Brassicaceae, considered to be a natural group of the mustard family (Schulz, 1923; Warwick and Black, 1994). The core Vellinae taxa are woody shrubs that share structural traits such as the possession of connate inner-stamen filaments, notched cotyledon apices, and a high chromosome base number of x = 17, otherwise unique in the family Brassicaceae (Gómez-Campo, 1981; Crespo et al., 2000). It has long been speculated that the woody genera of the core Vellinae might be palaeopolyploids of hybrid origin (Gómez-Campo, 1981; Warwick and Black, 1994; Warwick and Al-Shehbaz, 1998; Crespo et al., 2000); however, no attempts have been made to clarify the putative allopolyploid origin of Vella and its allies.
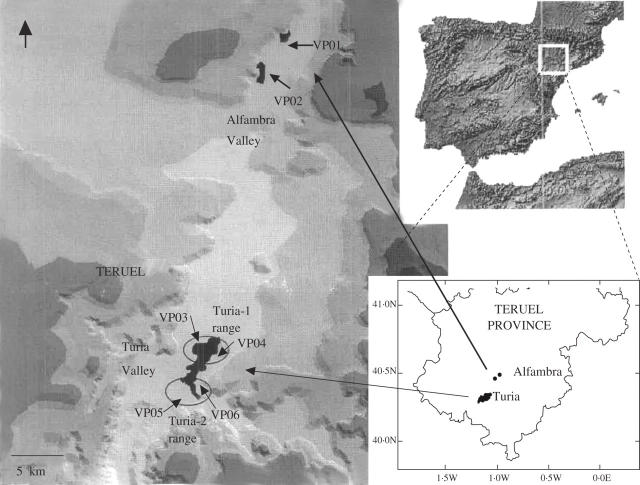
Vella pseudocytisus subsp. paui Gómez-Campo is one of the three described microtaxa of the V. pseudocytisus complex, which is characterized by its unarmed habit, spathulate leaves, and cochleariform fruit (Gómez-Campo, 1981, 1993). Vella pseudocytisus subsp. paui is a rare endemic taxon from eastern Spain, restricted to two limited and nearby habitats in the Alfambra and Turia valleys of Teruel province, in the autonomous community of Aragón. Its distribution area covers approx. 26 km2 (Fig. 1). This plant is a small camephyte nitrophilous shrub and differs from the others subspecies of V. pseudocytisus in the possession of rare tuberculate glaucous trichomes, though the plant is glabrous in all its main parts, including the edges of the leaves, the inflorescence axis and the valves of the fruit (Gómez-Campo, 1981, 1993).
Fig. 1.
Map of the area of distribution of Vella pseudocytisus subsp. paui and the locations of the populations studied. Alfambra valley: Cuevas Labradas (VP01) and Villaba Baja (VP02), Turia Valley: Turia-1 range, Villel-1 (VP03) and Villel-2 (VP04); Turia-2 range, Villastar-1 (VP05) and Villastar-2 (VP06). Map constructed with ARCVIEW GIS v3.2.
The chromosome counts given for the taxa of the V. pseudocytisus complex range from diploids with 2n = 34 (i.e. V. pseudocytisus subsp. glabrata Greuter, endemic to north-western Africa) to tetraploids with 2n = 68 (i.e. V. pseudocytisus subsp. pseudocytisus from central Spain) (Gómez-Campo, 1981). Two different ploidy levels have been reported for V. pseudocytisus subsp. paui. An initial study indicated that the populations were diploid with individuals showing 2n = 34 chromosomes (Gómez-Campo, 1981); however, a recent assay provided new counts on individuals with 2n = 68 chromosomes (Domínguez-Lozano et al., 2003) implying that the taxon could be tetraploid.
Vella pseudocytisus subsp. paui is a steppe plant that grows in an area characterized by continental Mediterranean climate. It ranges between 800 and 1200 m a.s.l. and lives on calcareous and gypsicolous substrates, associated with other typical species of open Mediterranean shrubland communities (Sainz-Ollero et al., 1996; Domínguez-Lozano et al., 2005). This endemic is considered to be a preglacial relict that was probably more widely distributed than at present. The present limited distribution reflects intense human and grazing pressures over many centuries (Sainz-Ollero et al., 1996).
Because of its scarcity and potential degree of endangerment, Vella pseudocytisus subsp. paui was included in the list of endangered plants of Aragón, under the category of ‘in danger of extinction’ (Decree 49 of 28 March 1995, BOA number 42). It has been also classified as ‘endangered’ (EN: B1+2abde) in two national catalogues of endangered plans, the red list of endangered vascular flora of Spain (VVAA, 2000), and in the atlas and red book of the endangered vascular flora of Spain (Bañares et al., 2003). In 2003, a recovery plan for this taxon was established by the regional Government of Aragón, which prioritised the study of its biology, demography and population genetics, and the establishment of a seed bank for the ex-situ conservation of this plant (Decree 92/2003).
Information on the structuring of intraspecific genetic variation in rare species is of great importance for the development of conservation strategies (Newton et al., 1999). Even if neutral markers are not expected to be linked to adaptive characters, levels of genetic diversity might indirectly reflect the fitness of populations or their adaptive potential (Ellstrand and Elam, 1993; Wise et al., 2002). Population genetic diversity and structure are crucial aspects for interpreting the evolutionary history and the ecological success of rare and threatened plants (Hamrick and Godt, 1996; Escudero et al., 2003). In this study the levels of genetic variability and the genetic structure of the critically endangered V. pseudocytisus subsp. paui were investigated through (a) multi-locus fingerprinting produced by the AFLP technique (Vos et al., 1995), which provides a large number of informative markers derived from loci widely dispersed throughout the nuclear genome, and (b) allozymic variation, reflecting population diversity based on the variability of genes that encode for specific enzymes. The use of both separate and combined hypervariable nuclear markers and allozyme methods has proven its potential use to address fine-scale population genetic and life-history studies of endemic and endangered plants (Travis et al., 1996; Palacios et al., 1999; Gaudeul et al., 2000; Keiper and McConchie, 2000; Lutz et al., 2000; Zhang et al., 2001; Segarra-Moragues and Catalán, 2002, 2003; Prentice et al., 2003; Rottenberg and Parker, 2003; Tero et al., 2003; López-Pujol et al., 2004).
Furthermore, allozyme analysis has been applied to the study of gametic progenies in V. pseudocytisus subsp. paui. The allozyme technique, due to its great capacity to genotype each individual (Wendel and Weeden, 1989) and to recover past hybridization signals (Liston et al., 1995), has been used extensively in the determination of levels of ploidy and gene duplication in plants (Gottlieb, 1977; Goldring et al., 1985; Wolko and Weeden, 1989; Segarra-Moragues and Catalán, 2002; Segarra-Moragues et al., 2004). A further aim of the present study was to identify the level and nature of ploidy in V. pseudocytisus subsp. paui. These data could provide evidence on population history and contribute to a more general understanding of the evolutionary processes implied in the putative hybrid origin of this plant and of other related taxa of the core subtribe Vellineae.
MATERIALS AND METHODS
Plant material
The study was undertaken on six populations of V. pseudocytisus subsp. paui covering the entire distribution of this taxon. The samples in each population were collected at distances of >5 m from each other to avoid sampling the same ramet more than once, given the reported vegetative spread of this taxon (Domínguez-Lozano et al., 2003, 2005). The populations are arranged into two valleys separated by 25 km (Fig. 1). The northern populations from the Alfambra valley, VP01 (Teruel, Cuevas Labradas) and VP02 (Teruel, Villalba Baja), cover a total area of approx. 9 km2, but are found in small patches in ravines separated by 2.5 km from each other (Fig. 1 and Table 1). The southern populations from the Turia valley, VP03 (Teruel, Villel-1), VP04 (Teruel, Villel-2), VP05 (Teruel, Villastar-1), and VP06 (Teruel, Villastar-2), cover a total area of approx. 16 km2 and are mainly distributed in fragmented patches at the borders of field crops (Fig. 1 and Table 1). The populations from the Turia valley are spatially close to each other but could be further subdivided into two microgeographical ranges (Fig. 1 and Table 1). The Turia-1 range includes the Villel-1 and Villel-2 populations, separated from each other by 1.6 km, whereas the Turia-2 range encompasses the Villastar-1 and Villastar-2 populations, separated from each other by 1.4 km and by <5 km from the Turia-1 range. Thirty individuals were sampled from each population except for the smaller population of Villel-2 from which only 12 individuals were available. The coordinates of each individual and the distances among them were measured using the Global Positioning System (GPS).
Table 1.
Population code, locality, geographical range, latitude and longitude coordinates, and number of individuals per population (n) of the six studied populations of Vella pseudocytisus subsp. paui
| Code | Locality | Range | Latitude | Longitude | n |
|---|---|---|---|---|---|
| VPO1 | Spain: Teruel: Cuevas Labradas | Alfambra | 40°26′N | −1°3′E | 30 |
| VPO2 | Spain: Teruel: Villalba Baja | Alfambra | 40°25′N | −1°4′E | 30 |
| VPO3 | Spain: Teruel: Villel-1 | Turia-1 | 40°14′N | −1°9′E | 30 |
| VPO4 | Spain: Teruel: Villel-2 | Turia-1 | 40°15′N | −1°7′E | 12 |
| VPO5 | Spain: Teruel: Villastar-1 | Turia-2 | 40°13′N | −1°1′E | 30 |
| VPO6 | Spain: Teruel: Villastar-2 | Turia-2 | 40°14′N | −1°8′E | 30 |
Allozyme analysis
Fresh leaves from a total of 162 individuals from six populations were sampled in the field and kept on ice until enzyme extraction. Leaf tissue was ground in cold extraction buffer consisting of 0.2 m Tris–HCl, pH 7.5, 2 mm EDTA, 0.12 m Na2S2O5, 1 m MgCl2, 80 mg mL−1 PVP and 40 µL mL−1 mercaptoethanol. Extracts were absorbed onto Whatman 3 MM paper wicks and subjected to electrophoresis on horizontal 10 % starch gels. Sixteen enzyme systems were assayed in three different types of gel and electrode buffers. Eleven of these were satisfactorily resolved: aspartate aminotransferase (AAT; EC 2.6.1.1), phosphoglucose isomerase (PGI; EC 5.3.1.9), phosphoglucose mutase (PGM; EC 5.4.2.2) and triosephosphate isomerase (TPI; EC 5.3.1.1), in lithium-borate pH 8.5 (electrode buffer: 0.19 m boric acid; 0.03 m lithium hydroxide; gel buffer 1 : 9 of electrode buffer and 0.05 m TRIS, 0.008 m acid citric monohydrate); aconitase (ACO; EC 4.2.1.3), esterase (EST, EC 3.1.1.–), malic enzyme (ME; EC 1.1.1.40), menadione reductase (MNR; EC 1.6.99), glucose-6-phosphate dehydrogenase (6-GDP; EC 1.1.1.49) in TRIS–borate pH 8.6 (electrode buffer: 1 m TRIS, 0.5 m boric acid, 0.02 m EDTA, adjusted to pH 8.6 with NaOH (pellets); gel buffer: 1 : 9 of electrode buffer); isocitrate dehydrogenase (IDH; EC 1.1.1.42), and shikimic dehydrogenase (SKD; EC 1.1.1.25), in morpholine-citrate pH 6.1 (electrode buffer: 0.04 m citric acid monohydrate adjusted to pH 6.1 with N-(3-aminopropyl)-morpholine; gel buffer 1 : 19 of electrode buffer solution). Gels were stained for band visualization following Wendel and Weeden (1989). The putative alleles were numbered consecutively commencing with the most anodal form.
The 11 enzyme systems were scored for 19 loci, of which only two (ME-1 and SKD-1) were invariable. The putative loci AAT-1, AAT-2, ACO-1, ACO-2, EST-1, EST-2, PGM-2, 6PGD-1, PGI-2, IDH-1, MNR-2 and TPI-2 presented a high number of bands per individual, ranging from three (6PGD-1, TPI-2) to nine bands (PGI-2). Neither the complexity of the phenotypic patterns nor the secondary structure characteristics of each enzymatic system correspond to the reported diploidy of V. pseudocytisus subsp. paui (Gómez-Campo, 1981, 1993). Because of the complex banding profiles, allozymic bands were scored as presence/absence for population genetic analyses. Three allozyme loci showed fixed heterozygosity in all (TPI-2) or part (PGI-2, PGM-2) of the studied individuals. These banding profiles indicated the possible allopolyploidy of V. pseudocytisus subsp. paui. To check for this hypothesis, two or three fresh leaves and 10–20 developed but not-opened floral buds were collected from the same individual in five distant plants from the Villalba Baja population (VP02; Fig. 1). The pollen grains were extracted from the anthers, homogenized on refrigerated plates and assayed simultaneously with the extract of the leaves from their respective mother plants for the two dimeric enzyme systems for which fixed heterozygosity has been detected previously (PGI and TPI).
DNA isolation and AFLP analysis
Genomic DNA from silica gel-dried leaves was extracted using the CTAB protocol of Doyle and Doyle (1987) with slight modifications. The quality of the extract was checked on 1 % TBE-agarose gel and the concentration was estimated by comparison with samples of known concentration. Samples were diluted to a final concentration of 100 ng µL−1. The AFLP technique (Vos et al., 1995) was carried out following the Invitrogen procedures according to the manufacturer's instructions. Digestion of 200 ng DNA samples was performed using EcoRI and MseI restriction enzymes at 37 °C for 2 h, followed by heat inactivation of the restriction endonucleases at 70 °C (15 min). A double–stranded adaptor ligation to the restriction site was applied for 2 h (20 °C). Pre-selective amplification was performed using the AFLP pre-amp primer mix®, followed by a three-nucleotide selective amplification with specific primer pairs. The products were separated on 6 % polyacrylamide gels and silver stained following Bassam et al. (1991).
Three (E-AAG/M-CTC, E-ACC/M-CAG and E-AGC/M-CTA) of the nine primer pair combinations tested in a pilot study that generated clear and reproducible bands and showed variation among and within populations were selected and used to assess the entire set of samples.
Data analysis
Binary data (presence/absence) matrices were constructed from different data sources: (a) from AFLP bands and (b) from allozyme patterns, because of the impossibility of coding individual genotypes with confidence. Nei's genetic diversity index (Nei, 1973) was calculated for each population and for the species as h = 1 – 1/m Σl Σu plu2, where plu is the frequency of the uth band in the lth locus and m is the total number of loci (Peever and Milgroom, 1994). As an additional estimation of the genetic diversity of each population, the percentages of both allozyme and AFLP polymorphic loci (%POP) and the numbers of rare (fr) and private (unique) fragments (fu) were calculated. Fragments were treated as ‘rare’ when frequencies were below 0.10 (Tribsch et al., 2002; Martínez-Ortega et al., 2004).
Population genetic structure was first analysed based on both allozyme and AFLP data by means of analysis of molecular variance (AMOVA; Excoffier et al., 1992) using ARLEQUIN v. 2.000 (Schneider et al., 2000). AMOVAs were conducted at two hierarchical levels: (1) considering all populations belonging to the same group (among populations and within individuals); and (2) considering different groupings of populations in order to test the existence of structure among different geographical divisions. Significance levels of the variance components estimated for each case were obtained by non-parametric permutation using 1000 replicates. AFLP markers detected a higher genetic structure among populations and geographical ranges from individual phenotypes than allozymes and, therefore, all subsequent analyses were exclusively based on AFLPs.
Population AFLP structure was also inferred through a Bayesian model-based clustering method using STRUCTURE v.2.0 (Pritchard et al., 2000). This software attempts to identify K populations or clusters, each of which is characterized by a set of allele frequencies at each locus, assigning probabilistically the sampled individuals to the hypothetical clusters. The probabilities for a range of values of K starting from 1 to 6 were examined. These analyses were based on an admixture ancestry model with correlated allele frequencies, using a burn-in period and a run length of the Markov Chain Monte Carlo of 105 and 106 iterations.
Genetic relationships among individuals were obtained through principal co-ordinates (PCO) analysis based on Dice distances computed with NTSYSpc v. 2.11a (Rohlf, 2002). A two-dimensional visualization was completed with the superposition of the minimum spanning tree (MST; Gower and Ross, 1969; Dunn and Everitt, 1982) on the corresponding PCO plot. Gene flow among populations was estimated according to FST statistics of Wright (1965), following the expression M = (1 – FST/2FST) for haplotypes implemented in ARLEQUIN, where Nm is the absolute number of migrants exchanged between two populations.
Mantel tests (Mantel, 1967) were used in three different ways to investigate: (1) the linear correlation between population genetic distances and geographical distances (R values), (2) the isolation-by-distance between pairs of populations which is a linear correlation between a matrix of the number of migrants per generation with a matrix of geographical distances (R values) (see Fig. 3), and (3) the autocorrelations between genetic and geographical distances among populations and among different geographical groups (RM values). For this purpose, the population genetic distance matrices were correlated to different binary matrices encoded according to the corresponding geographical relationship to be tested (Oden and Sokal, 1986; Stehlik et al., 2001) and the results were plotted (see Figs 4 and 5). Significance levels for all assayed Mantel tests were assessed through 1000 non-parametric permutations in NTSYS v.2.1. Three range correlograms were computed, one for each of three geographical areas under study (Alfambra, Turia1 and Turia2). Six population correlograms, one for each population, were constructed independently. Sequential Bonferroni-type corrections (Rice, 1989) were applied to test for significance in all multiple tests.
To investigate the relationship between AFLP-genetic distances as a function of the geographical distribution between pairs of individuals of V. pseudocytisus subsp. paui, estimates of autocorrelation per distance class were calculated following Gabrielsen et al. (1997) and Stehlik et al. (2001). A geographical distance matrix (km) between pairs of individuals, based on the GPS data, was constructed using the R-package v.4.0 d6 (Casgrain and Legendre, 2001). Ten distance classes were chosen for use in the analysis. The first distance class included all within-population distances (<1 km). The following classes were set at different distances, intending to contain similar numbers of pairwise comparisons: (1) 0 ≥ d < 1 km; (2) 1 ≥ d < 2 km; (3) 2 ≥ d < 3 km; (4) 3 ≥ d < 4 km; (5) 4 ≥ d < 19 km; (6) 19 ≥ d < 21 km; (7) 21 ≥ d < 22 km; (8) 22 ≥ d < 23 km; (9) 23 ≥ d < 24 km; (10) 24 ≥ d < 30 km. The geographical distance matrix was correlated with the Euclidean genetic distance matrix between pairs of individuals through 1000 non-parametric Mantel test permutations (RM values). The diameter of the genetic patch in V. pseudocytisus subsp. paui was deduced from the first x-axis intercept in the resulting correlogram (Escudero et al., 2003).
RESULTS
Nineteen putative allozyme loci were satisfactorily resolved across the studied individuals of V. pseudocytisus subsp. paui. Two of these loci (ME and SKD) were monomorphic. In most cases the remaining loci showed complex patterns of multiple banding profiles, consistent with a polyploid condition of this taxon. The monomeric enzyme systems ACO and PGM that usually generate one band for homozygotes and two bands for heterozygotes in diploid plant species (Wendel and Weeden, 1989) revealed up to five and six bands, respectively, in individuals of V. pseudocytisus subsp. paui. The PGM-2 locus also showed frequent diallelic heterozygous patterns in several individuals. The dimeric systems AAT and IDH (AAT1, AAT2 and IDH1) showed patterns of more than the three expected bands for heterozygotes, whereas other dimeric systems (PGI and TPI) showed fixed heterozygosity in their respective loci for most (PGI-2) or all (TPI-2) of the studied individuals. The complex systems MNR and EST, that could be di-, tetra-, or hexameric (Wendel and Weeden, 1989), showed up to five and eight bands per individual, respectively.
The PGI-2 and TPI-2 allozyme analysis of foliar and pollen grain tissues of five VP02 individuals showed a total correspondence between the sporophytic and the gametophytic profiles in the same individual, corroborating the polyploidy of V. pseudocytisus subsp. paui. All studied diallelic TPI-2 cases and all variable diallelic PGI-2 cases exhibited fixed three-banded profiles with enhanced staining of the heterodimeric band in both leaf and pollen grain individual samples.
Allozyme/AFLP diversity in V. pseudocytisus subsp. paui
The presence/absence matrix constructed for the 11 enzyme systems studied rendered 47 bands of which 19.14 % were monomorphic (Table 2). Only one of these bands was exclusive to the Alfambra valley. Populations VP01 and VP02, from the Alfambra valley, and VP06, from the Turia-2 range, each expressed a private band but in low frequency (<0.3) (Table 2). The highest number of rare bands was found in VP01 (fr = 7) and the lowest number in VP06 (fr = 2)
Table 2.
Genetic diversity values obtained from allozymes and AFLPs in the six studied populations of V. pseudocytisus subsp. paui
| Tzyme | TAFLP | fu zyme | fu AFLP | fr zyme | fr AFLP | hzyme | hAFLP | %Pzyme | %PAFLP | |
|---|---|---|---|---|---|---|---|---|---|---|
| VP01 | 33 | 133 | 1 | 1 | 7 | 10 | 0.523 | 0.530 | 63.62 | 62.50 |
| VP02 | 31 | 137 | 1 | 3 | 4 | 19 | 0.512 | 0.580 | 63.82 | 65.85 |
| VP03 | 28 | 126 | 0 | 1 | 4 | 7 | 0.557 | 0.603 | 59.57 | 63.94 |
| VP04 | 34 | 123 | 0 | 0 | 3 | 23 | 0.461 | 0.563 | 72.34 | 59.13 |
| VP05 | 30 | 135 | 0 | 0 | 5 | 9 | 0.546 | 0.522 | 63.00 | 64.90 |
| VP06 | 31 | 125 | 1 | 1 | 2 | 16 | 0.532 | 0.541 | 65.96 | 61.53 |
| Total | 47 | 208 | − | − | 9 | 23 | 0.552 | 0.581 | 80.86 | 91.83 |
Number of total bands (T) and number of private (fu) and rare (fr) alleles for each marker.
Genetic diversity estimators: Nei's genetic diversity index and percentage of polymorphic loci for allozyme (hzyme, %Pzyme) and AFLP (hAFLP, %PAFLP) data.
Population codes are those listed in Table 1.
The three AFLP primer combinations produced a total of 208 bands of which 8.17 % were monomorphic (Table 2). Two geographically private markers were found, one for each of the Alfambra and the Turia valleys. AFLP also detected a variable number of private markers for some populations though in low frequencies (<0.4): populations VP01, VP03 and VP06 exhibited one private marker each, whereas population VP02 exhibited three private markers. Rare AFLP fragments were more common than rare allozyme bands; VP04 and VP03 expressed the highest (fr = 23) and lowest (fr = 7) numbers of rare AFLP markers, respectively (Table 2).
Both allozyme and AFLP techniques provided 162 distinct multilocus phenotypes, one for each individual studied. AFLP detected slightly higher levels of genetic diversity than allozymes (Table 2). Although the allozymic diversity values could have been overestimated due to redundancy caused by heterodimeric and heteroduplex bands, the two data sets were congruent showing similar values of relatively high genetic diversity and high percentages of polymorphic loci (>59 %) across the populations of V. pseudocytisus subsp. paui. Allozyme values of genetic diversity ranged from 0.557 (VP03) to 0.461 (VP04) with a mean of 0.552, whereas AFLP values ranged from 0.603 (VP03) to 0.530 (VP01) with a mean of 0.581 (Table 2).
Genetic structure of V. pseudocytisus subsp. paui
The substantial within-population genetic variation revealed by both allozyme and AFLP techniques was evident in the AMOVA analysis (Table 3). Most of the total variation (>78 %) was found among individual within populations (P < 0.001), whereas a relatively small fraction of it (<22 %) (P < 0.001) was found among populations (Table 3A). In the hierarchical AMOVAs, the AFLP technique confirmed its greater precision in identifying higher levels of genetic population structure than that estimated by allozymes. The separation of the two close geographical ranges of Alfambra and Turia valleys corresponded to a significant value of 12.37 % (P < 0.001) of the total AFLP variance, in contrast to the 4.43 % non-significant value (P = 0.058) detected by allozymes (Table 3B). However, the most obvious structure was obtained for the partition of the variance into three geographical ranges (Alfambra vs. Turia-1 vs. Turia-2), for which AFLP also showed higher significant among-range values (19.72 % vs. 6.03 %) and lower significant within-range (among populations) values (5.25 % vs. 14.93 %) than allozymes (Table 3C).
Table 3.
Analysis of molecular variance (AMOVA) based on 162 phenotypes of Vella pseudocytisus subsp. paui assayed with allozymes and AFLP
| Allozymes |
AFLP |
|||
|---|---|---|---|---|
| Source of variation | % of the total variance | P-value | % of the total variance | P-value |
| (A) One group | ||||
| (V. pseudocytisus subsp paui) (VP01–VP06) | ||||
| Among populations | 20.00 | <0.001 | 21.89 | <0.001 |
| Within populations | 80.00 | <0.001 | 78.11 | <0.001 |
| (B) Two groups | ||||
| Alfambra valley (VP01–VP02) vs. Turia valley (VP03–06) | ||||
| Among groups | 4.43 | 0.058 | 12.37 | <0.001 |
| Among populations | 17.10 | <0.001 | 13.71 | <0.001 |
| Within populations | 78.47 | <0.001 | 73.92 | <0.001 |
| (C) Three groups | ||||
| Alfambra valley (VP01–VP02) vs. Turia-1 range (VP03–04) vs. Turia-2 range (VP05–06) | ||||
| Among groups | 6.03 | <0.001 | 19.72 | <0.05 |
| Among populations | 14.93 | <0.001 | 5.25 | <0.001 |
| Within populations | 79.04 | <0.001 | 75.03 | <0.001 |
Population codes are those listed in Table 1.
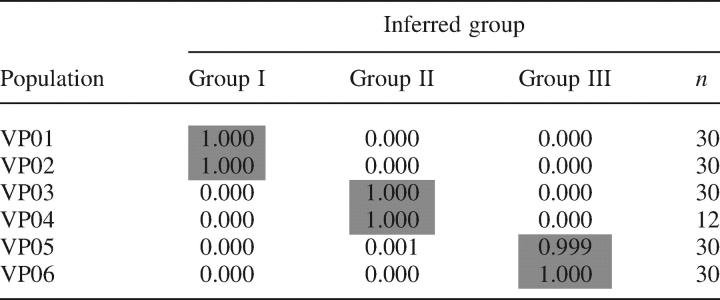
Estimation of individual membership to predefined groups using STRUCTURE revealed a marked AFLP geographical structure for K = 3 (Table 4), which correlated well with the three geographical groups previously characterized through AMOVA (Table 3C). All individuals from the two populations of the Alfambra valley (VP01 and VP02) clustered in one group (Group I; Table 4), whereas those from the Turia valley clustered into two different groups, corresponding to the Turia-1 (Group II) and Turia-2 (Group III) population cores (Table 4), showing high percentages of membership in all cases (>99.9 %).
Table 4.
Proportion of membership of individual AFLP multilocus phenotypes from each population of V. pseudocytisus subsp. paui (VP01–VP06) to each predefined group inferred by STRUCTURE (K = 3; Groups I–III)
Population codes and numbers of individuals per population are those indicated in Table 1.
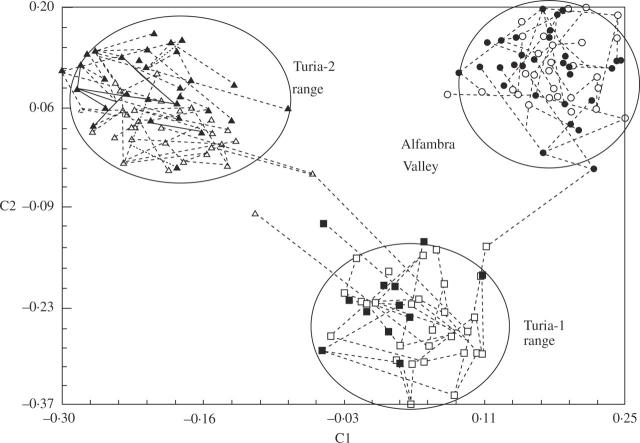
PCO analysis also detected a similar microspatial structure in the three separate clusters obtained after the bidimensional plotting of individual AFLP phenotypes (Fig. 2). Populations of the Alfambra valley clustered separately from those of the Turia-2 range and, to lesser extent, from those of the Turia-1 range along the first axis that accumulated 8.42 % of the total AFLP variance. The Turia-1 and Turia-2 ranges were further separated along the second axis that accounted for 6.54 % of the total variance (Fig. 2). The minimum spanning tree showed the high genetic similarity among individuals within populations and among populations within microgeographical areas (Fig. 2). Only one individual of the Turia-2 range (from population VP05) appeared more related to the Turia-1 range than to its own population group. No other individuals were found mixed among the three groups (Fig. 2).
Fig. 2.
Two-dimensional PCO plot of 162 individual AFLP phenotypes of Vella pseudocytisus subsp. paui. Axis 1 (8.42 %) and axis 2 (6.54 %) accounted for the 14.96 % of the total variance. Populations: Alfambra valley, Cuevas Labradas (open circles, VP01) and Villaba Baja (closed circles, VP02); Turia-1 range, Villel-1 (open squares, VP03) and Villel-2 (closed squares, VP04); Turia-2 range, Villastar-1 (open triangles, VP05) and Villastar-2 (closed triangles, VP06).
Isolation-by-distance among close geographical ranges in V. pseudocytisus subsp. paui
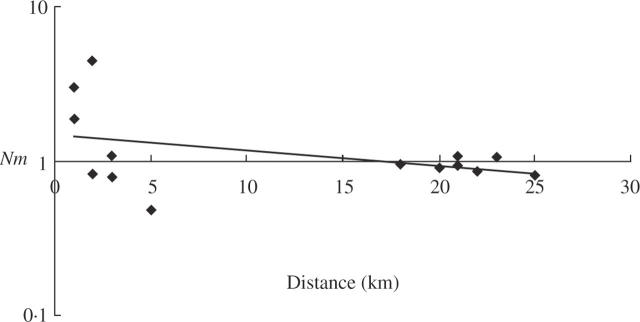
Despite the short geographical distances that separate the Alfambra and the Turia valleys (25 km; Fig. 1) and the Turia-1 and Turia-2 ranges (<5 km; Fig. 1), genetic isolation was detected among the three geographical areas. The Mantel test revealed a significant but moderate correlation between the genetic and the geographical distances (R = 0.560, P < 0.05). In the estimations based on the number of migrants, the Mantel correlation tests showed a significant negative value of r = –0.48 (P < 0.05) and the plotted values indicated that populations located at short geographical distances (<5 km) experience higher interpopulational gene flow than populations that are spatially separated by >20 km, which are therefore genetically isolated (Fig. 3). The two populations from the Alfambra valley exhibit the largest number of migrants per generation (4.45), followed by those from the Turia-2 range (3 migrants/generation), and then by those from the Turia-1 range (1.84 migrants/generation).
Fig. 3.
Relationship between geographical distances (x-axis in kilometres) and the number of migrants (Nm) (y-axis in logarithmic scale) between populations of Vella psudocytisus subsp. paui. Values above 1 are significant.
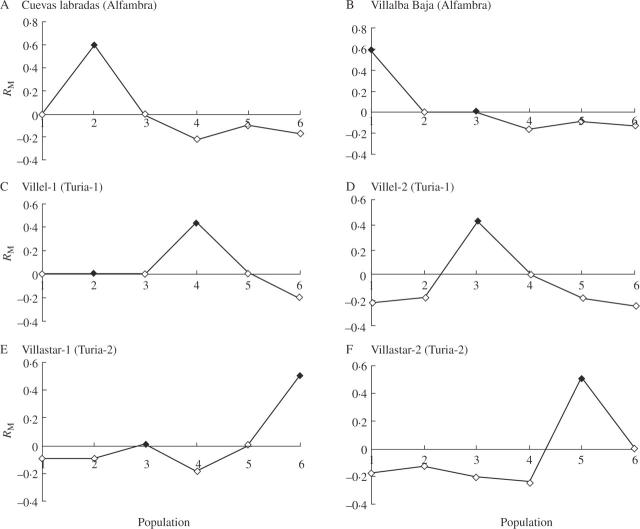
In regional Mantel correlograms, none of the correlation values within geographical ranges were significant (Alfambra: RM = 0.639, P = 0.077; Turia-1 core: RM = 0.392, P = 0.202; Turia-2 core: RM = 0.527, P = 0.110) and no significant correlations were detected among geographical ranges (results not shown). Similarly, population Mantel correlograms revealed non-significant correlations between populations within ranges (VP01 and VP02 in the Alfambra valley (RM = 0.593, P = 0.063), VP03 and VP04 in the Turia-1 range (RM = 0.434, P = 0.207) and VP05 and VP06 in the Turia-2 range (RM = 0.505, P = 0.131). Also, a non-significant correlation between the Villalba Baja (VP02, Alfambra valley) and the Villel-1 (VP03, Turia-1 range) populations was found (RM = 0.012, P = 0.341) (Fig. 4).
Fig. 4.
Mantel autocorrelograms of the six populations of Vella pseudocytisus subsp. paui studied compared with themselves and to the remaining populations. Closed diamonds indicate positive Mantel correlation. For details, see text or Oden and Sokal (1986), and for population codes see Table 1.
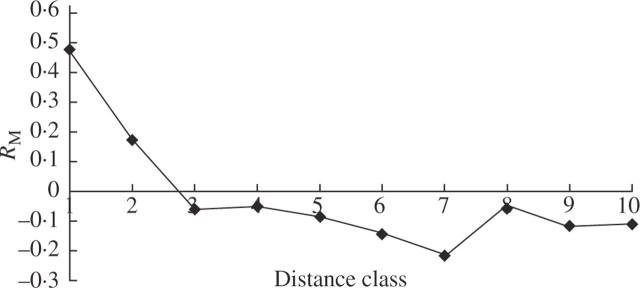
AFLP Mantel tests between genetic and geographic distance classes decreased from significantly positive RM values in distance classes 1 and 2 (separation between individuals <2 km) to significantly negative values in distance classes 3–10 (separation between individuals >3 km) (Fig. 5). The highest Mantel value was class 1 (within populations distances; RM = 0.476, P = 0.001), while the lowest Mantel value occurred in distance class 7 (RM = –0.212, P = 0.001). The correlogram showed a cline, structured by distance classes, as geographical distances were increased. Individuals located at short distances (<2 km) were more similar genetically than those separated by >3 km (Fig. 5). As the first intercept of the correlogram line with the x-axis is between 2 and 3 km, the size of the genetic patch in V. pseudocytisus subsp. paui would be around 2.5 km.
Fig. 5.
Mantel correlogram of RM values (y-axis) for different distance classes (x-axis) across the six populations studied in Vella pseudocytisus subsp. paui. Distance classes correspond to those indicated in the text. All values were significantly different from zero (P < 0.05) after Bonferroni corrections.
DISCUSSION
Palaeopolyploidy of V. pseudocytisus subsp. paui and of the core Vellinae group
Complex patterns of allozyme bands, like those detected in V. pseudocytisus subsp. paui, have been reported in a variety of angiosperms. These patterns have been viewed as the consequence of gene duplications (Gottlieb, 1977; Goldring et al., 1985; Wolko and Weeden, 1989; Williamson and Werth, 1999; Segarra-Moragues and Catalán, 2002), hybridization (Stebbins, 1985), or polyploidization (Weeden and Wendel, 1989; Liston et al., 1995; Segarra-Moragues et al., 2004). In the case of V. pseudocytisus subsp. paui the complex patterns might have resulted from single gene duplications, but it seems improbable that 12 out of the 17 studied variable loci were all duplicated. Moreover, the observed results are similar to the patterns obtained for other tetraploid plants (Hörandl and Grielhuber, 2002; López-Pujol et al., 2004). Therefore, it is more appropriate to interpret polyploidy of V. pseudocytisus subsp. paui as the factor responsible for its high number of allozymic bands rather than the independent duplication of many genes. Although previous results attributed a diploid level to this taxon (2n = 34; Gómez-Campo, 1981, 1993), our allozyme data support a tetraploid status (2n = 68; Domínguez-Lozano et al., 2003).
A further insight into the nature of the ploidy status of V. pseudocytisus subsp. paui was obtained from the allozymic analysis of gametic progenies. Allopolyploidy, implying the duplication of heterologous genomes from previously hybridised divergent taxa, is considered to be the most common speciation mechanism in higher plants (Stebbins, 1950, 1971; Grant, 1981; Masterson, 1994; Otto and Whitton, 2000). However, recent studies have enlarged the number of detected cases of autopolyploidy, where an increase in chromosome number was due to an increase in unreduced gametes within a single individual (Soltis and Soltis, 1993, 1999, 2000; Galloway et al., 2003). Fixed heterozygous patterns are widely interpreted as evidence of a hybrid origin of a plant (Weeden and Wendel, 1989; Wolko and Weeden, 1989; Liston et al., 1995) although they could also be interpreted as a result of independent gene duplication events. However, the heterozygous pattern observed in the sporophytic tissue of a mother plant would not be reproducible in all descendants of a conventional diploid that reproduces sexually (Segarra-Moragues et al., 2004). The congruence of the three-banded profiles found for the PGI-2 and TPI-2 loci in the sporophytic and gametophytic tissues of the heterozygous individuals of V. pseudocytisus subsp. paui studied strongly supports its allotetraploidy, because of the coinheritance of the two alleles in the same gamete. The common pattern observed in these two independent loci suggests duplicated disomic inheritance, typical of an allopolyploid, rather than an independent expression of each gene in the gamete pool of the anther, characteristic of diploid plants, or the polysomic segregations expected for an autotetraploid (Weeden and Gottlieb, 1979; Segarra-Moragues et al., 2004).
Regardless of the ploidy level detected, the allopolyploid nature of V. pseudocytisus subsp. paui suggests that a more ancestral hybridization event likely predated the divergence of the Vellinae core group. In the family Brassicaceae approximately 37 % of the taxa have increased genome sizes and have speciated through allopolyploidy (Koch, 2003). Some apparently diploid genera, like Brassica, are palaeopolyploids (Koch et al., 2003). Furthermore, a putative hexaploid ancestor has been proposed for Brassicaceae as all the species of this tribe, to which Vellineae belongs, show triplicated copies of a basic genome type (Lysak et al., 2005). The results of the present analysis concur with this scenario, leading to speculation on the hybrid palaeopolyploid origin of V. pseudocytisus subsp. paui and also on the core Vellineae genera (Boleum, Euzomodendron and Vella). Multiple-banded allozymic patterns and fixed heterozygosities for some loci/populations detected in V. pseudocytisus subsp. paui and in the closely related species Boleum asperum (Pers.) Desv. provided support for the hypothesis that a high chromosome base number for the core Vellineae taxa (x = 17) could be derived from hybridization and amphipolyploidization of ancestral lineages with low chromosome base numbers present today in the annual basal genera of the tribe, Carrichtera (x = 8) and Succowia (x = 9) (Gómez-Campo, 1981; Warwick and Black, 1994; Warwick and Al-Shehbaz, 1998; Crespo et al., 2000). More exhaustive analyses of other Vellineae core taxa would be required to confirm this hypothesis.
High levels of genetic diversity and spatial genetic structure in V. pseudocytisus subsp. paui
Levels of within-population genetic diversity and the spatial structuring of intraspecific diversity in plant species are determined by a range of ecological, demographic and historical factors (Hamrick and Godt, 1996). There is evidence that low levels of genetic diversity are common features of endemic plant species (Ayres and Ryan, 1999; Segarra-Moragues and Catalán, 2002; López-Pujol et al., 2003; Park, 2004), although several studies have also found high levels of genetic variability in narrow plant endemics (Ranker, 1994; Rossetto et al., 1995; Torres et al., 2003; López-Pujol et al., 2004; Xue et al., 2004). This is the case for V. pseudocytisus subsp. paui, for which the high degree of genetic diversity found was unexpected, in light of its narrow range and its geographic isolation from other conspecific taxa, the relatively low number of individuals per population, and the anthropogenic pressure that is currently operating on them. The allozyme (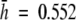 ) and AFLP (
) and AFLP (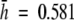 ) genetic diversity found in this taxon is higher than the diversity reported for other allogamous endemic and endangered plants studied with AFLPs [i.e. Astragalus cremnophylax var. cremnophylax
) genetic diversity found in this taxon is higher than the diversity reported for other allogamous endemic and endangered plants studied with AFLPs [i.e. Astragalus cremnophylax var. cremnophylax 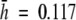 (Travis et al., 1996); Sticherus flabellatus
(Travis et al., 1996); Sticherus flabellatus 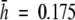 (Keiper and McConchie, 2000); Eryngium alpinum
(Keiper and McConchie, 2000); Eryngium alpinum 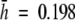 (Gaudeul et al., 2000)]. High levels of genetic variability have been interpreted as an indirect measure of the capability of a plant to adapt to its environment and to buffer against stochastic changes (Ellstrand and Elam, 1993; Paschke et al., 2002; Wise et al., 2002). The high genetic diversity values of V. pseudocytisus subsp. paui might be a consequence of the combined effects derived from its polyploidy, its biological traits, and the past history of its populations.
(Gaudeul et al., 2000)]. High levels of genetic variability have been interpreted as an indirect measure of the capability of a plant to adapt to its environment and to buffer against stochastic changes (Ellstrand and Elam, 1993; Paschke et al., 2002; Wise et al., 2002). The high genetic diversity values of V. pseudocytisus subsp. paui might be a consequence of the combined effects derived from its polyploidy, its biological traits, and the past history of its populations.
Polyploidy in plant species can buffer against genetic erosion resulting from processes of endogamy and genetic drift (Mahy et al., 2000). The high genetic diversity detected in Vella pseudocytisus subsp. paui can be explained by its tetraploidy, as reported for other highly variable polyploids (Soltis and Soltis, 1993; Hardy and Vekemans, 2001; López-Pujol et al., 2004). Allopolyploidy further counteracts the genetic depauperization of populations by the additive expression of different genomes in the same individual (Soltis and Soltis, 1993, 2000; Wendel, 2000). It has been demonstrated that allopolyploids have succeeded in the colonization of newly available niches because of their greater ability to adapt to extreme ecological habitats (Stebbins, 1985; Williamson and Werth, 1999; Mahy et al., 2000; Hedrén et al., 2001; Brochmann et al., 2004). It could be speculated, from a historical time scale, that the allotetraploidy of V. pseudocytisus subsp. paui is likely to have fostered its genetic variability facilitating its adaptation to the adverse climatic conditions of the Quaternary eastern Spain steppe habitat.
The breeding system and the dispersion of seeds are major biological factors that influence the levels of genetic variability and structure in plant species (Hamrick and Godt, 1989). Vella pseudocytisus subsp. paui reproduces sexually through a predominantly allogamous breeding system where pollination is mediated by insects of the families Apideae, Bombyliidae, Andrenidae, Sphingidae and Calliphoridae due to the double floral reward of pollen and nectar (Sainz-Ollero et al., 1996; Domínguez-Lozano et al., 2005). Our genetic data indicate that sexual allogamous reproduction may contribute to the maintenance of high levels of genetic diversity found in this taxon, and that clonal growth may be restricted to a few cases (Domínguez-Lozano et al., 2003, 2005). Allogamy, due to the recombination of different individuals' chromosomes, increases the levels of genetic variability, avoiding inbreeding depression in small populations (Lynch, 1991; Fenster and Galloway, 2000). Because of the limited dispersal capability of the seeds (Dominguez-Lozano et al., 2005), the connection between individuals of the same genetic patch or of nearby patches must occur through cross-pollination, securing a relatively large ratio of pollen interchange.
Although outcrossing perennials generally exhibit high levels of genetic diversity and low levels of population differentiation (Prentice et al., 2003; Franks et al., 2004), V. pseudocytisus subsp. paui showed a marked structure among the three regions where it grows at present. The Mantel correlograms revealed that the genetic similarity between populations increases at short distances (<2 km) and decreases at not very long distances (>5 km), showing a strong micro-geographical structure. This cline in structure is commonly found in a variety of plant species, but the negative correlations between genetic and geographical distances tend to span longer distances (Gabrielsen et al., 1997; Stehlik et al., 2001, 2002) than the ones found in V. pseudocytisus subsp. paui. Nonetheless, short-distance negative correlations have also been reported for other stenocorous plants that show a patchy microspatial distribution of different genetically related distance classes in conjunction with reproductive and ecological traits (Chung and Chung, 1999; Chung and Park, 2000; He et al., 2000; Lutz et al., 2000; Torres et al., 2003).
Most notable is the significant structure found between the two populations cores of V. pseudocytisus subsp. paui in the Turia valley, which are separated by <5 km (Fig. 1). PCO analysis revealed two differentiated groups, Turia-1 and Turia-2 (Fig. 2) that also correspond to the highest levels of percentage of variation among groups for both allozyme and AFLP revealed by the AMOVAs (Table 3). The estimates of gene flow between the two ranges are low in the best cases (Nm = 1 migrant/generation between Villel-1 and Villastar-1), which together with the absence of significant positive population genetic interactions among them (Fig. 5), indicates the existence of isolation by distance. In the Turia valley, the populations of both Turia-1 (Villel-1 and Villel-2) and Turia-2 (Villastar-1 and Villastar-2) are genetically differentiated from each other (Fig. 2) even at very short distances (<1.6 km), and show moderate levels of genetic interaction (Fig. 5). By contrast, in the Alfambra valley the populations of Cuevas Labradas and Villalba baja, separated by 3 km, present higher levels of gene flow (4 migrants/generation), a larger number of intermingled phenotypes (Fig. 2), and relatively higher levels of genetic interactions (Fig. 5). The difference between the high spatial genetic structure found within the Turia valley and the lower structure found within the Alfambra valley can be explained by the history of land use. Habitat fragmentation has been considerably more pronounced in the localities where V. pseudocytisus subsp. paui grows in the Turia valley than in those of the Alfambra valley. The Turia valley sites are located in high plateau steppes that have been ploughed for agricultural use over the centuries, implying a more prolonged history of disturbance than the Alfambra valley habitats that are located in ravines where no agriculture system has been implemented. The strong dispersal barriers created by habitat fragmentation and the ability of the plant to survive in the newly transformed habitat are factors that might have strengthened the genetic structure of V. pseudocytisus subsp. paui in the Turia-1 and Turia-2 ranges. This may also contribute to the ‘island’ distribution pattern typical of populations subjected to severe habitat loss (Saunders et al., 1991; Young et al., 1996).
Conservation implications
Steppe plants have received little attention in the conservation plans of most European countries (De Klem, 1997). However, the Spanish Regional Government of Aragón is developing conservation programmes for some endangered taxa from this habitat, like the special conservation plan for V. pseudocytisus subsp. paui (Decree 92/2003). This plan includes habitat restoration programmes and in-situ and ex-situ conservation strategies consisting of monitoring the demographic and genetic parameters of the populations and the establishment of germplasm banks for future reinforcements and reintroductions.
Based on the high number of private AFLP markers detected for each of the three geographical ranges where V. pseudocytisus subsp. paui lives, the strong genetic structure of all the populations studied, the genetic isolation of the two close valleys, and the marked micro-geographical genetic structure found in the Turia valley, V. pseudocytisus subsp. paui should be maintained in the category of ‘at risk of extinction’, equivalent to the ‘critically endangered (CR)’ IUCN category, that it currently possesses in the regional catalogue.
Because the main source of threat for V. pseudocytisus subsp. paui resides in anthropogenic disturbance, and despite the influence that land use might have caused to the strong microspatial genetic structure found in some of its populations, prolonged habitat modification coupled with subsequent population decline could damage the present genetic reservoir of this taxon. Therefore, the recovery plan for this plant should include the protection of its habitat. The ‘genetic reserves’ (López-Pujol et al., 2003) or ‘micro-reserves’ (Laguna et al., 2004) have proven to be an essential tool for effective protection of diverse endemic and endangered flora in recovery policies. Thus, it is recommended that small areas (1–2 ha) are established for the preservation of the three genetic ranges of V. pseudocytisus subsp. paui detected in this study.
In addition, conservation plans should consider the different levels of genetic differentiation between the Alfambra and Turia valleys. Given the genetic similarity and high levels of gene flow observed in the Alfambra valley, both populations of this latter area could be used to exchange individuals or seeds in case of population decline. In contrast, given the different genetic structure among the populations of the Turia valley, it is not advisable to introduce individuals or seeds from other distinct populations.
Several biological features and the status of the current populations of V. pseudocytisus subsp. paui facilitate the development of a management plan in order to maintain the levels of genetic diversity found in this taxon. The effective population size of V. pseudocytisus subsp. paui, its high potential for colonization, the high levels of genetic diversity among individuals, the high degree of outcrossing pollination (Sainz-Ollero et al., 1996; Domínguez-Lozano et al., 2005), and the high germination rate of seeds (Blanca et al., 1999; Maselli et al., 1999; Albert et al., 2002), are factors of importance in conservation that ensure the viability of recovery plans. Therefore it is recommended that a seed bank, which represents the genetic diversity of the four populations from the Turia valley sampled in this study, and at least one population from the Alfambra valley, is established. These seeds could serve as a potential source for reintroduction of individuals.
Acknowledgments
We thank José Gabriel Segarra-Moragues, Tom Ranker, Jose María Iriondo, Douglas Laing and two anonymous referees for their valuable comments and critical review of an earlier version of the manuscript. This work has been supported by an Aragón Government (DGA) project grant P2002/0290 to P. Catalán and by a DGA doctorate fellowship to E. Pérez-Collazos.
LITERATURE CITED
- Albert MJ, Iriondo JM, Pérez-García F. 2002. Effects of temperature and pretreatments on seed germination of nine semiarid species from NE Spain. Israel Journal of Plant Sciences 50: 103–112. [Google Scholar]
- Ayres DR, Ryan FJ. 1999. Genetic diversity and structure of the narrow endemic Wyethia reticulata and its congener W. bolanderi (Asteraceae) using RAPD and allozyme techniques. American Journal of Botany 86: 344–353. [PubMed] [Google Scholar]
- Bañares A, Blanca G, Güemes J, Moreno JC, Ortiz S. 2003. Atlas y libro rojo de la flora vascular amenazada de España. Madrid: Dirección General de Conservación de la Naturaleza.
- Bassam BJ, Caetano-Anolles G, Gresshoff PM. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Annals of Biochemistry 196: 80–83. [DOI] [PubMed] [Google Scholar]
- Blanca G, Cabezudo B, Hernández-Bermejo JE, Herrera CM, Molero-Mesa J, Muñoz J, et al. 1999. Libro rojo de flora silvestre amenazada de Andalucía. Tomo I. Especies en peligro de extinción. Sevilla, Junta de Andalucía: Consejería de Medio Ambiente.
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen AC, et al. 2004. Polyploidy in arctic plants. Biological Journal of the Linnean Society 82: 521–536. [Google Scholar]
- Casgrain P, Legendre P. 2001. The R package for multivariate and spatial analysis, version 4.0 d6. User′s Manual. Montreal: Departament des Sciences Biologiques, Université de Montreal.
- Charco J. 2001. Guía de los árboles y arbustos del norte de África. Madrid: Agencia Española de Cooperación Internacional.
- Chung MY, Chung MG. 1999. Notes on spatial genetic structure in populations of Cymbidium goeringii (Orchidaceae). Annales Botanici Fennici 36: 161–164. [Google Scholar]
- Chung MG, Park CW. 2000. Notes on spatial genetic structure in a hybrid population between Aconitum japonicum subsp napiforme and A. jaluense (Ranunculaceae). Annales Botanici Fennici 37: 243–247. [Google Scholar]
- Crespo MB, Lledó MD, Fay MF, Chase MW. 2000. Subtribe Vellinae (Brassiceae, Brassicaceae): a combined analisis of ITS nrDNA sequences and morphological data. Annals of Botany 86: 53–62. [Google Scholar]
- De Klem C. 1997. Comparative analysis of the effectiveness of legislation for the protection of wild flora in Europe. Nature and Environment, No. 88. Strasbourg: Council of Europe.
- Domínguez-Lozano F, Benito-García M, Sainz-Ollero H, Sánchez-de-Dios R. 2003. Vella pseudocytisus subsp. paui Gómez-Campo. In: Bañares A, Blanca G, Güemes J, Moreno JC, Ortiz S, eds. Atlas y libro rojo de la flora vascular amenazada de España. Madrid: Dirección General de Conservación de la Naturaleza, 872–873.
- Domínguez-Lozano F, Moreno-Saiz JC, Sainz-Ollero H. 2005. Biological properties of the endemic and threatened shrub in Iberia Vella pseudocytisus subsp. paui Gómez Campo (Cruciferae/Brassicaceae) and implications for its conservation. Journal for Nature Conservation 13: 17–30. [Google Scholar]
- Doyle J, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Dunn G, Everitt BSE. 1982. An introduction to mathematical taxonomy. Cambridge: Cambridge University Press.
- Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- Escudero A, Iriondo JM, Torres ME. 2003. Spatial analysis of genetic diversity as a tool for plant conservation. Biological Conservation 113: 351–365. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Galloway LF. 2000. Inbreeding and outbreeding depression in natural populations of Chamaecrista fasciculata (Fabaceae). Conservation Biology 14: 1406–1412. [Google Scholar]
- Franks SJ, Richards CL, Gonzales E, Cousins JE, Hamrick JL. 2004. Multi-scale genetic analysis of Uniola paniculata (Poaceae): a coastal species with a linear, fragmented distribution. American Journal of Botany 91: 1345–1351. [DOI] [PubMed] [Google Scholar]
- Gabrielsen TM, Bachmann K, Jakobsen KS, Brochmann C. 1997. Glacial survival does not matter: RAPD phylogeography of Nordic Saxifraga oppositifolia. Molecular Ecology 6: 831–842. [Google Scholar]
- Galloway LF, Etterson JR, Hamrick JL. 2003. Outcrossing rate and inbreeding depression in the herbaceous autotetraploid, Campanula americana. Heredity 90: 308–315. [DOI] [PubMed] [Google Scholar]
- Gaudeul M, Taberlet P, Till-Bottraud I. 2000. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Molecular Ecology 9: 1625–1637 [DOI] [PubMed] [Google Scholar]
- Goldring A, Zamir D, Degani C. 1985. Duplicated phosphoglucose isomerase genes in avocado. Theoretical and Applied Genetics 71: 491–494. [DOI] [PubMed] [Google Scholar]
- Gómez-Campo C. 1981. Taxonomic and evolutionary relationship in the genus Vella L. (Cruciferae). Botanical Journal of the Linnean Society 82: 165–179. [Google Scholar]
- Gómez-Campo C. 1993. Vella L. In Castroviejo S, Aedo C, Gómez-Campo C, Lainz M., Montserrat P, Morales R, et al., eds. Flora Iberica IV. Madrid: Real Jardín Botánico, 414–417
- Gottlieb LD. 1977. Evidence for duplication and divergence of the structural gene for phosphoglucose isomerase in diploid species of Clarkia. Genetics 86: 289–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower JC, Ross GJS. 1969. Minimum spanning tree and single-linkage cluster analysis. Applied Statistics 18: 54–64. [Google Scholar]
- Grant V. 1981. Plant speciation, 2nd edn. New York, NY: Columbia University Press.
- Hamrick JL, Godt MJW. 1989. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding, and genetic resources. Sunderland, MA: Sinauer, 43–63.
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London – Biological Sciences 351: 1291–1298. [Google Scholar]
- Hardy OJ, Vekemans X. 2001. Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evolution 55: 943–954. [DOI] [PubMed] [Google Scholar]
- He TH, Rao GY, You RL, Ge S, Hong DY. 2000. Spatial autocorrelation of genetic variation in three stands of Ophiopogon xylorrhizus (Liliaceae s.l.). Annals of Botany 86: 113–121. [Google Scholar]
- Hedrén M, Fay MF, Chase MW. 2001. Amplified fragment length polymorphisms (AFLP) reveal details of polyploid evolution in Dactylorhiza (Orchidaceae). American Journal of Botany 88: 1868–1880. [PubMed] [Google Scholar]
- Hörandl E, Greilhuber J. 2002. Diploid and autotetraploid sexuals and their relationships to apomicts in the Ranunculus cassubicus group: insights from DNA content and isozyme variation. Plant Systematics and Evolution 234: 85–100. [Google Scholar]
- Keiper FJ, McConchie R. 2000. An analysis of genetic variation in natural populations of Sticherus flabellatus [R. Br. (St John)] using amplified fragment length polymorphism (AFLP) markers. Molecular Ecology 9: 571–581. [DOI] [PubMed] [Google Scholar]
- Koch M. 2003. Molecular phylogenetics, evolution, and population biology in Brassicaceae. In: Sharma AK, Sharma A, eds. Plant genome: biodiversity and evolution, Vol. 1, Part A. Enfield, NH: Science Publishers, 1–35.
- Koch M, Al-Shehbaz IA, Mummenhoff K. 2003. Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae). Annals of the Missouri Botanical Garden 90: 151–171. [Google Scholar]
- Laguna E, Deltoro VI, Pérez-Botella J, Pérez-Rovira P, Serra L, Olivares A, et al. 2004. The role of small reserves in plant conservation in a region of high diversity in eastern Spain. Biological Conservation 119: 421–426. [Google Scholar]
- Liston A, St Hilary K, Wilson MV. 1995. Genetic diversity in populations of kincaid's lupine, host plant of fender's blue butterfly. Madroño 42: 309–322. [Google Scholar]
- López-Pujol J, Orellana MR, Bosch M, Simon J, Blanché C. 2003. Effects of habitat fragmentation on allozyme diversity and conservation status of the coastal sand dune plant Stachys maritima (Lamiaceae) in the Iberian Peninsula. Plant Biology 5: 504–512. [Google Scholar]
- López-Pujol J, Bosch M, Simon J, Blanché C. 2004. Allozyme diversity in the tetraploid endemic Thymus loscosii (Lamiaceae). Annals of Botany 93: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz E, Schneller JJ, Holderegger R. 2000. Understanding population history for conservation purposes: population genetics of Saxifraga aizoides (Saxifragaceae) in the lowlands and lower mountains north of Alps. American Journal of Botany 87: 583–590. [PubMed] [Google Scholar]
- Lynch M. 1991. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45: 622–629. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I. 2005. Chromosome triplication found across the tribe Brassiceae. Genome Research 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy G, Bruederle LP, Connors B, van Hofwegen M, Vorsa N. 2000. Allozyme evidence for genetic autopolyploidy and high genetic diversity in tetraploid cranberry, Vaccinium oxycoccos (Ericaceae). American Journal of Botany 87: 1882–1889. [PubMed] [Google Scholar]
- Mantel NA. 1967. The detection of disease clustering and generalized regression approach. Cancer Research 27: 209–220. [PubMed] [Google Scholar]
- Martínez-Ortega MM, Delgado L, Albach DC, Elena-Rosselló JA, Rico E. 2004. Species boundaries and phylogeographic patterns in cryptic taxa inferred from AFLP markers: Veronica subgen. Pentasepalae (Scrophulariaceae) in the Western Mediterranean. Systematic Botany 29: 965–986. [Google Scholar]
- Maselli S, Pérez-García F, Aguinagalde I. 1999. Evaluation of seed storage conditions and genetic diversity of four crucifers endemic to Spain. Annals of Botany 84: 207–212. [Google Scholar]
- Masterson J. 1994. Stomatal size in fossil plants – evidence for polyploidy in majority of angiosperms. Science 264: 421–424. [DOI] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, Allnut TR, Gillies ACM, Lowe AJ, Ennos RA. 1999. Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends in Ecology and Evolution 14: 140–145. [DOI] [PubMed] [Google Scholar]
- Oden NL, Sokal RR. 1986. Directional autocorrelation: an extension of spatial correlograms to two dimensions. Systematic Zoology 35: 608–617. [Google Scholar]
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Palacios C, Kresovich S, González-Candelas F. 1999. A population genetic study of the endangered plant species Limonium dufourii (Plumbaginaceae) based on amplified fragment length polymorphism (AFLP). Molecular Ecology 8: 645–657. [Google Scholar]
- Park KR. 2004. Comparisons of allozyme variation of narrow endemic and widespread species of Far East Euphorbia (Euphorbiaceae). Botanical Bulletin of Academia Sinica 45: 221–228. [Google Scholar]
- Paschke M, Abs C, Schmid B. 2002. Relationship between population size, allozyme variation, and plant performance in the narrow endemic Cochlearia bavarica. Conservation Genetics 3: 131–144. [Google Scholar]
- Peever TL, Milgroom MG. 1994. Genetic structure of Pyrenophora teres populations determined with random amplified polymorphic DNA markers. Canadian Journal of Botany 72: 915–923. [Google Scholar]
- Prentice HC, Malm JU, Mateu-Andrés I, Segarra-Moragues JG. 2003. Allozyme and chloroplast DNA variation in island and mainland populations of the rare Spanish endemic, Silene hifacensis (Caryophyllaceae). Conservation Genetics 4: 543–555. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranker TA. 1994. Evolution of high genetic variability in the rare Hawaiian fern Adenophorus periens and implications for conservation management. Biological Conservation 70: 19–24. [Google Scholar]
- Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. 2002. NTSYSpc, Numerical taxonomy and multivariate analysis system. Version 2.11a, User guide. New York, NY: Exeter software, 1–38.
- Rossetto M, Waver PK, Dixon KW. 1995. Use of RAPD analysis in devising conservation strategies for the rare and endangered Grevillea scapigera (Proteaceae). Molecular Ecology 4: 321–329. [DOI] [PubMed] [Google Scholar]
- Rottenberg A, Parker JS. 2003. Conservation of the critically endangered Rumex rothschildianus as implied from AFLP diversity. Biological Conservation 114: 299–303. [Google Scholar]
- Sainz-Ollero H, Franco Mújica F, Arias Torcal J. 1996. Estrategias para la conservación de la flora amenazada de Aragón. Zaragoza: Consejo de Protección de la Naturaleza de Aragón, 1–221.
- Saunders DA, Hobbs RJ, Margules CR. 1991. Biological consequences of ecosystem fragmentation: a review. Conservation Biology 5: 18–32 [Google Scholar]
- Schneider S, Roessli D, Excoffier L. 2000. Arlequin v. 2.000: a software for population genetics data analysis. Geneva: Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Schulz OE. 1923. Cruciferae-Brassicaceae. Part II. Subtribes Cakilinae, Zillinae, Vellinae, Savignyinae and Moricandiinae. In: Engler A, ed. Das Pflanzenreich. Heft 82–85. Wilhem Engelmann, Leipzig. 1–100.
- Segarra-Moragues JG, Catalán P. 2002. Low allozyme variability in the critically endangered Borderea chouardii and in its congener Borderea pyrenaica (Dioscoreaceae), two paleoendemic relicts from the central Pyrenees. International Journal of Plant Sciences 163: 159–166. [Google Scholar]
- Segarra-Moragues JG, Catalán P. 2003. Life history variation between species of the relictual genus Borderea (Dioscoreaceae): phylogeography, genetic diversity, and population genetic structure assessed by RAPD markers. Biological Journal of the Linnean Society 80: 483–498. [Google Scholar]
- Segarra-Moragues JG, Palop-Esteban M, González-Candelas F, Catalán P. 2004. Characterization of seven (CTT) (n) microsatellite loci in the Pyrenean endemic Borderea pyrenaica (Dioscoreaceae): remarks on ploidy level and hybrid origin assessed through allozymes and microsatellite analyses. Journal of Heredity 95: 177–183. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. 1993. Molecular data and the dynamic nature of polyploidy. Critical Reviews in Plant Sciences 12: 243–273. [Google Scholar]
- Soltis DE, Soltis PS. 1999. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2000. The role of genetics and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences of the USA 97: 7051–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants, 1st edn. New York, NY: Columbia University Press.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold.
- Stebbins GL. 1985. Polyploidy, hybridization, and the invasion of new habitats. Annals of Missouri Botanical Garden 72: 824–832. [Google Scholar]
- Stehlik I, Schneller JJ, Bachmann K. 2001. Resistance or emigration: response of the high-alpine plant Eritrichium nanum (L.) Gaudin to the ice age within the Central Alps. Molecular Ecology 10: 357–370. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Schneller JJ, Bachmann K. 2002. Immigration and in situ glacial survival of the low-alpine Erinus alpinus (Scrophulariaceae). Biological Journal of the Linnean Society 77: 87–103. [Google Scholar]
- Tero N, Aspi J, Siikmaki P, Jakalaniemi A, Tuomi J. 2003. Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Molecular Ecology 12: 2073–2085. [DOI] [PubMed] [Google Scholar]
- Torres E, Iriondo JM, Escudero A, Pérez C. 2003. Analysis of within-population spatial genetic structure in Antirrhinum microphyllum (Scrophulariaceae). American Journal of Botany 90: 1688–1695. [DOI] [PubMed] [Google Scholar]
- Travis SE, Manchinski J, Keim P. 1996. An analysis of genetic variation in Astragalus cremnophylax var. cremnophylax, a critically endangered plant, using AFLP markers. Molecular Ecology 5: 735–745. [DOI] [PubMed] [Google Scholar]
- Tribsch A, Schonswetter P, Stuessy TF. 2002. Saponaria pumila (Caryophyllaceae) and the Ice Age in the European Alps. American Journal of Botany 89: 2024–2033. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Lee TV, Hornes M, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VVAA. 2000. Lista roja de la flora vascular española (valoración según categorías UICN). Conservación Vegetal 6: 11–38. [Google Scholar]
- Warwick S, Al-Shehbaz IA. 1998. Generic evaluation of Boleum, Euzomodendron, and Vella (Brassicaceae). Novon 8: 321–325. [Google Scholar]
- Warwick S, Black LD. 1994. Evaluation of the subtribes Moricandiinae, Savignyinae, Vellineae, and Zillinae (Brassicaceae, tribe Brassiceae) using chloroplast DNA restriction site variation. Canadian Journal of Botany 72: 1692–1701. [Google Scholar]
- Weeden NF, Gottlieb LD. 1979. Distinguishing allozymes and isozymes of phosphoglucoisomerases by electrophoresis comparison of pollen and somatic tissues. Biochemistry Genetics 17: 287–296. [DOI] [PubMed] [Google Scholar]
- Weeden NF, Wendel JF. 1989. Genetics of plant isozymes. In Soltis DE, Soltis PS, eds. Isozymes in plant biology. London: Chapman and Hall, 46–72.
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225–249. [PubMed] [Google Scholar]
- Wendel JF, Weeden NF. 1989. Visualization and interpretation of plant isozymes. In Soltis DE, Soltis PS, eds. Isozymes in plant biology. London: Chapman and Hall, 5–45.
- Williamson PS, Werth CR. 1999. Levels and patterns of genetic variation in the endangered species Abronia macrocarpa (Nyctaginaceae). American Journal of Botany 86: 293–301. [PubMed] [Google Scholar]
- Wise CA, Ranker TA, Linhart YB. 2002. Modelling problems in conservation genetics with Brassica rapa. I. Genetic variation and fitness in plants under mild, stable conditions. Conservation Biology 16: 1542–1554. [Google Scholar]
- Wolko B, Weeden NF. 1989. Estimation of Lupinus genome polyploidy on the basis of isozymic loci number. Genetica Polonica 30: 165–171. [Google Scholar]
- Wright S. 1965. The interpretation of population structure by F-statistics with especial regard to systems of mating. Evolution 19: 395–420. [Google Scholar]
- Xue DW, Ge XJ, Hao G, Zhang CQ. 2004. High genetic diversity in a rare, narrowly endemic primrose species, Primula interjacens, by ISSR analysis. Acta Botanica Sinica 46: 1163–1169. [Google Scholar]
- Young A, Boyle T, Brown T. 1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution 11: 414–417. [DOI] [PubMed] [Google Scholar]
- Zhang LB, Comes HP, Kadereit JW. 2001. Phylogeny and quaternary history of the European montane/alpine endemic Soldanella (Primulaceae) based on ITS and AFLP variation. American Journal of Botany 88: 2331–2345. [PubMed] [Google Scholar]