Abstract
A central challenge for improving autoimmune therapy is preventing inflammatory pathology without inducing generalized immunosuppression. T helper 17 (TH17) cells, characterized by their production of interleukin-17, have emerged as important and broad mediators of autoimmunity. Here we show that the small molecule halofuginone (HF) selectively inhibits mouse and human TH17 differentiation by activating a cytoprotective signaling pathway, the amino acid starvation response (AAR). Inhibition of TH17 differentiation by HF is rescued by the addition of excess amino acids and is mimicked by AAR activation after selective amino acid depletion. HF also induces the AAR in vivo and protects mice from TH17-associated experimental autoimmune encephalomyelitis. These results indicate that the AAR pathway is a potent and selective regulator of inflammatory T cell differentiation in vivo.
Naïve CD4+ T cells differentiate into diverse effector and regulatory subsets to coordinate immunity to pathogens while establishing peripheral tolerance. Besides TH1 and TH2 effector subsets, which produce interferon-γ (IFN-γ) and interleukin-4 (IL-4), respectively, naïve T cells can differentiate into proinflammatory T helper 17 (TH17) cells or tissue-protective induced T regulatory (iTreg) cells (1, 2). TH17 cells are key regulators of autoimmune inflammation; characteristically produce IL-17 (IL-17A), IL-17F, and IL-22; and differentiate in the presence of inflammatory cytokines, such as IL-6 or IL-21, together with transforming growth factor–β (TGF-β) (1, 2).
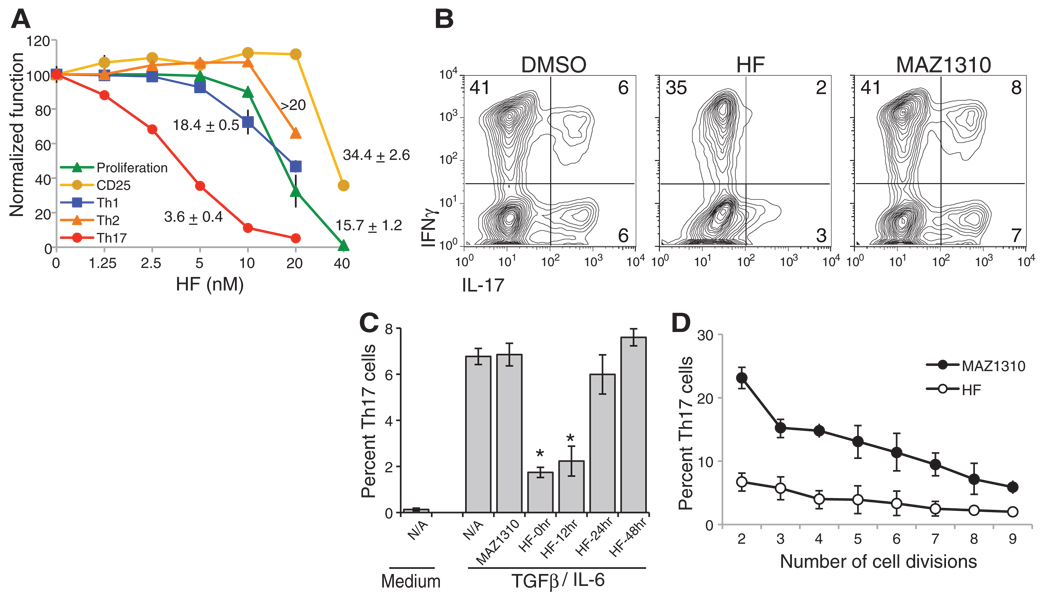
The small molecule halofuginone (HF) is a derivative of the plant alkaloid febrifugine (3). HF has shown therapeutic promise in animal models of fibrotic disease and a clinical trial for scleroderma (3–5), but its mechanism of action is unclear. To investigate whether HF could modulate T cell differentiation, we stimulated murine T cells to induce TH1, TH2, iTreg, or TH17 differentiation and treated these cells with HF or an inactive derivative, MAZ1310 (fig. S1, A and B) (6). HF selectively inhibited the development of TH17 cells with a median inhibitory concentration (IC50) of 3.6 ± 0.4 nM (Fig. 1A and fig. S2A). Low concentrations of HF that impaired TH17 differentiation did not influence TH1, TH2, or iTreg differentiation (Fig. 1A and fig. S2A) and had no impact on T cell receptor (TCR)–induced cytokine secretion by naïve T cells (fig. S2B). HF also repressed IL-17 expression by human T cells without influencing IFN-γ production (Fig. 1B). Consistent with a previous report (7), 10-fold higher concentrations of HF broadly impaired lymphocyte function (Fig. 1A and fig. S2C). Inhibition of TH17 differentiation by HF was most pronounced when added within the first 24 hours of culture (Fig. 1C), was stereospecific (fig. S2D), and was not cytotoxic below 100 nM (fig. S2E). Although HF treatment delayed S-phase entry within 24 hours of TCR activation, these T cells recovered thereafter, showing no defect in expansion kinetics between days 2 and 4 postactivation (fig. S3). Moreover, HF suppressed TH17 differentiation, irrespective of the number of cell divisions completed (Fig. 1D), and reduced TH17 differentiation when IFN-γ and IL-4, cytokines that inhibit TH17 differentiation (8), were neutralized by antibodies (fig. S4A).
Fig. 1.
Selective inhibition of TH17 differentiation by HF. (A) Carboxyfluorescein diacetate succinimidyl ester (CFSE)–labeled T cells were activated in the presence of dimethyl sulfoxide (DMSO), 40 nM MAZ1310, or titrating concentrations of HF. CFSE dilution and the percentages of cells expressing CD25, IFN-γ+ IL-4− (TH1 cells), IL-4+ IFN-γ− (TH2 cells), or IL-17+ IFNγ− (TH17 cells) cells are displayed as mean values ± SD normalized to MAZ1310-treated cells. IC50 values are listed next to corresponding lines ± SD. (B) Intracellular cytokine expression in primary human T cells treated with DMSO, HF, or MAZ1310. (C) Differentiating TH17 cell cultures were incubated with HF at the indicated times after activation. Intracellular cytokine expression was determined on day four. The mean percentage of TH17 cells is shown ± SD (error bars). *P < 0.005, relative to MAZ1310-treated T cells. (D) CFSE-labeled T cells were differentiated to TH17 in the presence of HF or MAZ1310. After intracellular cytokine staining, CFSE peaks were gated and mean percentages of IL-17+ T cells within each peak are displayed ± SD (error bars). All data represent at least three similar experiments.
HF inhibited Il17a and Il17f mRNA production without affecting the expression of IL-2 and tumor necrosis factor, cytokines expressed by all effector T cells (fig. S4B). HF treatment did not affect the induction of RORγt and RORα, two orphan nuclear receptors induced by TH17 polarizing cytokines that mediate lineage commitment (9, 10) (fig. S4C). Ectopic expression of RORγt in T cells did not override the inhibitory effects of HF on TH17 differentiation (fig. S4D), confirming that RORγt is not sufficient to drive the effector function of TH17 cells (11).
HF did not directly inhibit signaling induced by TGF-β or IL-6, the two principal cytokines that instruct TH17 differentiation. Although high concentrations (>50 nM) of HF were reported to impair TGF-β signaling in fibroblasts (4), low doses of HF that repress TH17 differentiation inhibited neither TGF-β–induced R-Smad2 phosphorylation (fig. S5A) nor a variety of other lymphocyte responses to TGF-β (fig. S5, B to D) (12). In contrast, the type 1 TGF-β receptor kinase inhibitor SB-431542 (fig. S1C) abrogated all responses to TGF-β (fig. S5). Additionally, HF did not inhibit early IL-6–induced STAT3 phosphorylation (where STAT proteins are signal transducers and activators of transcription) (fig. S6), but it did reduce sustained STAT3 activation beginning 12 hours poststimulation (fig. S6), indicating that HF indirectly modulates factors that maintain STAT3 signaling. Consistent with decreased STAT3 activity (13), HF-treated TH17 cells displayed a reciprocal increase in Foxp3 expression (fig. S7A). However, retroviral expression of FOXP3 in T cells did not decrease TH17 differentiation (fig. S7B), and HF repressed IL-17 expression in T cells lacking Foxp3 (fig. S7C). Thus, changes in Foxp3 expression are not necessary or sufficient for the effects of HF on TH17 differentiation.
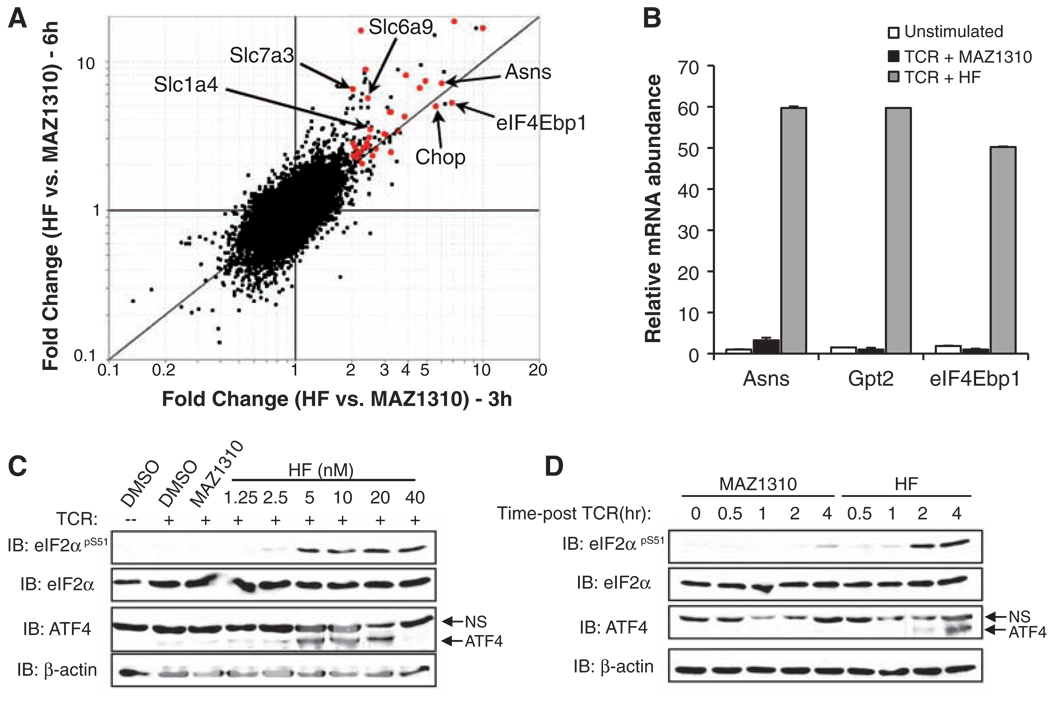
HF-treated T cells stimulated in TH17 polarizing conditions for 3 or 6 hours showed differential expression of 81 annotated genes, the majority of which were up-regulated (Fig. 2A and table S1). Among HF-inducible transcripts, many were functionally associated with amino acid transport and biogenesis, as well as protein synthesis (table S1), a pattern characteristic of an amino acid starvation response (AAR, also called general amino acid control in yeast) (14). The AAR pathway is physiologically induced by unaminoacylated (i.e., uncharged) tRNAs, which accumulate during amino acid insufficiency and bind to the protein kinase GCN2 (15, 16). Activated GCN2 phosphorylates and inhibits eukaryotic translation initiation factor 2A (eIF2α), leading to a transient reduction in protein synthesis, while enhancing the translation of ATF4 (fig. S8), a transcription factor that activates stress-induced gene expression (15–17). HF treatment activated ATF4 target genes (Fig. 2A; fig. S9A, red and blue dots, respectively; and table S2) (14, 17), including Asns, Gpt2, and eIF4Ebp1, as confirmed by quantitative real-time fluorescence polymerase chain reaction(QPCR) (Fig. 2B).
Fig. 2.
HF activates the AAR pathway. (A) Histogram of microarray data from differentiating TH17 cells treated with HF or MAZ1310 for 3 or 6 hours. Red dots indicate transcripts increased greater than twofold by HF-treatment at both 3 and 6 hours. Text and arrows denote several defined AAR genes (27). (B) Quantitative real-time PCR was performed on cDNA from unstimulated T cells or those activated for 4 hours in the presence of MAZ1310 or HF. Asns, Gpt2, or eIF4Ebp1 mRNA expression was normalized to Hprt levels and are shown as mean values ± SD. (C) Unstimulated or TCR-activated T cells were treated with DMSO, 40 nM MAZ1310, or HF and were lysed for Western blotting after 4 hours. (D) Activated T cells were lysed at the indicated times after treatment with MAZ1310 or HF. ATF4 protein is indicated by an arrow. NS, nonspecific band. Microarray data were generated from three biological replicates. Other data represent two to three experiments.
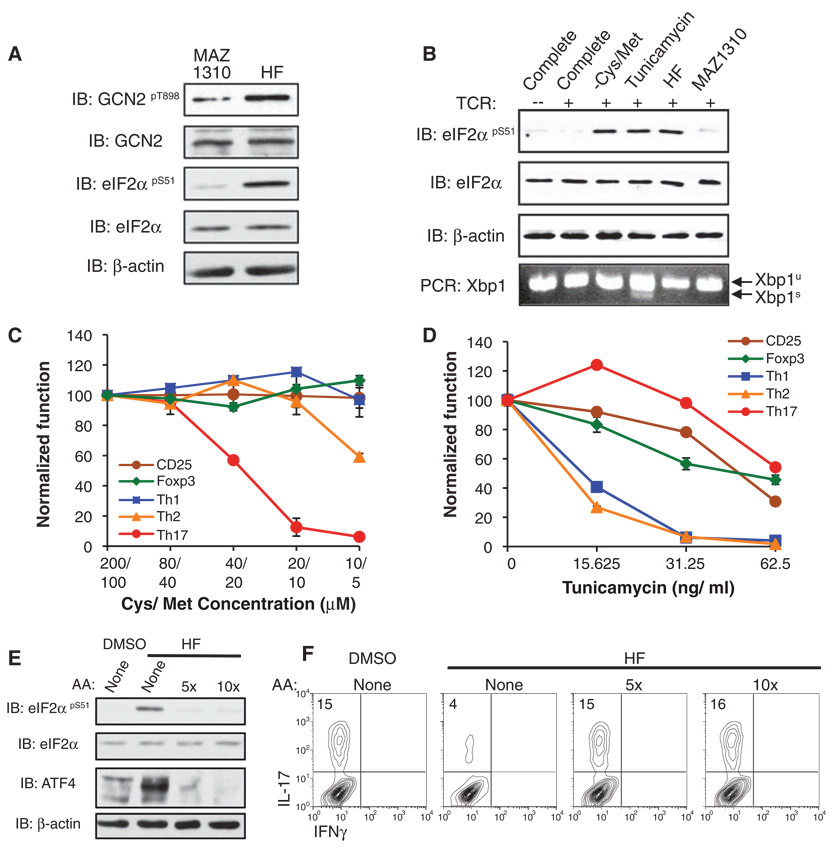
HF treatment rapidly induced eIF2α phosphorylation and ATF4 expression (Fig. 2, C and D) and did so independent of stimulation and polarization conditions (fig. S9B), indicating that AAR activation by HF is not restricted to TH17 cells. Phosphorylation of eIF2α and ATF4 expression can be initiated by multiple upstream kinases through a shared signaling cassette termed the integrated stress response (fig. S8) (15). GCN2 autophosphorylation was activated in response to HF treatment (Fig. 3A), indicating that HF activates the AAR pathway. Furthermore, neither HF nor depletion of cysteine and methionine (Cys/Met) activated IRE-1–dependent splicing of the transcription factor Xbp-1, a response characteristic of endoplasmic reticulum (ER) stress (18), in contrast to a known inducer of ER stress, tunicamycin (Fig. 3B) (17). Microarray analyses indicated that HF does not activate gene expression associated with ER or oxidative stress (e.g., GRP78/BiP, calreticulin) (table S1) (18, 19). HF also activated the AAR in fibroblasts and epithelial cells (fig. S10) (20), establishing that AAR activation by HF is not limited to T cells. HF activated the AAR without concomitantly altering signaling through the nutrient sensor mTOR as determined by p70-S6K phosphorylation (fig. S11) (21).
Fig. 3.
Regulation of TH17 differentiation by amino acids. (A) Lysates from TCR-activated T cells treated with MAZ1310 or HF for 4 hours were analyzed by Western blotting. (B) Unstimulated or activated T cells were cultured in complete medium, medium lacking Cys/Met (-Cys/Met), or complete medium containing tunicamycin, HF, or MAZ1310 and lysed after 4 hours for Western blot analysis. Xbp-1 splicing assay was performed on isolated cDNA (6). (C) Activated T cells were cultured without cytokines or polarized to TH1, TH2, iTreg, or TH17 cells. Titrating concentrations of Cys/Met are indicated. The percentages of TH1, TH2, and TH17 cells, and those expressing CD25 or Foxp3, were determined as in Fig. 1A and are displayed as mean values ± SD, normalized to cells cultured in complete medium. (D) T cells differentiated as in (C) were treated with tunicamycin as indicated. T cell activation and differentiation was determined as in (C). (E) Activated T cells were treated with DMSO or HF. Some cultures were supplemented with 5× or 10× amino acids (6), and lysates were analyzed after 4 hours by Western blotting. (F) T cells activated in TH17 polarizing conditions were treated with DMSO or HF plus amino acids, as indicated, and stained for intracellular cytokine expression. These data represent three experiments.
We next tested whether AAR activation induced by selective amino acid deprivation (fig. S12, A and B) could mimic the effects of HF on T cell differentiation. Decreasing Cys/Met concentrations in differentiating T cell cultures impaired TH17 cell development in a dose-dependent manner without affecting CD25 up-regulation or the differentiation of TH1, TH2, or iTreg cells (Fig. 3C and fig. S12C). As with HF, amino acid restriction reduced TH17 differentiation, independent of cell survival or proliferation (fig. S12D). Deprivation of leucine, or treatment with l-tryptophanol, an inhibitor of tryptophanyl-tRNA charging (fig. S12E), also diminished TH17 differentiation, indicating that AAR activation restricts TH17 differentiation in a manner not specific to individual amino acids. In contrast to amino acid deprivation, tunicamycin-induced ER stress suppressed the differentiation of both TH1 and TH2 cells at doses that did not affect TH17 differentiation (Fig. 3D and fig. S12C). To establish whether AAR activation is required for the inhibition of TH17 differentiation by HF, we added excess free amino acids to abrogate activation of the AAR in HF-treated cells. Under these conditions HF failed to induce eIF2α phosphorylation, up-regulate ATF4 protein expression, or prevent TH17 differentiation (Fig. 3, E and F). Thus, activation of the AAR by HF is both necessary and sufficient for the repression of TH17 differentiation.
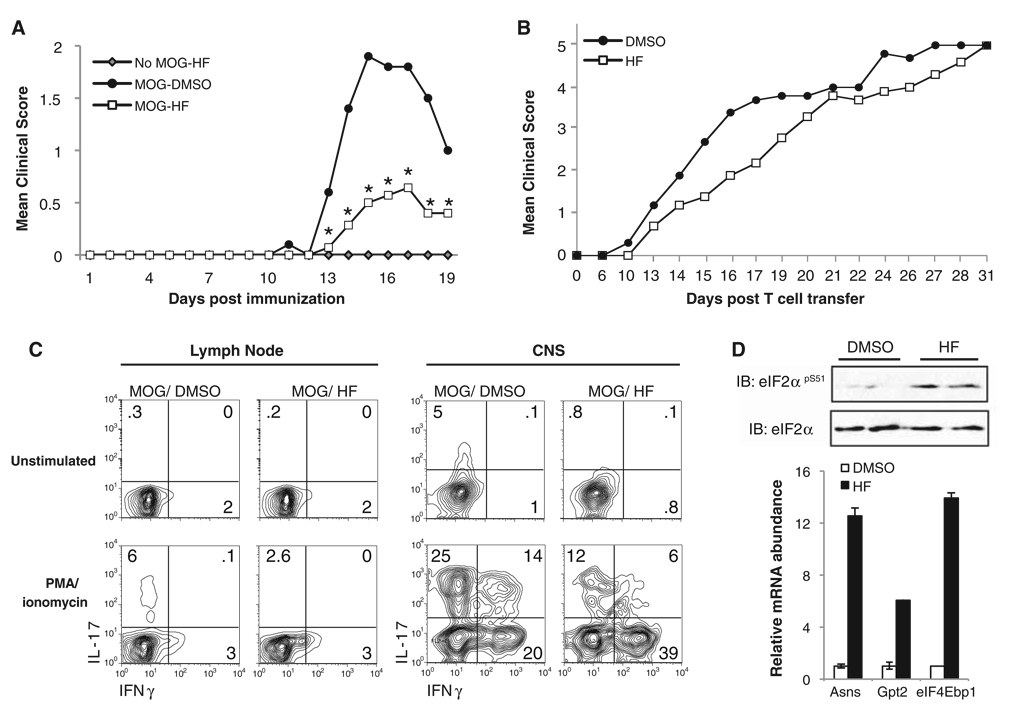
Selective inhibition of TH17 differentiation in vivo may have broad therapeutic implications. We used two distinct models of experimental autoimmune encephalomyelitis (EAE) to investigate whether systemic HF treatment could inhibit TH17 differentiation and associated autoimmune inflammation in mice. Adjuvant-driven EAE was induced by immunizing wild-type mice with the immunodominant myelin antigen MOG33-55 emulsified in complete Freund’s adjuvant (CFA) and was associated with infiltration of both TH17- and IFN-γ–expressing TH1 cells into central nervous system (CNS) tissue (fig. S13A). This model of EAE is sensitive to modulation of TH17 responses (9, 22, 23). Low-dose HF treatment significantly reduced both the frequency and severity of adjuvant-driven EAE (Fig. 4A and fig. S13B). In contrast, passive EAE was initiated by transferring myelin proteolipid protein (PLP)–specific T cells into recombination activating gene-2−/− (Rag2−/−) hosts (24), and this disease was associated with a predominant TH1 response in the CNS (fig. S13C). HF-treated recipients developed passive EAE similar to control animals (Fig. 4B), indicating that HF specifically blunts autoimmune inflammation associated with TH17 differentiation without inducing global immunosuppression. Both TH1 and TH17 cells are capable of inducing EAE upon adoptive transfer (25), but only adjuvant-driven, and not passive, EAE initiates a substantial TH17 response. Thus, the protective specificity of HF in EAE is probably due to selective inhibition of TH17, but not TH1 differentiation.
Fig. 4.
HF inhibits TH17-associated autoimmune inflammation in vivo. (A) Wild-type mice were immunized with phosphate-buffered saline or MOG33-55 emulsified in CFA and treated daily with DMSO or HF, and disease was monitored (6). (B) After PLP-specific T cell transfer, recipients were treated daily with DMSO or HF, and disease was monitored. Mean EAE scores are displayed. (C) (Left) Paraaortic lymph nodes from MOG-immunized mice treated with DMSO or HF were harvested on day 6. Mononuclear cells were cultured without (top) or with (bottom) PMA and ionomycin and stained for intracellular cytokines. (Right) Mononuclear cells isolated from CNS tissue of DMSO-treated (score = 2) or HF-treated (score = 0) mice were cultured and stained for intracellular cytokines as above. TCRβ+ CD4+ cells were gated on for analyses. (D) (Top) Splenocyte lysates were prepared from DMSO- or HF-treated mice and analyzed by Western blotting. (Bottom) AAR-associated gene expression (Asns, Gpt2, eIF4Ebp1) was analyzed by QPCR on cDNA from spleens of mice treated with DMSO or HF. Transcript levels were normalized to Hprt and are displayed as mean expression ± SD (error bars) from triplicate samples. EAE data are cumulative from three independent experiments; other data represent two to three similar experiments.
Protection from adjuvant-driven EAE by HF was associated with fewer TH17 cells, both in the periphery before disease onset and the CNS during active disease (Fig. 4C). TH17 cells initiate mononuclear cell recruitment into the CNS during adjuvant-driven EAE (22, 26). HF treatment reduced T cell infiltration into the CNS (fig. S14) but did not change the proportion of TH1 cells in the periphery or CNS (fig. S14). Moreover, splenocytes from HF-injected mice displayed increased eIF2α phosphorylation and expression of AAR-associated transcripts (Fig. 4D). Thus, HF treatment activates the AAR and selectively impairs both TH17 differentiation and autoimmune inflammation in vivo.
In conclusion, HF selectively inhibits TH17 differentiation and associated autoimmune inflammation via the cytoprotective AAR pathway. Endogenous amino acid restriction has been suggested to regulate inflammation (27). Indoleamine 2,3-dioxygenase (IDO), an IFN-γ–induced enzyme expressed by dendritic cells (DCs), metabolizes tryptophan, causes local amino acid depletion, and inhibits the proliferation of bystander T cells via the AAR (28). Local IDO expression also has been reported to expand, convert, and directly activate Foxp3+ Tregs (29, 30). Thus, AAR activation may protect against pathophysiologic inflammation by enforcing the tolerogenic effects of IDO-expressing DCs and concomitantly blunting TH17 differentiation. Although the mechanism by which AAR activation constrains TH17 differentiation remains unclear, these results highlight a previously unknown link between the AAR pathway and TH17-mediated immune-pathology.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/324/5932/1334/DC1
Materials and Methods
Figs. S1 to S14
Tables S1 and S2
References
References and Notes
- 1.Korn T, Bettelli E, Oukka M, Kuchroo VK. Annu. Rev. Immunol. 2009;27:485. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 2.Dong C. Nat. Rev. Immunol. 2008;8:337. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 3.Pines M, Nagler A. Gen. Pharmacol. 1998;30:445. doi: 10.1016/s0306-3623(97)00307-8. [DOI] [PubMed] [Google Scholar]
- 4.Gnainsky Y, et al. Cell Tissue Res. 2007;328:153. doi: 10.1007/s00441-006-0330-1. [DOI] [PubMed] [Google Scholar]
- 5.Pines M, Snyder D, Yarkoni S, Nagler A. Biol. Blood Marrow Transplant. 2003;9:417. doi: 10.1016/s1083-8791(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 6.Materials and methods are available as supporting material on Science Online.
- 7.Leiba M, et al. J. Leukocyte Biol. 2006;80:399. doi: 10.1189/jlb.0705409. [DOI] [PubMed] [Google Scholar]
- 8.Park H, et al. Nat. Immunol. 2005;6:1133. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XO, et al. Immunity. 2008;28:29. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov II, et al. Cell. 2006;126:1121. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, et al. Nat. Immunol. 2007;8:967. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 12.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Annu. Rev. Immunol. 2006;24:99. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 13.Yang XO, et al. J. Biol. Chem. 2007;282:9358. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 14.Deval C, et al. FEBS J. 2009;276:707. doi: 10.1111/j.1742-4658.2008.06818.x. [DOI] [PubMed] [Google Scholar]
- 15.Wek RC, Jiang HY, Anthony TG. Biochem. Soc. Trans. 2006;34:7. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 16.Harding HP, et al. Mol. Cell. 2000;6:1099. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 17.Harding HP, et al. Mol. Cell. 2003;11:619. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 18.Lee AH, Iwakoshi NN, Glimcher LH. Mol. Cell. Biol. 2003;23:7448. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ron D, Walter P. Nat. Rev. Mol. Cell Biol. 2007;8:519. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 20.Kamberov Y. thesis. Harvard University; 2008. [Google Scholar]
- 21.Fingar DC, Blenisc J. Oncogene. 2004;23:3151. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 22.Serada S, et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9041. doi: 10.1073/pnas.0802218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langrish CL, et al. J. Exp. Med. 2005;201:233. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldner H, Collins M, Kuchroo VK. J. Clin. Invest. 2004;113:990. doi: 10.1172/JCI19388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroenke MA, Carlson TJ, Andjelkovic AV, Segal BM. J. Exp. Med. 2008;205:1535. doi: 10.1084/jem.20080159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reboldi A, et al. Nat. Immunol. 2009;10:514. doi: 10.1038/ni.1716. [DOI] [PubMed] [Google Scholar]
- 27.Mellor AL, Munn DH. Nat. Rev. Immunol. 2008;8:74. doi: 10.1038/nri2233. [DOI] [PubMed] [Google Scholar]
- 28.Munn DH, et al. Immunity. 2005;22:633. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Park MJ, et al. Arthritis Res. Ther. 2008;10:R11. doi: 10.1186/ar2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puccetti P, Grohmann U. Nat. Rev. Immunol. 2007;7:817. doi: 10.1038/nri2163. [DOI] [PubMed] [Google Scholar]
- 31.D. Littman and A. Rudensky provided reagents for this work, and K. Otipoby prepared recombinant TAT-Cre. We thank J. Hill and K. Leatherbee for technical assistance with microarray data analysis and members of the Rao lab for critical discussion. This work was supported by grants from NIH (to A.R., D.U., and M.W.) and the Juvenile Diabetes Research Foundation (to A.R.). M.S.S. was supported by the Irvington Institute fellowship program of the Cancer Research Institute. D.P.C. was supported by a fellowship from the Portuguese Foundation for Science and Technology. S.K. was supported by an NIH training grant (T32). Further support was provided by a P01 grant to K Rajewsky. M.S.S., A.R., T.K., and M.W. have filed patent applications concerning the use of HF and its derivatives to inhibit TH17 cell differentiation. Array data can be found at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15624 with the accession number of GSE15624.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.