Abstract
The tripartite motif (TRIM) family member, TRIM21, is an E3 ubiquitin ligase for IRF3 and IRF8 that functions in both innate and acquired immunity. It is also an autoantigen known as Ro52/SS-A. The function of TRIM21 in vivo, however, has remained elusive. We generated Trim21−/− mice with the Trim21 gene replaced by an EGFP reporter. EGFP expression analyses showed that Trim21 was widely expressed in many tissues, with the highest levels in immune cells. Studies of Trim21−/− embryonic fibroblasts demonstrated that TLR-mediated induction of proinflammatory cytokines, including IL-1β, IL-6, TNFα and CXCL10, was consistently upregulated relative to wild-type cells. Reporter analyses demonstrated that TLR-mediated NF-κB activation was higher in Trim21−/− cells than in wild-type cells, likely accounting for their enhanced cytokine expression. In contrast, functional analyses of immune cells from Trim21−/− mice revealed no abnormalities in their composition or function, even though ubiquitylation of IRF3 and IRF8 was impaired. These results suggested possible redundancies in activities mediated by TRIM21. In keeping with this concept, we found that a number of TRIM family members were upregulated in Trim21−/− cells. Taken together, these findings demonstrate that TRIM21 plays a previously unrecognized role in the negative regulation of NF-κB-dependent proinflammatory cytokine responses, and suggest that multiple TRIM proteins contribute to the maintenance of functional equilibrium in inflammatory responses, in part through functional redundancy.
Keywords: autoantigen, TRIM family, EGFP, NF-κB, proinflammatory cytokines, Toll-like receptors
Introductions
The tripartite motif (TRIM)3 family, which includes TRIM21, numbers over 60 proteins in humans with homologues identified in many species ranging from primates to nematodes (1–3). TRIM proteins exhibit a wide range of activities, including the regulation of innate and adaptive immune responses. Some TRIM proteins are capable of inhibiting the replication of HIV and other retroviruses at various stages of the virus life cycle (3, 4). Underscoring their role in host defense, many TRIM proteins are induced following stimulation of cells with type I and type II IFNs (3, 5).
TRIM21, a ~52 kDa protein also known as Ro52/SS-A, is expressed broadly in many cell types (6). It predominantly localizes to the cytoplasm, although some is found in the nucleus. Similar to other TRIM proteins, TRIM21 expression is stimulated by IFNs, which prompts increased nuclear localization of the protein (7, 8). TRIM21 has long been known as an autoantigen recognized by Abs in sera of patients with systemic lupus erythematosus (SLE) and Sjögren’s syndrome/lymphoepithelial sialadenitis (SS/LESA), chronic, systemic autoimmune diseases of unknown cause (9, 10). The human TRIM21 gene maps to the p15 region of chromosome 11 associated with an increased risk of SLE (11). Allelic polymorphisms of TRIM21 are also linked to the susceptibility to these autoimmune diseases (12). TRIM21 transcript levels are increased in PBMCs of patients with SLE and SS/LESA, further suggesting a role for TRIM21 in autoimmune diseases (13).
TRIM21 is comprised of several distinct domains— a RING finger, B-box and coiled-coil domain in the N-terminal region followed by a PRY/SPRY domain in the C-terminus. These domains represent signature motifs conserved among many TRIM proteins (1–3). The RING domain contains two cross-braced zinc fingers that denote a ubiquitin E3 ligase motif. Indeed TRIM21 is a ubiquitin ligase that conjugates ubiquitin molecules onto itself and various substrate proteins (14–18). We showed previously that TRIM21 ubiquitylates IFN regulatory factor (IRF) 8, an IFN-inducible transcription factor, and that this modification is associated with enhanced IL-12p40 transcription in macrophages (8). TRIM21 also ubiquitylates IRF3 to promote its destabilization, resulting in the downregulation of IFN-β expression in fibroblasts (19). Structural studies have demonstrated that the PRY/SPRY domain of TRIM21 functions as an immunoglobulin Fc receptor, binding the Fc region of IgG with high affinity, suggestive of a role for this protein in immune regulation (20, 21). The PRY/SPRY domain of other TRIM proteins is responsible for innate protection against infection by retroviruses, including HIV, as it interferes with uncoating of viral envelope proteins (4, 22). TRIM21 may also play a role in T-cell responses, as suggested by the observation that IL-2 production is increased upon exogenous expression of TRIM21 in Jurkat T cells stimulated by anti-CD28 Ab (23).
Despite increasing understanding of structural and molecular aspects of TRIM21, its physiological role in vivo has remained obscure. To gain knowledge on the in vivo function of TRIM21, we constructed and studied Trim21−/− mice in which Trim21 exons are replaced by an enhanced GFP (EGFP) reporter gene. These mice allowed us to study not only the function of TRIM21, but also to detail its expression patterns among various cells in vivo.
Our analyses showed that Trim21 is expressed broadly in various organs in vivo, with particularly high levels in some immune cells. Functional analysis unraveled a distinct abnormality in Trim21−/− embryonic fibroblasts (EFs), as evidenced by elevated expression of several proinflammatory cytokines in response to TLR signaling. Enhanced cytokine induction was at least partly accounted for by enhanced NF-κB activation in Trim21−/− cells, indicating a previously unappreciated role for TRIM21 in the negative regulation of NF-κB activity. In contrast, Trim21−/− EFs exhibited no defect in IFN-α/β induction, even though ubiquitylation of IRF3 was significantly reduced. Moreover, the development, number and distribution of subsets of immune cells in tissues of Trim21−/− mice were essentially normal. Further defying expected roles for TRIM21 in innate and adaptive immunity, functional tests performed with macrophages, dendritic cells (DCs), T and B lymphocytes revealed comparable activity in Trim21+/+ and Trim21−/− cells. We found that multiple TRIM proteins structurally related to TRIM21 were constitutively upregulated in Trim21−/− EFs, and that their expression was further increased after TLR and IFN stimulation. Together, our results point to the presence of a compensatory mechanism that coordinately makes up the loss of TRIM21 to maintain functional equilibrium in Trim21−/− cells, which is likely to reflect the contribution of multiple TRIM proteins.
Materials and Methods
Antibodies
Abs used in the study were to TRIM21/Ro52 (M-20), IRF3, IRF8 and ubiquitin (Santa Cruz Biotechnology), GFP (Roche), β-actin (Sigma-Aldrich), and IL-4 and IL-12 (BioLegend). Anti-CD4-allophycocyanin (APC), anti-CD11b-APC, anti-CD19-APC, anti-CD25-APC, anti-CD44-PE, anti-CD5-biotin, anti-NK-1.1-biotin and anti-F4/80-biotin Abs were from BioLegend. Other Abs, PerCP-Cy5.5-conjugated streptavidin and isotype-matched control Abs used for flow cytometric analyses were purchased from BD Biosciences.
Generation of Trim21−/− mice
A non-conditional knockout of Trim21 was generated by knocking an EGFP reporter into the locus (Ozgene Pty Ltd, Canning Vale, WA, Australia; Fig. 1A). The coding sequence for EGFP was inserted in-frame into exon 3 replacing the ATG start codon of Trim21, with transcription stopped by the SV40 polyA signal. A PGK-neo cassette floxed by LoxP sites was inserted downstream of exon 3. Following homologous recombination of the targeting vector in C57BL/6 ES cells and establishment of germline transmission, the PGK-neo cassette was deleted using Cre recombinase. Selective breeding was used to eliminate the Cre gene. Mice were genotyped by PCR using primers FW (5′-CCTTGGCATTATTTGGGGGA-3′) and RV (5′-CTCCATGCTTCATGCAGTGC-3′) for the wild-type allele. The mutated allele was identified with the same forward primer and RV (5′-GCGGATCTTGAAGTTCACCT-3′).
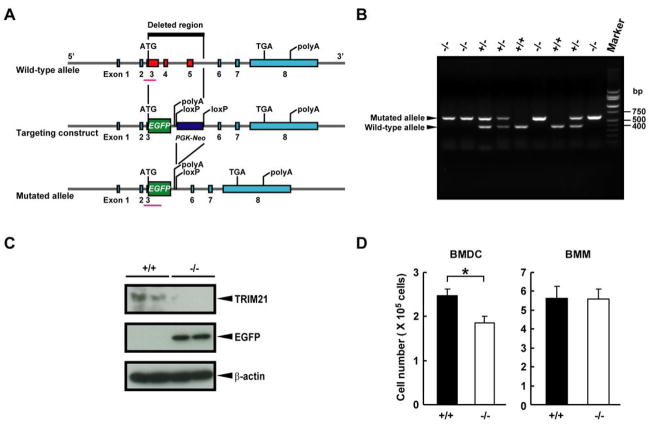
Figure 1. Generation of Trim21−/− mice.
A, A schematic diagram of the wild-type allele, the targeting construct and the mutated allele. B, Littermate pups from a Trim21+/− cross were genotyped by PCR of genomic DNA from tails. The position of bands to detect the mutated and the wild-type alleles are marked by arrows. C, Western blot analysis was performed with extracts from Trim21+/+ and Trim21−/− spleens using anti-TRIM21 Ab and anti-GFP Ab diluted at 1:5000 and 1:1000, respectively. β-Actin was used as a loading control. D, Suppression of BMDC production from Trim21−/− mice. 1×106 BM cells from Trim21+/+ or Trim21−/− mice were cultured with Flt3L for 8 days (for BMDC) or M-CSF for 5 days (for BMM). *, p < 0.05 as determined by Student’s t test.
Mice and cell culture
All animal work conformed to protocols approved by the animal care and use committees of NICHD and NIAID. Ifnar1−/− mice were obtained from J. Durbin (Ohio State University) through H. Young (National Cancer Institute). Fms-like tyrosine kinase 3 ligand (Flt3L)-mediated bone marrow (BM)-derived dendritic cells (BMDCs) were prepared as previously described (24). For BM-derived macrophage (BMM) preparation, BM cells (1×107 cells in a 15 cm dish) were cultured with 20 ng/ml M-CSF (Invitrogen) for 5 days and used as BMMs after washing away nonadherent cells. BMMs were primed for 24 h with 100 U/ml IFN-γ (Invitrogen). Peritoneal macrophages were obtained from mice injected i.p. with 3 ml of 3% thioglycollate (Difco) 4 days prior to harvest. Macrophages were cultured in DMEM supplemented with 10% FBS. EFs were prepared from day 13 embryos and cultured in DMEM supplemented with 10% FBS. BMDCs, primed BMMs or EFs were infected with Newcastle disease virus (NDV) or stimulated with poly(I:C) (100 μg/ml; Amersham Biosciences), Salmonella minnesota-derived LPS (200 ng/ml; Sigma-Aldrich), CpG (ODN1826; 1 μg/ml; Operon Biotechnologies), murine IL-1β (10 ng/ml; Peprotech) or murine TNFα (10 ng/ml; Peprotech). In some experiments, unprimed BMMs or EFs were stimulated with murine IFN-β (100 U/ml; PBL InterferonSource) or murine IFN-γ (100 U/ml).
Flow cytometric analysis and cell separations
Cells were blocked with anti-mouse FcγR Ab (CD16/CD32, 2.4G2) and stained with indicated Abs. Stained cells were analyzed on FACSCaliber (Becton Dickinson) and data were analyzed by FlowJo software (Tree Star). Viable cells were gated using forward and side scatter parameters. For cell separations using MACS technology (Miltenyi Biotec), single cell suspensions were mixed with microbeads conjugated with anti-CD4 or anti-CD8 Ab and sorted according to the manufacturer’s instructions.
T-cell activation
Purified CD4+ T cells were stimulated with Dynabeads Mouse CD3/CD28 T Cell Expander (Invitrogen) in 24-well plates (5 × 105 cells/well) in RPMI1640 with 10% FBS and murine IL-2 (10 ng/ml; Peprotech) for 2 days. To prepare Th1-skewed CD4+ cells, murine IL-12 (20 ng/ml; Peprotech) and anti-IL-4 Ab (10 μg/ml) were added to the cultures, whereas murine IL-4 (20 ng/ml; Peprotech) and anti-IL-12 Ab (10 μg/ml) were added for the preparation of Th2-skewed cells. Beads were removed on day 2 and the cells were expanded in appropriate medium containing IL-2 for a total of 5 days. Cells were re-stimulated with Dynabeads Mouse CD3/CD28 T Cell Expander or 50 ng/ml PMA and 2 μM ionomycin in 96-well plates (2 × 105 cells/well). For testing naïve CD4+ or CD8+ T cells, cells were stimulated with Dynabeads Mouse CD3/CD28 T Cell Expander without IL-2.
B-cell experiments
For proliferation assay, splenic B cells were purified by the Dynal mouse B cell negative isolation kit (Invitrogen). Cells were labeled with CFSE (Invitrogen) and cultured in the medium with 10 μg/ml LPS, 1 μg/ml CpG, or 20 μg/ml anti-IgM plus 1 μg/ml anti-CD40 Abs for 3 days. The cells were stained with 7-aminoactinomycin D and analyzed by flow cytometry. For IgM secretion assay, purified B cells were stimulated with 10 μg/ml LPS or 1 μg/ml CpG for 3 days. IgM concentrations in supernatants were measured by ELISA (see below).
Quantitative real time reverse transcriptase PCR (qPCR)
Total RNA was extracted with RNeasy Mini Kit and RNase-Free DNase Kit (Qiagen), and cDNA was prepared with Superscript II enzyme (Invitrogen) according to manufacturer’s protocols. Amplification of sample cDNA was monitored with SYBR Green (Fast SYBR Green Master Mix; Applied Biosystems) in combination with 7500 Fast Real-Time PCR (Applied Biosystems), according to the manufacturer’s instructions. Transcript levels were normalized by amount of hypoxanthine phosphoribosyltransferase mRNA. The RT2 Profiler PCR Array System (SuperArray Bioscience) was used to detect a panel of IFN-stimulated genes.
Cytometric bead array (CBA) and ELISA
The amounts of cytokines in culture supernatants or sera were measured using the BD CBA Mouse Th1/Th2 Cytokine Kit and Mouse Inflammation Kit in combination with the BD CBA Software (BD Biosciences). To detect antigen-specific Abs, 96-well high-binding plates were coated with 100 μl of 50 μg/ml (4-hydroxy-3-nitrophenyl) acetyl (NP)23-BSA (Biosearch Technologies) and blocked with BSA. Plates were incubated with serially diluted sera for 2 h, followed by HRP-labeled anti-mouse Ig isotype Abs (Jackson Immunoresearch Laboraotories). Sera from mice immunized with NP-keyhole limpet hemocyanin (KLH; Biosearch Technologies) twice were used for generating a standard curve. The plates were developed with Sigma Fast OPD substrate (Sigma) and read at 450 nm.
Western blot analysis
Cells were lysed in lysis buffer (25 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 0.1% NP-40 and 10% glycerol) or RIPA buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS and 1% NP-40) in the presence of a protease inhibitor cocktail (Roche). Alternatively, tissues were homogenized in lysis buffer containing the protease inhibitor cocktail, incubated at 4°C for 12 h, and supernatants were collected after centrifugation. Proteins separated on 4–12% SDS-PAGE (Invitrogen) were transferred onto PVDF membranes (Millipore) and reacted with indicated Abs. Secondary detection was carried out with HRP-conjugated anti-rabbit IgG, anti-mouse IgG (both from Amersham Bioscience) or anti-goat IgG (Abcam), followed by chemiluminescence development with ECL reagent (Pierce Biotechnology).
Ubiquitylation assay
EFs were stimulated with 100 μg/ml poly(I:C) for 24 h and BMMs with 100 U/ml murine IFN-γ overnight followed by 1 μg/ml CpG for 24 h. Cultures were treated with 25 μM MG-132 (Calbiochem) for 2 h before harvest. Extracts were immunoprecipitated with anti-IRF3 or anti-IRF8 Ab bound to protein A/G agarose beads (Santa Cruz Biotechnology). Immunoprecipitates were analyzed by western blot using anti-ubiquitin Ab as described (8).
Dual luciferase reporter assay
EFs were transiently transfected with pBL firefly luciferase vector carrying 3 copies of NF-κB motifs upstream of Igκ (25) and pRL-TK Renilla luciferase vector (Promega) using SuperFect Transfection Reagent (Qiagen). Twenty-four hours later, cells were stimulated with 100 μg/ml of poly(I:C) or 200 ng/ml of S. minnesota-derived LPS for 3, 6 or 12 h. Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) in VICTOR3 Multilabel Counter (PerkinElmer).
In vivo experiments
Mice were injected i.p. with 30 μg/g of LPS from E. coli serotype O55:B5 (Sigma). Sera collected 4 h later were tested for cytokine production by CBA as described above. For Ab production studies, mice were immunized i.p. with 50 μg of alum-precipitated NP-KLH. Mice were bled 14, 28 and 42 days after immunization and serum NP-specific Abs were quantified by ELISA.
Results
Trim21−/− mice are viable and fertile
To study the function of TRIM21 in vivo, we generated a constitutive null mutation of the Trim21 gene in the mouse by inserting an EGFP reporter gene in frame and under the control of the endogenous regulatory sequences of the gene (Fig. 1A). Heterozygous Trim21+/− mice with the C57BL/6 background were crossed to generate homozygous Trim21−/− mice. They were identified by PCR analysis of tail DNA using the primers corresponding to exon 3 and EGFP sequences (pink line in Fig. 1A). As shown in Fig. 1B, the former primers detected a 410 bp fragment of the wild-type allele, while the latter primers detected a 640 bp sequence of the mutant allele. Genotyping of 236 pups from Trim21+/− crosses showed that the alleles segregated among the offspring with expected Mendelian ratios. Furthermore, homozygous null mice did not express Trim21 transcripts, confirming proper Trim21 gene disruption in Trim21−/− mice. In Supplemental Fig. 1, Trim21 transcripts from BMMs were tested by RT-PCR using five primer sets corresponding to various regions of the Trim21 gene, including exons 6–8 that were not deleted in our Trim21−/− mice. None of the primer sets produced detectable bands from Trim21−/− mice, while all five primer sets produced corresponding transcripts in wild-type mice. Western blot analysis of spleen tissues verified the absence of TRIM21 protein and the presence of the EGFP protein in Trim21−/− mice (Fig. 1C).
The general appearance and body weights were indistinguishable among Trim21−/−, Trim21+/− and Trim21+/+ mice. Gross and histological features of thymus, lung, heart, liver, spleen, kidney, intestine and lymph nodes (LNs) were comparable for mice of all three genotypes. In addition, Trim21−/− mice were fertile and lived a normal life span. These data indicate that disruption of Trim21 gene had no obvious effects on embryonic development, adult body development or fertility.
Trim21−/− mice exhibit normal development of lymphocytes, myeloid and dendritic cells
Based on previous reports demonstrating functional activities of TRIM21 in hematopoietic cells, it seemed possible that mice deficient in TRIM21 expression might exhibit altered development, size or function of different hematopoietic compartments (8, 13, 23). However, total cell numbers were comparable in spleens, BMs, LNs and thymuses of Trim21−/− and Trim21+/+ mice. In addition, flow cytometric analyses of surface markers revealed no significant difference in the numbers and proportions of T cells, B cells, macrophages, granulocytes and DCs in these tissues (Supplemental Table I).
Since IRF8 is ubiquitylated by TRIM21, and is required for the development of macrophages and some DC subsets, we investigated whether the generation of BMDCs and BMMs in vitro was altered in mice deficient in TRIM21 (26, 27). The number of CD11c+ DCs generated from Trim21−/− BM was ~ 25% lower than that from Trim21+/+ BM (Fig. 1D), however, the number of BMM generated from BM of mice of both genotypes was very similar. This suggests that TRIM21 may exert a non-redundant role in the generation of DCs under some conditions. Nevertheless, the frequencies of CD11cintB220+ plasmacytoid DC (pDC) and CD11c+B220− conventional DC (cDC) in spleen were similar in Trim21+/+ and Trim21−/− mice (data not shown). In addition, expression of CD8α and MHC II on BMDCs and expression of CD11b, F4/80 (EMR1), MHC II and CD86 on BMMs was indistinguishable on Trim21+/+ and Trim21−/− cells before and after stimulation with CpG or IFN-γ (data not shown).
Analysis of EGFP reporter expression reveals high Trim21 expression in immune cells
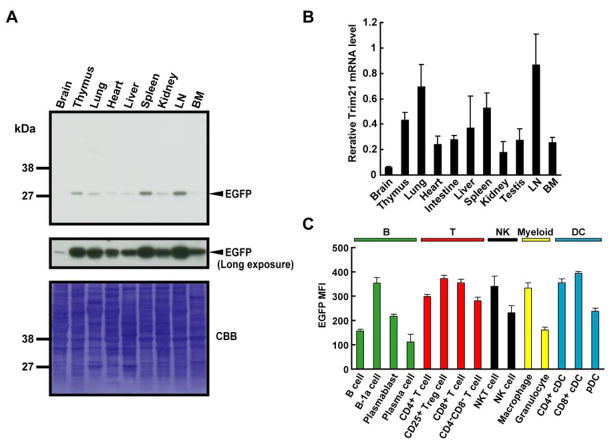
Although Trim21 is thought to be ubiquitously expressed, its precise distribution patterns and the levels among various tissues have not been precisely quantified (1). The availability of the EGFP reporter mice allowed us to examine this issue in detail. Western blot analyses showed that EGFP was expressed at the highest levels in LN, spleen and thymus with lower levels present in kidney, heart and liver (Fig. 2A). EGFP levels were very low, near the limits of detection, in brain, although a previous report indicated otherwise (1). Trim21 mRNA levels (Fig. 2B) mirrored the data from western blot analyses (Fig. 2A). In sum, Trim21, while widely expressed, is present at particularly at high levels in hematopoietic tissues.
Figure 2. Trim21 expression in tissues and hematopoietic cells.
A, Forty microgram of extracts from indicated tissues from Trim21+/− mice were immunoblotted with anti-GFP Ab. Coomassie brilliant blue (CBB) staining was used for loading control. B, Trim21 mRNA levels in Trim21+/+ tissues were quantified by qPCR. Values represent the mean for 3 mice ± SEM. C, Mean florescence intensity (MFI) of EGFP in leukocyte subsets. Trim21−/− splenocytes were analyzed by flow cytometry. Cells were classified according to the following markers: B cells; CD19+B220+, B-1a cells; CD19+CD5+, plasmablasts; CD138+B220+, plasma cells; CD138+B220−, CD4+ T cells; CD3+CD4+CD8−, CD25+ regulatory T (Treg) cells; CD4+CD25+, CD8+ T cells; CD3+CD4−CD8+, CD4−CD8− T cells; CD3+CD4−CD8−, NKT cells; NK-1.1+CD3+, NK cells; NK-1.1+CD3−, macrophages; CD11b+F4/80+, granulocytes; CD11b+F4/80−Gr-1high, CD4+ cDCs; CD11c+CD4+CD3−, CD8+ cDCs; CD11c+CD8+CD3− and pDCs; CD11c+B220+CD19−. Values are the means of 4 mice ± SEM.
We then assessed the levels of Trim21 expression among spleen cell subsets. Flow cytometric analyses showed that Trim21 expression was generally high in T cells, NKT cells, macrophages and DCs, but lower in B cells and granulocytes, although the levels varied for cells in each type (Fig. 2C). Among B-cell subsets, B-1a cells expressed relatively high levels of Trim21, while plasma cells showed much lower levels of expression. Within DC subsets, Trim21 levels were higher in the cDCs than pDCs.
Changes in expression of Trim21 during lymphocyte development
The observation that in secondary lymphoid tissues Trim21 was expressed at differing levels in subsets of cells belonging to different hematopoietic lineages suggested that levels of expression might change during the early stages of B- and T-cell differentiation. We examined this possibility by using flow cytometry to quantify EGFP expression in: 1) defined developmental stages during B-cell differentiation in the BM as well as transitional and mature B-cell subsets in the spleen (Fig. 3A); and 2) progressive stages of T-cell differentiation in the thymus (Fig. 3B).
Figure 3. Changes in Trim21 expression levels during lymphocyte development.
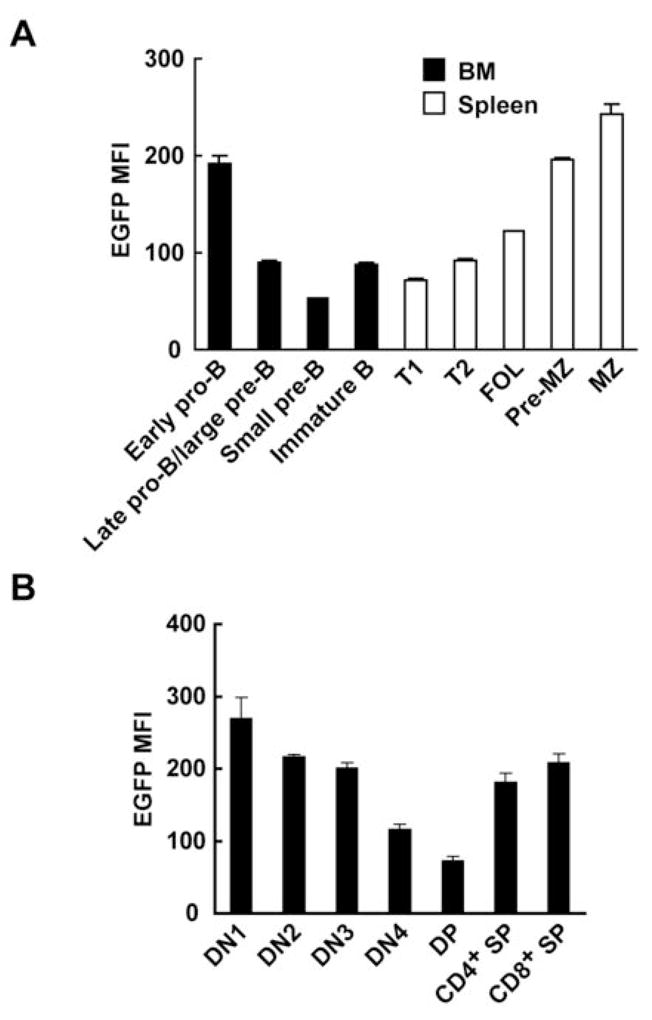
A, EGFP MFI was measured in each stage of B-cell development in Trim21−/− BM and spleen by flow cytometry. Cells were classified by surface markers: early pro-B cells; B220lowIgM−CD43+CD24−, late pro-B or large pre-B cells; B220lowIgM−CD43+CD24+, small pre-B cells; B220lowIgM−CD43−CD24+, immature B cells; B220lowIgM+, T1 cells; B220highIgMhighCD23−CD21−, T2 cells; B220highIgMhighCD23+CD21−, FOL cells; B220highIgMlowCD23+CD21int, pre-MZ cells; B220highIgMhighCD23+CD21high and MZ cells; B220highIgMhighCD23−CD21high. Values are the means of 2 mice ± SEM. B, EGFP MFI was analyzed in each stage of T-cell development in Trim21−/− thymus with the following markers: DN1 cells; CD4−CD8−CD44+CD25−, DN2 cells; CD4−CD8−CD44+CD25+, DN3 cells; CD4−CD8−CD44−CD25+, DN4 cells; CD4−CD8−CD44−CD25−, DP cells; CD3+CD4+CD8+, CD4+ SP cells; CD3+CD4+CD8− and CD8+ SP cells; CD3+CD4−CD8+. Values are the means of 4 mice ± SEM.
In the B-cell lineage, the highest levels of expression in the BM were seen with early pro-B cells with significantly lower and somewhat comparable levels seen in pre-B and immature B cells. In spleen, expression levels in transitional 1 (T1) and T2 cells were similar to those observed with pre-B and immature B cells in the BM, with levels in follicular (FOL) B cells being only slightly higher. Interestingly, the level of Trim21 expression in marginal zone (MZ) B cells was over twice that of FOL B cells with the difference presaged by levels in pre-MZ cells that were more than twice that of T1 and T2 cells. It is noteworthy that the levels of expression seen in MZ B cells approached those seen in peritoneal B-1a cells with which they have considerable functional overlap (data not shown). In the thymus, levels of Trim21 expression decreased progressively during the double-negative (DN) stages of differentiation and were lowest in double-positive (DP) cells. Levels were substantially higher and comparable in intrathymic single positive (SP) cells, but these levels were lower than those of SP cells in the periphery (Fig. 2C).
These observations demonstrated that the levels of Trim21 expression varied substantially during the early development of both the B- and T-cell lineages as well as in mature peripheral populations. Since the total numbers and subset distribution of cells in different lineages did not differ significantly between Trim21+/+ and Trim21−/− mice, these data also indicate that the observed differences in expression levels are probably irrelevant to normal processes of hematopoietic differentiation and are more likely to reflect the abundance of transcription factors that regulate its expression.
Induction of Trim21 by TLR and IFN signaling
IFN-inducible expression of Trim21 has been extensively documented in various cell types (6, 8, 28–31). Here, we extended these studies to examine the effects of viral infection, TLR engagement and IFN stimulation on Trim21 expression quantified by qPCR in BMDCs, BMMs, and EFs. To increase the sensitivity of detecting Trim21 expression in macrophages, BMMs were primed with IFN-γ prior to these stimulations (8, 32). Trim21 mRNA was rapidly induced in these cells following infection with NDV or stimulation with the TLR ligands poly(I:C), CpG and LPS (Fig. 4A and data not shown). Parallel studies of EGFP transcripts in infected or stimulated cells from Trim21−/− mice revealed comparable changes in terms of kinetics and magnitude (data not shown). This indicates that heightened expression of Trim21 transcripts in response to these stimuli is likely to be due to transcriptional induction, rather than increased mRNA stability or some other posttranscriptional mechanism. Flow cytometric analysis of EGFP expression also showed that Trim21 was induced to comparable levels following stimulation of both pDC and cDC (Fig. 4B and data not shown). While all TLR ligands tested stimulated Trim21 expression in BMDCs and IFN-γ-primed BMMs, in EFs LPS and poly(I:C), but not CpG, stimulated Trim21 expression (Fig. 4C and data not shown). CpG did not induce Trim21 in EFs, probably due to the lack of TLR9 expression in these cells. As expected, IFNs, both type I IFN-β and type II IFN-γ, stimulated expression of Trim21 and EGFP transcripts in unprimed BMMs and EFs of Trim21+/+ and Trim21−/− mice, respectively. As assessed by either qPCR (Fig. 4D) or flow cytometry (Fig. 4E), IFN-β was more potent than IFN-γ in stimulating Trim21 expression. Lastly, CpG stimulation of BMDCs prepared from mice lacking type I IFN receptors (Ifnar1−/−) did not induce Trim21 expression (Fig. 4F), consistent with previous studies indicating that TLR-induced type I IFNs accounted for Trim21 expression (5). Interestingly, two potent NF-κB inducers, IL-1β and TNFα, did not induce Trim21 expression in EFs, identifying signal-specific features of Trim21 induction (data not shown).
Figure 4. Induction of Trim21 by NDV infection, TLR and IFN stimulation.
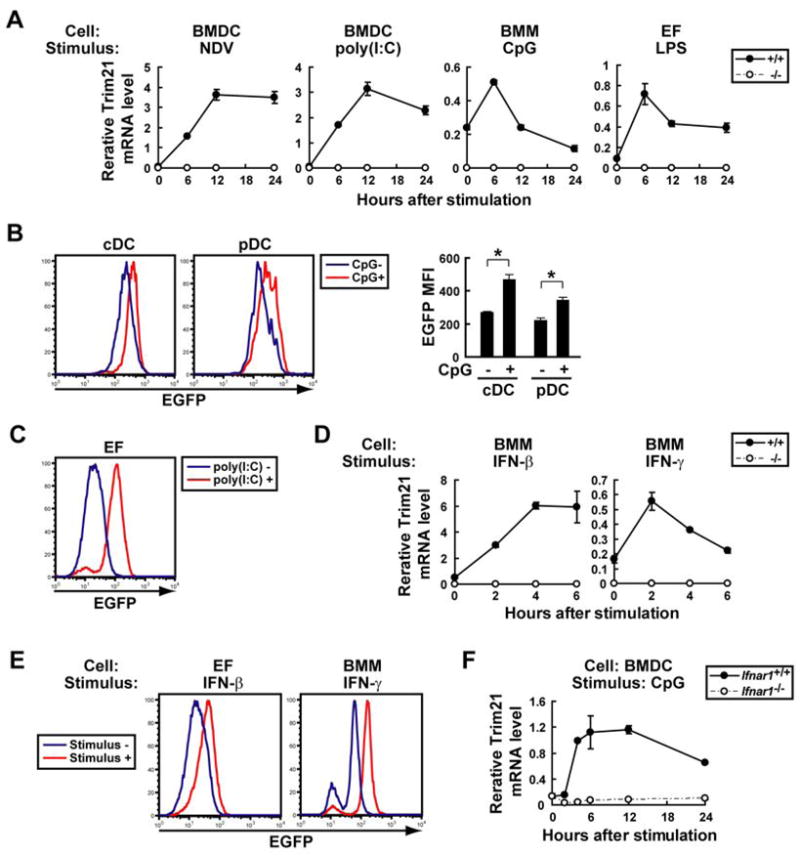
A, Trim21+/+ and Trim21−/− BMDCs, IFN-γ-primed BMMs, and EFs were infected with NDV or stimulated with TLR ligands, poly(I:C), CpG or LPS, and Trim21 mRNA was quantified by qPCR. Data are representatives of at least three independent experiments. B, EGFP MFI was measured for cDC (CD11c+B220−) and pDC (CD11c+B220+) harvested from Trim21−/− BMDCs with or without CpG stimulation for 24 h. Values are the means of three experiments ± SEM. *, p < 0.05. C, EGFP MFI was measured for Trim21−/− EFs cultured with or without poly(I:C) for 24 h. D, Unprimed Trim21+/+ and Trim21−/− BMMs were stimulated with IFNs, and Trim21 mRNA was quantified by qPCR. Data are representatives of at least three independent experiments. E, EGFP MFI was measured for Trim21−/− EFs or unprimed BMMs stimulated with or without IFN-β or IFN-γ, respectively, for 24 h. F, Ifnar1+/+ and Ifnar1−/− BMDCs were stimulated with CpG, and Trim21 mRNA was quantified by qPCR.
Elimination of Trim21 expression does not affect lymphocyte activation
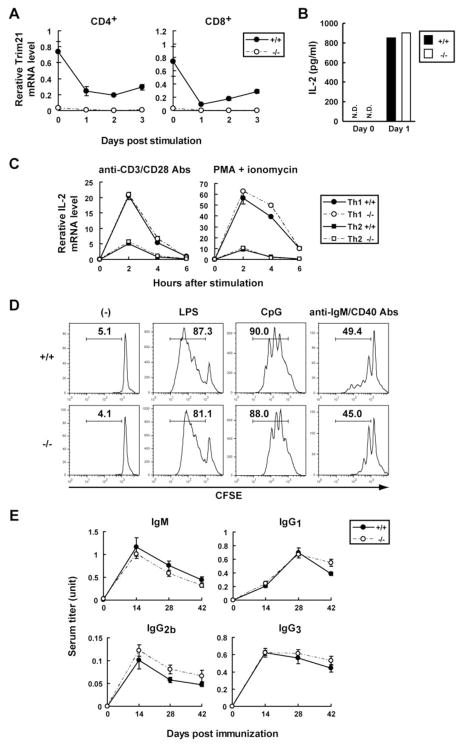
In light of reports that TRIM21 has a role in adaptive immune responses (13, 23), we proceeded to study the activity of T and B lymphocytes from Trim21−/− mice. Initial qPCR analyses of naïve CD4+ and CD8+ T cells stimulated with anti-CD3 plus anti-CD28 Abs revealed that Trim21 expression was markedly reduced for 3 days following activation (Fig. 5A). In further studies, comparisons of stimulated CD4+ or CD8+ T cells from Trim21+/+ and Trim21−/− mice showed that they produced comparable levels of IL-2 protein (Fig. 5B and data not shown). This result was somewhat unexpected in view of previous studies showing that ectopic expression of TRIM21 enhanced IL-2 production in T cells (23). Furthermore, the proliferative responses of both T-cell subsets from Trim21+/+ and Trim21−/− mice were essentially indistinguishable (data not shown).
Figure 5. The absence of Trim21 expression does not affect T-cell IL-2 production or B-cell prolifereation.
A, Naïve CD4+ T cells or CD8+ T cells from Trim21+/+ and Trim21−/− mice were stimulated with anti-CD3 and anti-CD28 Abs, and Trim21 mRNA was quantified by qPCR. B, IL-2 concentrations in the above cultures were measured by CBA. N.D., not detected. C, Th1 (circle)- or Th2 (square)-skewed CD4+ T cells from Trim21+/+ and Trim21−/− mice were re-stimulated with anti-CD3 plus anti-CD28 Abs or PMA plus ionomycin, and IL-2 mRNA was quantified by qPCR. D, Trim21+/+ and Trim21−/− B cells stained with CFSE were cultured with or without stimulant (LPS, CpG or anti-IgM plus anti-CD40 Abs) for 3 days. The percentages of divided cells are indicated in each panel. Data are representative of two experiments. E, Trim21+/+ and Trim21−/− mice (n = 6) were immunized with NP-KLH in alum and NP-specific IgM, IgG1, IgG2b and IgG3 Abs were detected by ELISA.
We also examined the cytokine profiles exhibited by stimulated Th1 and Th2 cells. Naïve CD4+ T cells were cultured in Th1- or Th2-skewing conditions and then re-stimulated with Abs to CD3 and CD28 or with PMA plus ionomycin. As shown in Fig. 5C, there were no significant differences in the levels of IL-2 transcripts produced by cells from Trim21−/− and Trim21+/+ mice. In additional studies, we found that levels of IFN-γ, IL-4, IL-5, TNFα and IL-10 transcripts expressed by these cells were comparable for mice of both genotypes (data not shown). Similarly, granzyme B expression by CD8+ T cells from Trim21−/− and Trim21+/+ mice was comparable (data not shown).
It was reported previously that overexpression of TRIM21 in a B-cell line inhibited proliferation (13). This prompted us to investigate the proliferative responses of B cells from Trim21+/+ and Trim21−/− mice to stimulation with LPS, CpG or Abs to IgM and CD40 using cells labeled with CFSE; no differences were seen (Fig. 5D). In addition, production of IgM by B cells from Trim21−/− and Trim21+/+ mice stimulated with LPS or CpG were indistinguishable (data not shown).
Finally, we examined the Ab responses of mice of both genotypes immunized with NP-KLH in alum. Anti-NP Ab titers were measured for IgM, IgG1, IgG2b and IgG3 on day 0, 14, 28 and 42 by ELISA. There were essentially no differences between Trim21+/+ and Trim21−/− mice for the time course or level of anti-NP Abs for any of the isotypes examined (Fig. 5E). Moreover, flow cytometric analysis of splenocytes from immunized mice revealed comparable development of germinal center B cells and plasma cells in Trim21−/− and Trim21+/+ mice (data not shown).
Enhanced expression of proinflammatory cytokines by Trim21−/− EFs, but not by Trim21−/− BMDCs or BMMs
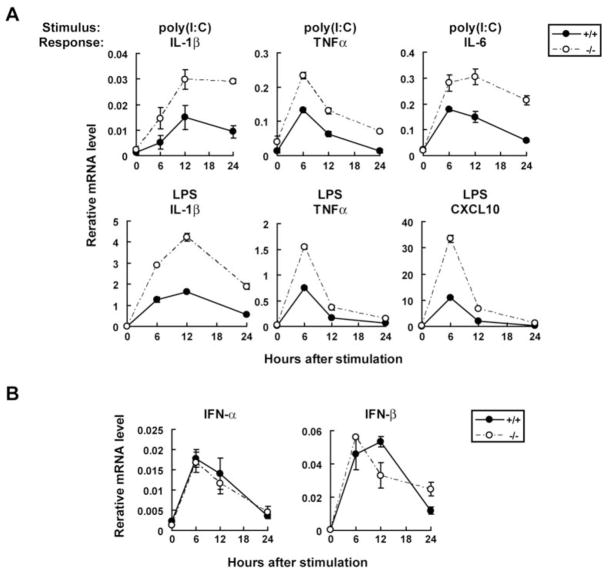
Previous reports indicated that TRIM21 regulates transcription of type I IFN and IL-12p40 in fibroblasts and macrophages through ubiquitin modifications of IRF3 and IRF8, respectively (8, 19). To expand on these studies, we investigated cytokine induction in EFs, BMDCs and BMMs from Trim21+/+ and Trim21−/− mice. The data in Fig. 6A illustrate the transcript levels for IL-1β, TNFα, IL-6 and a chemokine, CXCL10, expressed by EFs from Trim21−/− and Trim21+/+ mice stimulated with poly(I:C) or LPS and assayed 6 to 24 h later. Transcripts for each of these factors were rapidly induced by both poly(I:C) and LPS, with peak expression at 6 or 12 h, followed by a decline, and with LPS proving to be a more potent stimulus than poly(I:C). Strikingly, transcript levels for each gene were higher in EFs from Trim21−/− than Trim21+/+ mice in response to both poly(I:C) and LPS at all time points tested. Similar results were obtained with separate independent preparations of EFs. These proinflammatory cytokines are, for the most part, induced as a consequence of NF-κB activation downstream from TLR signaling (33, 34). Poly(I:C) can also activate NF-κB through retinoic acid-inducible gene I (RIG-I)/melanoma differentiation-associated gene 5 (MDA5) signaling (35). Since both TLR and RIG-I/MDA-5 pathways also stimulate type I IFN transcription by activating IRF3 and IRF7, we next asked whether induction of IFN-α and IFN-β is altered in Trim21−/− EFs. Contrary to our expectations, poly(I:C) and LPS induced comparable levels of IFN transcription in EFs from Trim21−/− and Trim21+/+ mice (Fig. 6B). The pattern of enhanced induction of proinflammatory cytokines but not of type I IFNs by cells from Trim21−/− mice suggests that TRIM21 might regulate NF-κB activation downstream from TLR and RIG-I/MDA5 pathways (see below).
Figure 6. Upregulation of proinflammatory cytokine expression in Trim21−/− EFs.
A, Trim21+/+ and Trim21−/− EFs were stimulated with poly(I:C) or LPS, and transcripts for indicated cytokines were quantified by qPCR. Data are representatives of three independent experiments. B, Trim21+/+ and Trim21−/− EFs were stimulated with poly(I:C), and type I IFN transcripts were measured by qPCR.
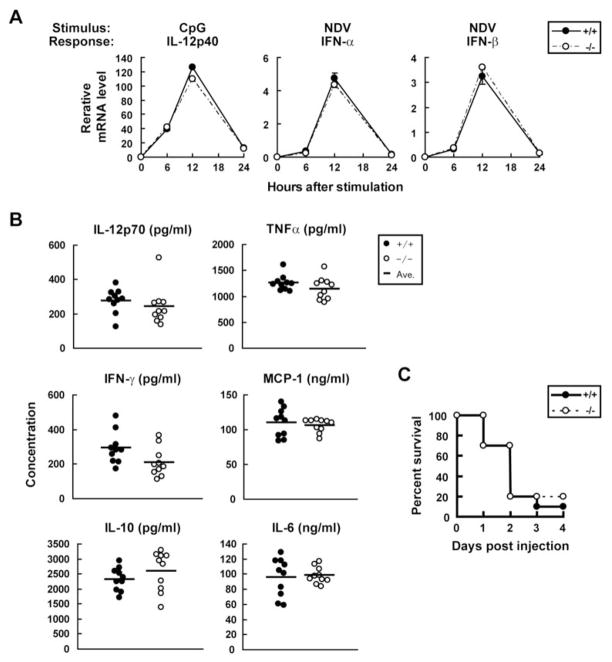
Because macrophages and DCs respond to TLR and RIG-I/MDA5 signaling by expressing many cytokines at high levels, we next examined IFN-γ-primed BMMs and BMDCs infected with NDV or stimulated with poly(I:C), LPS or CpG for expression of the factors studied above as well as IL-12p40. Surprisingly, cells from mice of either genotype exhibited no notable differences in induction of IL-1β, TNFα, IL-6, CXCL10, IFN-α and IFN-β (Fig. 7A and data not shown). IL-12p40 induction was also comparable for BMMs and BMDCs from Trim21−/− and Trim21+/+ mice in response to all stimuli tested (Fig. 7A and data not shown).
Figure 7. Cytokine induction in vivo and in vitro.
A, Trim21+/+ and Trim21−/− BMDCs were stimulated with CpG or infected with NDV, and transcripts for indicated cytokines were measured by qPCR. Data are representatives of three independent experiments. B, In vivo cytokine induction by LPS. Age- and gender-matched Trim21+/+ and Trim21−/− mice (n = 10) were injected with 30 μg/g of LPS and cytokine levels in sera were measured by CBA 4 h after injection. C, Trim21+/+ and Trim21−/− mice (n = 10) were injected with LPS as above and their survival was monitored.
Since above data suggested that inflammatory responses are increased in some but not all cell types from Trim21−/− mice, we studied whether elimination of TRIM21 might alter the responses of mice to an in vivo challenge with LPS. For this study, mice were injected i.p. with LPS and cytokine levels in sera were measured 4 h later. The results showed that the levels of TNFα, MCP-1, IL-6, IL-12p70, IFN-γ and IL-10 protein were greatly increased and to comparable levels in sera of both Trim21−/− and Trim21+/+ mice (Fig. 7B). Subsequently, most of the mice succumbed to endotoxic shock within 4 days after LPS injection. The survival curve was essentially identical in Trim21−/− and Trim21+/+ mice (Fig. 7C), indicating that the absence of TRIM21 did not alter systemic sensitivity to LPS.
Elimination of Trim21 expression does not impair IFN-stimulated gene expression
In view of the fact that Trim21 transcription is potently induced by both type I and type II IFNs, it was of interest to test whether TRIM21 regulates expression of IFN-inducible genes. To this end, we tested EFs or unprimed BMMs from Trim21+/+ and Trim21−/− mice stimulated with IFN-β or IFN-γ for transcript levels of 36 type I IFN-inducible genes and 23 type II IFN-inducible genes. As presented in Supplemental Fig. 2, all genes examined were expressed at comparable levels by cells of Trim21−/− and Trim21+/+ mice at both 6 h and 24 h after IFN addition. These data indicate that TRIM21 does not have a non-redundant function in IFN-stimulated gene expression.
Enhanced NF-κB promoter activity in Trim21−/− EFs
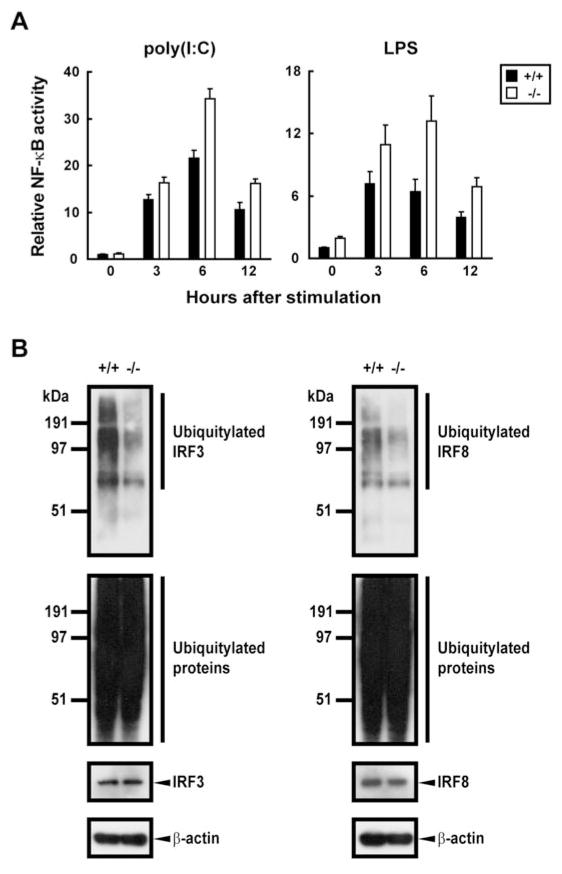
In studies described above, we showed that a series of proinflammatory cytokines were expressed at elevated levels in EFs, but not in macrophages and DCs of Trim21−/− mice. These data suggested that TRIM21 may play a previously unidentified role in regulating proinflammatory cytokine expression in EFs by modulating NF-κB activation. They also suggest that the function of TRIM21 is redundant in BMMs and BMDCs and that its activity is compensated in Trim21−/− cells in a pathway- and cell type-dependent fashion.
To study NF-κB activation in Trim21−/− EFs, we performed promoter analyses using a NF-κB-driven luciferase reporter for cells stimulated with LPS or poly(I:C). As shown in Fig. 8A, LPS- and poly(I:C)-induced luciferase activity was significantly higher in Trim21−/− cells than in Trim21+/+ cells at 3, 6 and 12 h after stimulation. These data support the idea that the elevated proinflammatory cytokine expression in Trim21−/− cells stimulated with TLR ligands can be accounted for by enhanced NF-κB activity.
Figure 8. Non-redundant role of Trim21 in the regulation of NF-κB promoter activity and ubiquitylation of IRF3 and IRF8.
A, EFs transfected with pBL driven by 3 copies of NF-κB binding site and pRL-TK were stimulated with 100 μg/ml poly(I:C) or 200 ng/ml LPS for 3, 6 or 12 h. Luciferase activity was measured at the indicated hours. Reporter activities were normalized by Renilla luciferase activities. B, Extracts from Trim21+/+ and Trim21−/− EFs stimulated with poly(I:C) were precipitated with anti-IRF3 Ab and blotted with anti-ubiquitin Ab (left top panel). The left lower panels show total ubiquitylated proteins and un-ubiquitylated IRF3 in total extracts, respectively. Extracts from Trim21+/+ and Trim21−/− BMMs stimulated with IFN-γ/CpG were precipitated with anti-IRF8 Ab and blotted with anti-ubiquitin Ab (right top panel). The right lower panels represent western blot of total extracts. β-Actin was used as a loading control.
Reduced ubiquitylation of IRF3 and IRF8 in Trim21−/− cells
Previous studies using established cell lines documented that TRIM21 regulates IFN and IL-12p40 expression by mediating ubiquitylation of IRF3 and IRF8 (8, 19). However, ubiquitylation of IRF3 by TRIM21 has recently been questioned (36). In view of the absence of discernible defects in type I IFNs and IL-12p40 induction in Trim21−/− cells, suggestive of functional redundancy, it was of importance to determine whether IRF3 and IRF8 were ubiquitylated by TRIM21 in primary cells and whether this activity was lost in cells of Trim21−/− mice. Whole cell extracts prepared from poly(I:C)-stimulated EFs or from IFN-γ/CpG-stimulated BMMs were precipitated using Ab for IRF3 or IRF8 and blots were probed for ubiquitin conjugates using anti-ubiquitin Ab. Immunoblots of anti-IRF3-precipitated material from Trim21+/+ and Trim21−/− EFs (Fig. 8B, left panels) showed that the amounts of ubiquitylated IRF3 and associated proteins were greatly reduced in Trim21−/− cells relative to Trim21+/+ cells (left top panel). Immunoblot analysis of total extracts (left bottom panels) confirmed that ubiquitylation of total proteins and the levels of IRF3 were similar in these cells. Similarly, immunoblots of anti-IRF8 Ab precipitates from stimulated BMMs showed that the amounts of ubiquitin-conjugated IRF8 and associated proteins were significantly reduced in Trim21−/− as compared to Trim21+/+ cells (Fig. 8B, right top panel) while total ubiquitylated proteins and levels of IRF8 were the same (right bottom panels). These results demonstrated first, that IRF3 and IRF8 are true substrates for TRIM21-mediated ubiquitylation in vivo, and second, that the ubiquitin ligase activity of TRIM21 for these substrates is non-redundant. This suggests that there is a mechanism of compensation that ensures normal IFN and IL-12p40 induction in TRIM21-deficient cells.
Enhanced expression of other Trim genes in Trim21−/− cells
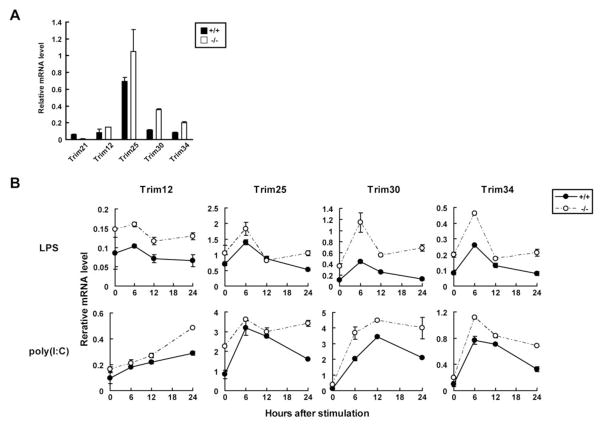
The Trim21 gene maps to a region of mouse chromosome 7 that is also occupied by 10 other Trim family members. A similar “TRIM gene cluster” exists on human chromosome 11. We were interested in the possibility that expression of some of these genes may be upregulated as a result of TRIM21 deficiency, thereby compensating for the lack of TRIM21 function in some cells. We therefore quantified expression of Trim12, Trim25, Trim30 and Trim34 transcripts in Trim21+/+ and Trim21−/− EFs before and after stimulation by LPS and poly(I:C). All the Trims, except for Trim25, map to chromosome 7 and are induced by IFNs (5). We included Trim25 (chromosome 11 in mouse, chromosome 17 in human), since it plays a role in RIG-I-mediated IFN induction (37). Data in Fig. 9A and Fig. 9B showed that transcript levels of these genes were all higher in Trim21−/− than Trim21+/+ cells both prior to and after stimulation. Given that type I IFN induction levels did not differ in Trim21−/− and Trim21+/+ cells (see Fig. 6B), increased expression of these Trim genes in Trim21−/− cells was not due to increased type I IFN secretion and secondary stimulation. These results demonstrated that the lack of TRIM21 was associated with elevated expression of multiple Trim genes in a type I IFN-independent manner, possibly compensating for the functional deficiency caused by the elimination of TRIM21 expression. It is possible that the lack of robust phenotypes in Trim21−/− mice may be due, at least in part, to substitution of TRIM21 functions by these other TRIM family members.
Figure 9. Increased expression of transcripts for TRIM family proteins in Trim21−/− EFs.
A, Constitutive transcript levels for indicated Trim members in Trim21+/+ and Trim21−/− EFs were measured by qPCR. Data are representatives of three independent experiments. B, Above transcripts were measured after stimulation with LPS or poly(I:C) for the indicated times.
Discussion
Members of the TRIM family proteins have been found to play a large number of distinct but sometimes overlapping roles in innate and acquired immunity with responses triggered by IFNs as well as signals from TLRs and other pattern recognition receptors (2, 3, 38). TRIM21, like many other family members, is an E3 ubiquitin ligase, that has been shown by in vitro studies to target IRF3, IRF8 and IgG for ubiquitylation with distinct functional outcomes (8, 16, 19). In this study, we generated a Trim21-null allele that permitted us to determine the levels and pattern of Trim21 expression at the single cell level by virtue of an EGFP knockin, as well as to assess the effects of TRIM21 deficiency in mice homozygous for the null allele.
Expression studies showed that Trim21 is constitutively transcribed in all tissues and cell types examined but with highly divergent levels of expression. Highest expression was seen in cells of the immune system with particularly high levels in T cells, macrophages and DCs, where the expression was further augmented by stimulation with IFNs and TLR ligation. Interestingly, during both T- and B-cell development, expression of Trim21 decreased during successive stages of differentiation within the thymus and BM, but increased again in later stages of development in the periphery.
We anticipated that these major changes in Trim21 expression associated with distinct hematopoietic lineages and stages of differentiation would be associated in Trim21−/− mice with readily detectable numeric or functional changes in specific subsets of hematopoietic cells. Remarkably, essentially none were found. The only exception was a reduction in the number of DCs that could be generated in vitro from BMs of adult TRIM21-deficient mice. We conclude that TRIM21 is dispensable for the development of immune cells under normal conditions. Since there were no gross or histologically discernable abnormalities in the development of other organ systems, fertility or life span of Trim21−/− mice, the gene is generally superfluous to almost all cell types under normal conditions.
In keeping with this perspective, functional studies of immune cells in Trim21−/− mice revealed no abnormalities for macrophages, DCs, T cells or B cells. Cytokine responses of macrophages and DCs from Trim21−/− and Trim21+/+ mice to stimulation with NDV, IFNs or TLR ligands were indistinguishable. Similarly, no abnormality was found in activation/proliferation and Ab production by Trim21−/− T and B cells in vitro or in vivo. Given the number of reports documenting a role for TRIM21 in innate and adaptive immune responses, these results indicate that compensatory mechanisms can obscure the functional role of TRIM21 in immune cells (see below) (8, 13, 19, 23). It would thus seem that some of the previously reported activities of TRIM21 may be partly attributed to the ectopic expression strategies used in these studies. It should be stated here that additional, more sophisticated functional analyses may reveal abnormalities in Trim21−/− mice in the future.
In contrast to our studies of immune cells, analyses of EFs identified distinct differences between cells obtained from Trim21−/− and normal mice. First, a number of proinflammatory cytokines were expressed at elevated levels in TRIM21-deficient cells following stimulation with LPS or poly(I:C). The enhanced expression of these cytokines may be expected to have a significant impact on inflammatory responses in these mice, as fibroblasts play an active role in conditioning local cellular and cytokine production by modulating leukocyte behavior. For example, in rheumatoid arthritis, synovial fibroblasts contribute to persistent inflammation in the synovium by recruiting immune cells through the secretion of cytokines and chemokines (39).
Evidence garnered from promoter-reporter analyses indicated that elevated induction of cytokines and chemokines in Trim21−/− EFs can be ascribed to enhanced NF-κB activity. NF-κB is activated in response to TLR and RIG-I/MDA5 signaling and plays a central role in eliciting inflammatory responses (33, 40). IL-1β, IL-6, TNFα and CXCL10, with elevated transcripts in Trim21−/− cells, are direct targets of NF-κB and all partake in inflammatory processes (41). NF-κB is activated by a cascade of events that result in the activation of TNF receptor-associated factor 6 (TRAF6) and IκB kinase (IKK) α/β, which leads to the degradation of IκBα and liberation of p65 and p50 (40). Many proteins have been identified that act within the TLR/RIG-I/MDA5 cascade to promote NF-κB activation, some through ubiquitin ligase activity, others through phosphorylation (34, 40, 42). Interestingly, there are a number of proteins that negatively regulate NF-κB activation, which also act within the cascade. For example, TRIM30α promotes the degradation of TAB2 and TAB3 by acting in the ubiquitin-proteasome pathway, and prevents TRAF6 autoubiquitylation to reduce NF-κB activation and cytokine production (43). In addition, TRIM27 is reported to inhibit both canonical and noncanonical IKKs to inhibit activation of NF-κB as well as IRF3, leading to reduced activities of promoters with NF-κB- and IRF3-binding sites (44). Cylindromatosis (CYLD), a deubiquitylating enzyme, is another negative regulator of NF-κB activation (42, 45, 46). It targets several proteins within the NF-κB activation pathway, including TRAF6 and IKKγ/Nemo and interferes with biological processes mediated by NF-κB, including inflammation and apoptosis. With the results presented in this paper, TRIM21 may be added to the list of negative regulators of NF-κB activation, and is likely to be involved in attenuating proinflammatory responses in fibroblasts.
Although the precise mechanism by which TRIM21 regulates NF-κB activation in EFs is so far not clear, TRIM21 as an E3 ubiquitin ligase may play a role in the ubiquitylation of some proteins within the NF-κB activation pathway (8, 18, 19). We have recently observed that TRIM21 interacts with p62, also called sequestosome 1, in addition to IRF8 in macrophages after stimulation with IFN-γ and TLR ligands (47). p62 is a scaffold/adaptor protein that carries a ubiquitin binding domain and it regulates NF-κB activity both positively and negatively by affecting the function of TRAF6 and CYLD (48, 49). Thus, TRIM21 might possibly regulate NF-κB activity through p62. It would be of interest to test this possibility in the future. Regardless of the mechanisms involved, it is interesting to note that NF-κB activity is increased in cells of patients with SLE, finding that may be relevant to the proposed role of TRIM21 in the pathogenesis of autoimmune diseases (13, 50).
It is important to stress here that expression of IFN-inducible genes by EFs or BMMs from Trim21−/− mice stimulated with IFN-β or of IFN-γ was completely normal. This result was unexpected for several reasons. First, Trim21 is strongly induced by TLR and IFN signaling and this induction stimulates TRIM21 protein to translocate from the cytoplasm to the nucleus, indicative of functional participation of TRIM21 in signaling (8, 30). Secondly, TRIM21 is known to ubiquitylate IRF3 and regulate type I IFN induction in fibroblasts (19). In agreement with this role, TLR-stimulated IRF3 ubiquitylation was substantially reduced in Trim21−/− EFs. These results support the notion that there is a compensatory mechanism that allows Trim21−/− cells to express IFN-responsive genes at normal levels in response to stimulation with type I or type II IFNs. This mechanism appears not to be based on restoration of IRF3 ubiquitylation, but rather by alteration of some other signaling pathway. Type I IFN induction and NF-κB activation are triggered by a common pathway of TLR and RIG-I/MDA5 signaling, and events downstream of TRAF6 activation, such as activation of IKKα/β and IKKi/TBK1, are responsible for a separate mode of NF-κB and IRF3/7 activation (33, 34). Given that NF-κB activation was not compensated for in Trim21−/− EFs, the mechanism that restore IFN induction in Trim21−/− cells may lie in a step downstream from TRAF6 activation.
Our observation that a series of TRIM proteins are upregulated in Trim21−/− EFs before and after TLR stimulation is highly significant and supports the idea that these proteins are likely to be part of the compensatory mechanisms for TRIM21 deficiency in Trim21−/− cells. In view of the fact that Trim21, Trim12, Trim30 and Trim34 are closely linked genetically and that all were upregulated in Trim21−/− cells, the Trim gene cluster on chromosome 7 may have arisen to support their functional redundancy. It is reasonable to assume that multiple TRIM proteins are expressed in specific cell types in order to maintain functional equilibrium. In the immune system, TRIM proteins may form an even more extensive network than in EFs to support tight redundancy. The observation that IRF8 ubiquitylation induced by IFN/TLR stimulation was impaired while IL-12p40 induction was intact in Trim21−/− macrophages is also consistent with the idea that the compensation is achieved at the level of functional outcome, not at the level of an individual regulatory event. This leads us to suggest that many TRIM proteins contribute to the establishment of extensive functional redundancy, which compensates for the absence of TRIM21 activity in immune cells of Trim21−/− mice.
In conclusion, we describe a previously unappreciated role for TRIM21 in the negative regulation of NF-κB activation, resulting in a reduction in proinflammatory cytokine induction in fibroblasts after TLR stimulation. In addition, our analysis indicates the existence of potent compensatory mechanisms that effectively make up the loss of TRIM21 in Trim21−/− fibroblasts. This compensatory mechanism may partly be attributable to multiple, related TRIM proteins that coordinately integrate their expression and activities to attain functional redundancy.
Supplementary Material
Acknowledgments
We thank members of Ozato and Morse laboratories for reagents, suggestions and critical discussions of this work. Especially, we thank Dr. P. Tailor for his expert advice.
Footnotes
Abbreviations used in this paper: TRIM, tripartite motif; SLE, systemic lupus erythematosus; SS, Sjögren’s syndrome; LESA, lymphoepithelial sialadenitis; IRF, IFN regulatory factor; EGFP, enhanced GFP; EF, embryonic fibroblast; DC, dendritic cell; APC, allophycocyanin; Flt3L, Fms-like tyrosine kinase 3 ligand; BM, bone marrow; BMDC, BM-derived dendritic cell; BMM, BM-derived macrophage; NDV, Newcastle disease virus; qPCR, quantitative real time reverse transcriptase PCR; CBA, cytometric bead array; NP, (4-hydroxy-3-nitrophenyl) acetyl; KLH, keyhole limpet hemocyanin; LN, lymph node; pDC, plasmacytoid DC; cDC, conventional DC; T, transitional; FOL, follicular; MZ, marginal zone; DN, double-negative; DP, double-positive; SP, single-positive; RIG-I, retinoic acid-inducible gene I; MDA5, melanoma differentiation-associated gene 5; TRAF6, TNF receptor-associated factor 6; IKK, IκB kinase; CYLD, cylindromatosis; CBB, Coomassie brilliant blue; MFI, mean florescence intensity; Treg, regulatory T.
Disclosures
The authors have no financial conflict of interest.
This work was supported by the Intramural Research Program of the NIH, National Institute of Child Health and Human Development and National Institute of Allergy and Infectious Diseases. R.Y. and T.A. were supported in part by the JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH.
References
- 1.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. Embo J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 3.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. TRIM E3 Ligases Interfere with Early and Late Stages of the Retroviral Life Cycle. PLoS Pathog. 2008;4:e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajsbaum R, Stoye JP, O’Garra A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol. 2008;38:619–630. doi: 10.1002/eji.200737916. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes DA, Ihrke G, Reinicke AT, Malcherek G, Towey M, Isenberg DA, Trowsdale J. The 52 000 MW Ro/SS-A autoantigen in Sjogren’s syndrome/systemic lupus erythematosus (Ro52) is an interferon-gamma inducible tripartite motif protein associated with membrane proximal structures. Immunology. 2002;106:246–256. doi: 10.1046/j.1365-2567.2002.01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strandberg L, Ambrosi A, Espinosa A, Ottosson L, Eloranta ML, Zhou W, Elfving A, Greenfield E, Kuchroo VK, Wahren-Herlenius M. Interferon-alpha Induces Up-regulation and Nuclear Translocation of the Ro52 Autoantigen as Detected by a Panel of Novel Ro52-specific Monoclonal Antibodies. J Clin Immunol. 2007 doi: 10.1007/s10875-007-9157-0. [DOI] [PubMed] [Google Scholar]
- 8.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, 3rd, Ozato K. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Chetrit E, Chan EK, Sullivan KF, Tan EM. A 52-kD protein is a novel component of the SS-A/Ro antigenic particle. J Exp Med. 1988;167:1560–1571. doi: 10.1084/jem.167.5.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salomonsson S, Dorner T, Theander E, Bremme K, Larsson P, Wahren-Herlenius M. A serologic marker for fetal risk of congenital heart block. Arthritis Rheum. 2002;46:1233–1241. doi: 10.1002/art.10232. [DOI] [PubMed] [Google Scholar]
- 11.Frank MB, Itoh K, Fujisaku A, Pontarotti P, Mattei MG, Neas BR. The mapping of the human 52-kD Ro/SSA autoantigen gene to human chromosome 11, and its polymorphisms. Am J Hum Genet. 1993;52:183–191. [PMC free article] [PubMed] [Google Scholar]
- 12.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinosa A, Zhou W, Ek M, Hedlund M, Brauner S, Popovic K, Horvath L, Wallerskog T, Oukka M, Nyberg F, Kuchroo VK, Wahren-Herlenius M. The Sjogren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Eversole T, Lee DJ, Sontheimer RD, Capra JD. Protein-protein interactions between native Ro52 and immunoglobulin G heavy chain. Scand J Immunol. 1999;49:620–628. doi: 10.1046/j.1365-3083.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang YS, Yang MC, Wang B, Weissler JC. Autoantigen Ro52 directly interacts with human IgG heavy chain in vivo in mammalian cells. Mol Immunol. 2000;37:591–602. doi: 10.1016/s0161-5890(00)00068-7. [DOI] [PubMed] [Google Scholar]
- 16.Takahata M, Bohgaki M, Tsukiyama T, Kondo T, Asaka M, Hatakeyama S. Ro52 functionally interacts with IgG1 and regulates its quality control via the ERAD system. Mol Immunol. 2008;45:2045–2054. doi: 10.1016/j.molimm.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Sabile A, Meyer AM, Wirbelauer C, Hess D, Kogel U, Scheffner M, Krek W. Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol Cell Biol. 2006;26:5994–6004. doi: 10.1128/MCB.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada K, Kamitani T. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem Biophys Res Commun. 2006;339:415–421. doi: 10.1016/j.bbrc.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 19.Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci U S A. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Si Z, Vandegraaff N, O’Huigin C, Song B, Yuan W, Xu C, Perron M, Li X, Marasco WA, Engelman A, Dean M, Sodroski J. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci U S A. 2006;103:7454–7459. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii T, Ohnuma K, Murakami A, Takasawa N, Yamochi T, Iwata S, Uchiyama M, Dang NH, Tanaka H, Morimoto C. SS-A/Ro52, an autoantigen involved in CD28-mediated IL-2 production. J Immunol. 2003;170:3653–3661. doi: 10.4049/jimmunol.170.7.3653. [DOI] [PubMed] [Google Scholar]
- 24.Tsujimura H, Tamura T, Gongora C, Aliberti J, Reis e Sousa C, Sher A, Ozato K. ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood. 2003;101:961–969. doi: 10.1182/blood-2002-05-1327. [DOI] [PubMed] [Google Scholar]
- 25.Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993;7:1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 26.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 27.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O’Shea JJ, Singh H, Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 28.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmerer JM, Lesinski GB, Radmacher MD, Ruppert A, Carson WE., 3rd STAT1-dependent and STAT1-independent gene expression in murine immune cells following stimulation with interferon-alpha. Cancer Immunol Immunother. 2007;56:1845–1852. doi: 10.1007/s00262-007-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strandberg L, Ambrosi A, Espinosa A, Ottosson L, Eloranta ML, Zhou W, Elfving A, Greenfield E, Kuchroo VK, Wahren-Herlenius M. Interferon-alpha induces up-regulation and nuclear translocation of the Ro52 autoantigen as detected by a panel of novel Ro52-specific monoclonal antibodies. J Clin Immunol. 2008;28:220–231. doi: 10.1007/s10875-007-9157-0. [DOI] [PubMed] [Google Scholar]
- 31.Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, Chow JM, Gri G, Carra G, Gerosa F, Wolf SF, Dzialo R, Trinchieri G. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang K, Shi HX, Liu XY, Shan YF, Wei B, Chen S, Wang C. TRIM21 Is Essential to Sustain IFN Regulatory Factor 3 Activation during Antiviral Response. J Immunol. 2009;182:3782–3792. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 37.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 38.Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–8980. doi: 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- 39.Ritchlin C. Fibroblast biology. Effector signals released by the synovial fibroblast in arthritis. Arthritis Res. 2000;2:356–360. doi: 10.1186/ar112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 41.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Israel A. NF-kappaB activation: Nondegradative ubiquitination implicates NEMO. Trends Immunol. 2006;27:395–397. doi: 10.1016/j.it.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, Sun S, Xiang C, Sun B. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 44.Zha J, Han KJ, Xu LG, He W, Zhou Q, Chen D, Zhai Z, Shu HB. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IkappaB kinase family members. J Immunol. 2006;176:1072–1080. doi: 10.4049/jimmunol.176.2.1072. [DOI] [PubMed] [Google Scholar]
- 45.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 46.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 47.Kim JY, Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J Immunol. 2009;182:2131–2140. doi: 10.4049/jimmunol.0802755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 49.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Wong HK, Kammer GM, Dennis G, Tsokos GC. Abnormal NF-kappa B activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J Immunol. 1999;163:1682–1689. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.