Abstract
The incidences of alveolar/bronchiolar adenomas and carcinomas in cumene-treated B6C3F1 mice were significantly greater than those of the controls. We evaluated these lung neoplasms for point mutations in the K-ras and p53 genes that are often mutated in humans. K-ras and p53 mutations were detected by cycle sequencing of PCR-amplified DNA isolated from paraffin-embedded neoplasms. K-ras mutations were detected in 87 % cumene-induced lung neoplasms, and the predominant mutations were exon 1 codon 12 G to T transversions and exon 2 codon 61 A to G transitions. P53 protein expression was detected by immunohistochemistry in 56 % cumene-induced neoplasms and mutations were detected in 52 % neoplasms. The predominant mutations were exon 5, codon 155 G to A transitions and codon 133 C to T transitions. No p53 mutation and one of 7 (14 %) K-ras mutation was detected in spontaneous neoplasms. Cumene-induced lung carcinomas showed loss of heterozygosity (LOH) on chromosome 4 near the p16 gene (13 %) and on chromosome 6 near the K-ras gene (12 %). No LOH was observed in spontaneous carcinomas or normal lung tissues examined. The pattern of mutations identified in the lung tumors suggests that DNA damage and genomic instability may be contributing factors to the mutation profile and development of lung cancer in mice exposed to cumene.
Keywords: K-ras, p53, cumene, lung, mouse
Introduction
Lung cancer is the most frequently diagnosed cancer in the world and the most common cause of the cancer mortality worldwide. The high mortality is largely due to the late stage of diagnosis and the poor response to therapy. The need to develop better diagnostic techniques and therapies is urgent. Mouse models have been utilized for studying carcinogenesis of human lung cancers and many of the major genetic alterations detected in human lung cancers have also been identified in mouse lung tumors.
Mouse alveolar/bronchiolar adenomas and carcinomas, which are common spontaneous and chemical induced lung tumors in mice, are similar to human adenocarcinomas in histomorphology and molecular characteristics including activation of the K-ras gene (25, 27). The patterns of mutations in cancer genes, such as ras and p53, have been found to aid in the understanding of tumorigenesis in rodents and humans (7, 21, 24, 28). For example, in some neoplasms, the profile of activating mutations in ras genes or inactivating mutations in the p53 gene are specific for particular chemicals and differ from those detected in spontaneous neoplasms (38, 39).
Cumene, or isopropylbenzene, is a constituent of crude oil used primarily for the production of phenol and acetone. It is a good solvent for fats and resins and has been suggested as a replacement for benzene in many industrial applications. The annual production of cumene in the US is high and increasing with 4.49 billion pounds produced in 1993 (9). Being a major commodity chemical, there is a high potential for many workers to be exposed to cumene. The most probable route of human exposure is inhalation of contaminated air (16) from cumene evaporated into the environment during production and processing from petroleum refining, combustion of petroleum products, and use of a variety of products containing cumene.
National Toxicology Program (NTP) 2-year whole-body inhalation studies revealed that treatment of B6C3F1 mice with cumene significantly increased the incidence of alveolar/bronchiolar adenomas and carcinomas in all groups of exposed males and females. Cumene is not genotoxic which was demonstrated by several studies involving bacterial and mammalian cells in culture and in in vivo studies involving mice and rats (10, 26, 43). In vitro cell transformation assays using BALB/3T3 mouse embryo cells and unscheduled DNA synthesis assays using rat primary hepatocytes yielded conflicting results regarding a cumene effect that were not reproducible. Cumene was weakly positive with no clear dose response for the induction of micronuclei in rat bone marrow at doses ranging from 78 to 2500 mg/kg intraperitoneally (26).
The goal of this study was to evaluate both spontaneous and cumene-induced lung neoplasms for mutations in the K-ras and p53 genes, which are considered important in the pathogenesis of human lung cancer. Another goal was to identify chemical specific mutations that might serve as biomarkers of occupational exposure with possible relevance to human health.
Materials and Methods
Lung Neoplasms
Male and female B6C3F1 mice were exposed to 0, 125 (females only), 250, 500, or 1000 (males only) ppm cumene (50 animals each group) by whole-body inhalation, 6hr/day, 5 days/week for 2 years (NTP, Cas Number 98-82-8). Husbandry and experimental procedures were in compliance with the requirements set forth by the Public Health Service’s Guide for the Care and Use of Laboratory Animals. Incidences of cumene-induced lung tumors were evaluated statistically using the poly-3 test. At necropsy, tissues were fixed in 10% neutral-buffered formalin, routinely processed, embedded in paraffin, sectioned to a thickness of 5 um, and stained with hematoxylin and eosin. Subsequently, 5 unstained serial sections, 10 um thick, were prepared from paraffin blocks containing alveolar/bronchiolar adenomas or carcinomas. In order to isolate adequate amounts of DNA, neoplasms greater than 1 mm in diameter were identified for analysis. Fifty- two cumene-induced alveolar/bronchiolar neoplasms (6 adenomas and 46 carcinomas), 7 spontaneously occurring carcinomas, and 6 normal lung tissues were evaluated for K-ras mutations in exons 1 and 2 (codons 12, 13 and 61) and p53 mutations in exons 5–8.
DNA Isolation, Amplification, and Cycle Sequencing
DNA was isolated and extraced from paraffin-embedded sections containing lung neoplasms and normal lung tissues and amplified by PCR. Details of the use of nested primers for K-ras and p53 genes have been described previously (20, 37). Positive DNA controls for K-ras and p53 mutations and controls lacking DNA were run with all sets of reactions. PCR products were purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). The purified samples were sequenced utilizing a cycle sequencing kit (US Biochemical, Cleveland, OH), which incorporated [α-33p]-dideoxynucleotide triphosphate (ddNTP) terminators (A, C, G, T) into the sequencing products. Detected mutations were confirmed by repeat analysis, starting from amplification of the original DNA extract.
Immunohistochemistry for p53 Protein
Alveolar/bronchiolar adenomas and carcinomas were examined for p53 protein expression by immunohistochemical analysis, using an avidin-biotin-peroxidase detection system (Vectastain Rabbit Elite Kit, Vector Laboratory, Burlingame, CA). The immunohistochemical staining for expression of mutant p53 protein was performed as previously described (12, 13, 15, 23, 39). A 1:300 dilution of the primary polyclonal rabbit antibody CM-5 (Vector Laboratories), which detects accumulation of the mutant p53 in rodents, was used. Normal rabbit serum (Vector Laboratories) was used as the negative control in place of the primary antibody. Tissue specimens from a p53 transgenic mouse (mutation in codon 135 of p53) served as the positive control.
Loss of Heterozygosity (LOH) Analysis
Microsatellite marker Mts 1 near the p16 tumor suppressor gene on chromosome 4 was used to amplify polymorphic regions between strain C57BL/6 (B) and C3H/He (H) mice (Jackson Laboratories, Bar Harbor, Maine) to identify portions of tumors with LOH at the p16 locus. The Mts 1 forward (5′-GA TTT CTA CGG AAA GCC CTG-3′) and reverse (5′-TAT TGT GCA TTT GTG TGT CTG G-3′) primers were located 2395 and 2172 bp upstream of the translational start site, respectively. PCR amplification was performed on 40 cumene-induced lung neoplasms (36 carcinomas, 4 adenomas) and 7 spontaneous B6C3F1 carcinomas. Controls lacking DNA were included with all amplifications. The PCR products were resolved on 4% NuSieve (FMC BioProducts, Rockland, ME.)- agarose (3:1) to separate B and H alleles.
The inner PCR products from paraffin-embedded plus 8 frozen treated neoplasms (7 carcinomas & 1 adenoma) were labeled by incorporation with [α-33p]-dATP (MP/ICN Biomedicals, rvine, CA), using single stranded conformation polymorphism (SSCP) analysis to distinguish between the H and B alleles on a 0.5X MDE (Cambrex Bio Science, Rockland, ME) gel at 3W for 15 hours at room temperature.
To determine LOH in 50 cumene-induced lung neoplasms (46 carcinomas, 4 adenomas) and 7 spontaneous carcinomas at marker D6MCO12 near the K-ras gene on chromosome 6 in B6C3F1 mice, SSCP analysis was utilized. PCR amplification with [α-33p] - labeled and unlabeled dATP was performed as described previously (39). The MDE gel was electrophoresed at 3W at room temperature for 15 hours to separate the H and B alleles. The gels were dried and exposed to X-ray film overnight.
Results
Mutation Analysis
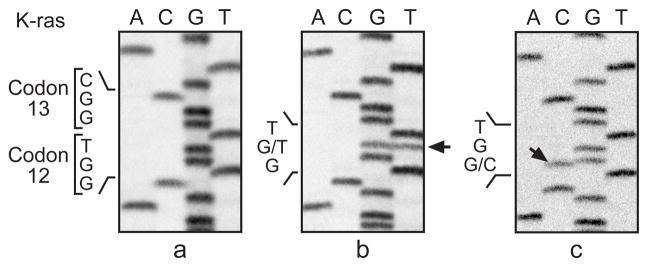
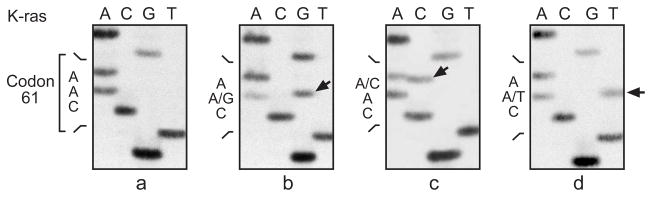
The incidence of alveolar/bronchiolar adenomas and carcinomas in male and female treated groups was significantly greater than those of the controls (Table 1). A higher frequency of K-ras mutations (45/52, 87 %) was observed in the cumene-induced lung neoplasms, as compared to spontaneous lung neoplasms from control animals (historical 33/117, 28 %; concurrent 1/7, 14 %) (Table 2). These mutations were observed in 91% (41/45) of males and 57% (4/7) of females. K-ras mutations were detected in 67% (4/6) of the adenomas (1 in codon 13 and 3 in codon 61) and in 89% (41/46) of the carcinomas. The predominant K-ras mutations were codon 12 GTT (G to T transversions) and codon 61 CGA (A to G transitions). Three codon 12 CGT mutations and one codon 61 CTA mutation were found in the cumene-induced neoplasms, but none were found in spontaneous lung neoplasms (0/124, concurrent controls combined with historical controls) (Table 2; Fig. 1a & 2). There was no significant increase in the incidence of mutations at codon 13 in the cumene-induced lung neoplasms.
Table 1.
Incidences of Cumene-lnduced Lung Tumors in a 2-Year Inhalation Study of B6C3F1 Micea
| Lung Tumors | Males | Females | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 ppm | 250 ppm | 500 ppm | 1000 ppm | 0 ppm | 125 ppm | 250 ppm | 500 ppm | |
| Alveolar/Bronchiolar Adenoma | 13/50 (26%) | 31/50* (62%) | 31/50* (62%) | 29/50* (58%) | 1/50 (2%) | 26/50* (52%) | 36/50* (72%) | 38/50* (76%) |
| Alveolar/Bronchiolar Carcinoma | 9/50 (18%) | 19/50* (38%) | 32/50* (64%) | 33/50* (66%) | 3/50 (6%) | 16/50* (32%) | 20/50* (40%) | 34/50* (68%) |
| Alveolar/Bronchiolar Adenoma or Carcinoma | 19/50 (38%) | 38/50* (76%) | 42/50* (84%) | 43/50* (86%) | 4/50 (8%) | 31/50* (62%) | 42/50* (84%) | 46/50* (92%) |
Male and female B6C3F1 mice were exposed to 0, 125 (females only), 250, 500, or 1000 (males only) ppm cumene by whole-body inhalation, 6hr./day, 5 days/week for 2 years (NTP TR 542, 2007).
Significantly different (p<0.001) from the control group by the poly-3 test.
Table 2.
K-ras Mutations in Lung Neoplasms of B6C3F1 Mice in a 2-Year Inhalation Study of Cumene
| Treatment ppm | Activate K-ras(%) | Codon12(GGT) (GAT) (TGT) (GTT) (CGT) | Codon13(GGC) (CGC) | Codon 61 (CAA) (CGA) (CAT) (CAC) (CTA) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | ||||||||||
| Historicala | 33/117 (28) | 14 | 5 | 1 | 0 | 6 | 2 | 4 | 1 | 0 |
| Concurrent | 1/7 (14) | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Cumeneb | 45/52 (87) | 6 | 5 | 11 | 3 | 4 | 13 | 0 | 2 | 1 |
| 125 | 1/4 (25) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 250 | 10/13 (77) | 0 | 0 | 1 | 2 | 0 | 5 | 0 | 2 | 0 |
| 500 | 17/18 (94) | 4 | 1 | 6 | 0 | 2 | 4 | 0 | 0 | 0 |
| 1000 | 17/17 (100) | 2 | 3 | 4 | 1 | 2 | 4 | 0 | 0 | 1 |
historical spontaneous lung neoplasms from control B6C3F1 mice (Hong et al., 2007).
Male and female B6C3F1 mice were exposed to 0, 125 (females only), 250, 500, or 1000 (males only) ppm cumene by whole-body inhalation, 6hr./day, 5 days/week for 2 years.
Fig. 1.
Fig. 1a. Identification of K-ras mutations in codon 12 of lung neoplasms from B6C3F1 mice exposed to cumene by whole-body inhalation for 2 years. Sequencing panels are from left to right: (a) normal K-ras codon 12 sequence GGT: (b) mutated codon 12 sequence GTT from animal No. 819 carcinoma; (c) mutated codon 12 sequence CGT from animal No. 808 carcinoma. Arrows point to mutant bands.
Fig. 1b. SSCP analysis at marker D6MCO12 to assess LOH on chromosome 6 near K-ras gene in cumene-induced carcinomas. Lanes 3–5 represent normal (N) lungs from B6C3F1 (F1), C3H (H) and C57BL/6 (B) inbred mouse strains. Lane 2 (animal No. 644 carcinoma) showed loss of C3H (H) allele: lane 6 (animal No. 846 carcinoma) showed loss of C57 (B) allele; lane 1 (animal No. 828 carcinoma) appeared normal with both alleles present.
Fig. 2.
Identification of K-ras codon 61 mutations in cumene-induced lung neoplasms from B6C3F1 mice by cycle sequencing of amplified exon 2. Sequencing panels are from left to right: (a) normal codon 61 sequence CAA; (b) mutated sequence CGA from animal No. 604 adenoma; (c) mutated sequence CAC from animal No. 430 carcinoma; (d) mutated sequence CTA from animal No. 824 carcinoma. Arrows point to mutant bands.
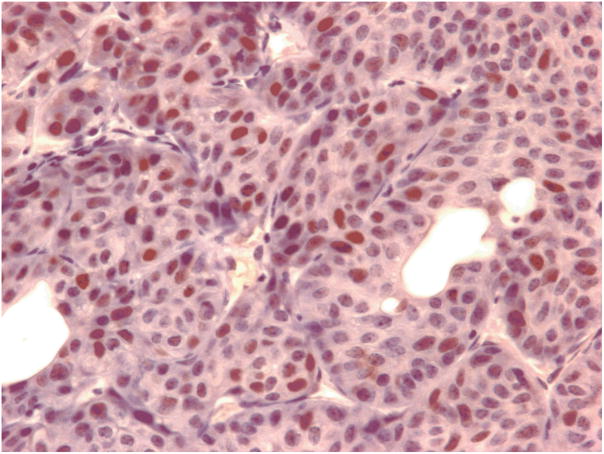
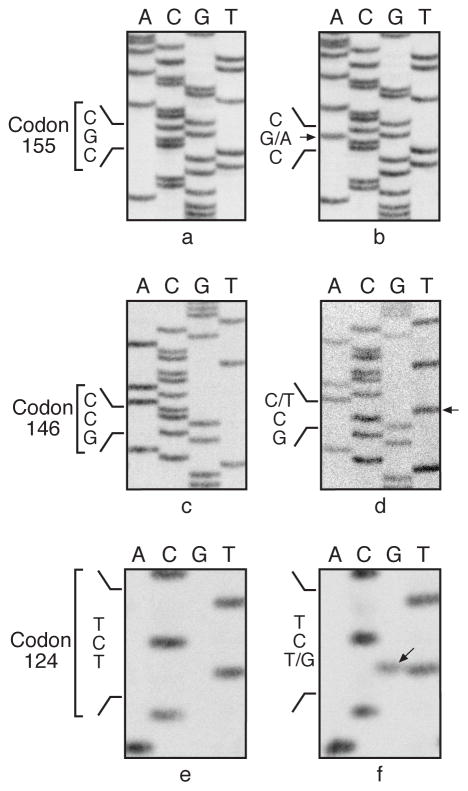
p53 protein expression was detected by immunohistochemistry in 29/52 (56%) of cumene-induced lung neoplasms (Fig. 3), and mutations were identified in 27/52 (52%) of the neoplasms. p53 protein expression and gene mutation were not detected in the 7 spontaneous carcinomas or in the 6 normal lung tissues except one spontaneous carcinoma with positive p53 protein expression (Table 3). The predominant p53 mutations were identified in exon 5 (24/27, 89 %) and in exon 7 (3/27, 11%) (Table 3; Fig. 4). No mutation was detected at exons 6 and 8 in the samples examined.
Fig. 3.
Alveolar/bronchiolar carcinoma from a male mouse exposed to 1000 ppm cumene. Note p53 protein expression (brown chromogen) in the nucleus of malignant epithelial cell, 400x.
Table 3.
p53 Mutations in Lung Neoplasms of B6C3F1 Mice in a 2-Year Inhalation Study of Cumenea
| Treatment ppm | Activate p53b(%) | IHC Positive No. (%) | Exon5 | Exon7 |
|---|---|---|---|---|
| Control | 0/7 (0) | 1/7 (14) | 0 | 0 |
| Cumene | 27/52 (52) | 29/52 (56) | 24 | 3 |
| 125 | 0/4 (0) | 1/4 (25) | 0 | 0 |
| 250 | 5/13 (38) | 6/13 (46) | 4 | 1 |
| 500 | 11/18 (38) | 8/18 (44) | 10 | 1 |
| 1000 | 11/17 (65) | 14/17 (82) | 10 | 1 |
Male and female B6C3F1 mice were exposed to 0, 125 (females only), 250, 500, or 1000 (males only) ppm cumene by whole-body inhalation, 6hr./day, 5 days/week for 2 years.
No mutation detected for p53 at exons 6 & 8 in the samples examined.
Fig. 4.
Examples of sequencing p53 exon 5 mutations in lung neoplasms from B6C3F1 mice exposed to cumene. Sequencing panels a, c and e are normal p53 sequences and panels b, d and f are mutated sequences at indicated codons from animal No. 402 carcinoma, animal No. 840 carcinoma, and animal No. 601 carcinoma. The wild-type allele was not detected in the sequencing reaction shown in panel (d).
The p53 mutations were observed in 58% (26/45) of males and 14% (1/7) of females. Mutations were detected in 50% (3/6) of the adenomas and 52% (24/46) of the carcinomas. There was a dose-dependent increase in incidence of K-ras and p53 mutations, however, a similar mutation spectrum of both mutations was detected in cumene-induced neoplasms regardless of whether the neoplasms were adenomas or carcinomas.
LOH Analysis
LOH on chromosome 6 near K-ras gene was observed in 6/50 (12 %; 4 –H & 2 –B) treated carcinomas, 0/6 of adenomas, and 0/7 of the spontaneous carcinomas examined (Fig. 1b).
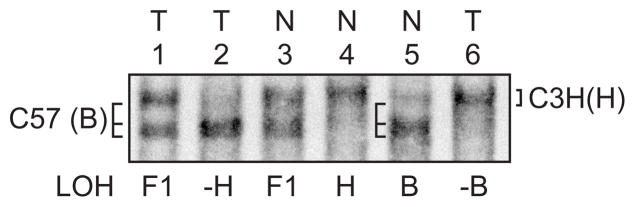
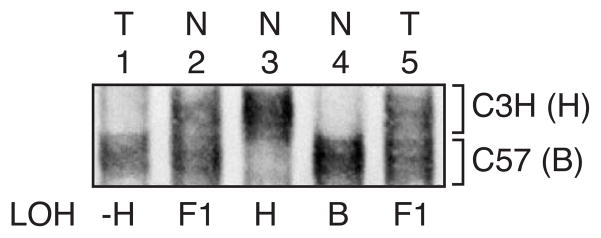
LOH of the C3H/He allele (H) was observed in 5/40 (13 %) cumene-induced carcinomas, 1/6 of adenomas, and 0/7 of the spontaneous carcinomas examined at the microsatellite marker on chromosome 4 near the p16 gene (Fig. 5).
Fig. 5.
SSCP analysis at Mts 1 to assess LOH on chromosome 4 near p16 tumor suppressor gene in cumene-induced carcinomas. Lanes 2–4 represent normal (N) lungs from B6C3F1 (F1), C3H (H) and C57 (B) inbred mouse strains. Lane 1 (animal No. 324 carcinoma) showed loss of the C3H (H) allele; lane 5 (animal No. 834 carcinoma) appeared normal with both alleles present.
Discussion
There was a high frequency (87%) of K-ras mutations in cumene-induced alveolar/bronchiolar neoplasms compared to that in spontaneous alveolar/bronchiolar neoplasms from untreated B6C3F1 mice (28% historical database; 14% concurrent controls). The predominant mutations were K-ras codon 12 G to T transversions (GGT to GTT, 21%) and codon 61 A to G transitions (CAA to CGA, 25%), which clearly differed from those identified in untreated control mice (0.008% & 2%, respectively). Point mutations at codon 12 of the K-ras gene are activating mutations, rendering ras insensitive to the down-regulatory action of GTPase activating proteins, thereby locking the protein in the active state and promoting cellular transformation (2).
The high frequency and pattern of K-ras mutations in mouse lung tumors may directly depend on the nature of the chemical carcinogen or its metabolites. In our study, the development of lung neoplasms in B6C3F1 mice exposed to cumene may involve multiple carcinogenic processes including direct DNA damage and/or indirect DNA damage. Metabolites of cumene may have caused DNA adducts and subsequent point mutations. Side-chain oxidation of cumene (isopropylbenzene) is rapid and extensive and occurs in both hepatic and extra-hepatic tissues including the lung (35), with the secondary alcohol 2-phenyl-2-propanol being the principal metabolite in rats (30, 43) and humans (25, 43). The C-isopropyl bonds are readily cleaved, and the remaining electrophilic carbon moiety may form DNA adducts and cause subsequent DNA damage. Previous studies showed that the related benzene is carcinogenic and genotoxic (1, 6, 40, 44).
Alternatively indirect damage from oxidative stress may have contributed to the mutations. G to T transversions are commonly detected DNA base changes associated with active oxygen species and are consistent with 8-OH-G adducts produced during oxidative damage to DNA (17, 18, 36, 42). In other studies, exposure of B6C3F1 mice to ozone or vanadium pentoxide is thought to result in the generation of hydroxyl radicals which induce a G to T transversion at codon 12 of K-ras gene (3, 37). Interestingly, G to T transversion in K-ras codon 12 is the most common mutation detected in human adenocarcinomas (32). In human lung tumors, K-ras mutations appear to correlate with DNA adducts of benzo(a)pyrene and are associated with smoking (34). It is possible that smoking in combination with cumene exposure in humans may have an additive effect on K-ras mutations.
While K-ras appears to play a critical role in mouse and human lung adenocarcinoma formation, its function is complex. Zhang et al. (46) proposed that wild-type K-ras may be a mouse lung tumor suppressor gene. Further LOH analysis in cumene-induced lung neoplasms demonstrsted allelic loss on chromosome 6 near the K-ras gene in 6 carcinomas and on chromosome 4 near p16 tumor suppressor gene in 5 carcinomas. Allele loss of p16 has been detected in human cancers (4, 19) and mouse lung tumors (29).
A high frequency (52%) of p53 mutations were detected in cumene-induced alveolar/bronchiolar neoplasms which were correlated with increased p53 protein expression (56%) by immunohistochemistry. The presence of p53 protein expression without p53 gene mutation could be due to mutations outside exons 5–8 or possibly due to alterations of other proteins downstream of p53 (5). The predominance of cumene-induced alveolar/bronchiolar neoplasms that contain p53 mutations may provide a selective advantage for unregulated growth and the avoidance of apoptosis (5, 8, 28, 33). A study of aflatoxin-B1 (AFB1)-induced mouse lung tumors found a high proportion (>70%) of tumors with p53 accumulation and mutations (41), and lung tumors of mice exposed transplacentally to AZT also had a high proportion (84 %) of p53 mutations. (14). Other studies such as methylene chloride-induced mouse lung tumors (11), ozone-induced, and vanadium pentoxide-induced mouse lung tumors (3, 37) exhibited no or low frequency of p53 mutations. Unlike the mostly random mutation pattern for the AFB1-induced tumors (41), the cumene-induced tumors displayed specific p53 mutations. P53 mutations were only observed in exon 5 (24/27, 89%) and exon 7 (3/27, 11%). The mutations observed in the p53 gene in cumene-exposed mice clearly imply this genetic event is related to chemical exposure, since these mutations were not detected in spontaneous tumors.
In conclusion, the patterns of K-ras and p53 mutations identified in the cumene-induced lung tumors suggest that DNA damage and genomic instability may be the contributing factors to the mutation profile and development of lung cancer in these mice. The molecular alterations identified in the cumene-induced lung neoplasms appear to affect the same pathways as those reported in human lung cancer, suggesting that the response in the mouse may be of relevance to humans. Futher research of the cumene mouse lung tumors by microarray analysis underscore the complexity of lung cancer where in addition to genetic factors, signal transduction pathways and epigenetic mechanisms are also contributing factors to the carcinogenesis process (45).
Acknowledgments
The authors thank Dr. Darlene Dixon and Dr. Mark Cesta for their critical review of this manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
References
- 1.Abemethy DJ, Kleymenova EV, Rose J, Recio L, Faiola B. Human CD34+ hematopoietic progenitor cells are sensitive targets for toxicity induced by 1,4- benzoquinone. Toxicol Sci. 2004;79:82–89. doi: 10.1093/toxsci/kfh095. [DOI] [PubMed] [Google Scholar]
- 2.Chad AE, Geoff C. The importance of being K-ras. Cellilar Signalling. 2000;12:425–434. doi: 10.1016/s0898-6568(00)00084-x. [DOI] [PubMed] [Google Scholar]
- 3.Devereux TR, Holliday W, Anna C, Ress N, Roycroft J, Sills RC. Map kinase activation correlates with K-ras mutation and loss of heterozygosity on chromosome 6 in alveolar bronchiolar carcinomas from B6C3F1 mice exposed to vanadium pentoxide for 2 years. Carcinogenesis. 2002;23:1737–1743. doi: 10.1093/carcin/23.10.1737. [DOI] [PubMed] [Google Scholar]
- 4.de Vos S, Miller CW, Takeuchi S, Gombart AF, Cho SK, Koeffler HP. Alterations of CDKN2 (p16) in non-small cell lung cancer. Genes Chromosomes cancer. 1995;14:164–170. doi: 10.1002/gcc.2870140303. [DOI] [PubMed] [Google Scholar]
- 5.Greenblatt MS, Bonnett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 6.Gut I, Nedelcheva V, Soucek P, Stopka P, Tichavska B. Cytochromes p450 in benzene metabolism and involvement of their metabolites and reactive oxygen species in toxicity. Environ Health Perspect. 1996;104(Suppl 6):1211–1218. doi: 10.1289/ehp.961041211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris CC. P53 : at the crossroads of molecular carcinogenesis and risk assessment. Science. 1993;262:1980–1981. doi: 10.1126/science.8266092. [DOI] [PubMed] [Google Scholar]
- 8.Harris CC. P53 tumor suppressor gene: from the basic research laboratory to the clinic—an abridged historical perspective. Carcinogenesis. 1996;17:1187–1198. doi: 10.1093/carcin/17.6.1187. [DOI] [PubMed] [Google Scholar]
- 9.Hazardous Substance Database. Cumene Chemistry and Engineering News. 15. Vol. 72. Toxicology & Environmental Health Information Program, National Library of Medicine; Bethesda, MD: 2000. p. 11. 4/11/94. [Google Scholar]
- 10.Hazardous Substance Data Bank (HSDB) Cumene. Toxicology Information Program, National Library of Medicine; Bethesda, MD: 2003. [Google Scholar]
- 11.Hegi ME, Soderkvist P, Foley JF, Schoonhoven R, Swenberg JA, Kari F, Maronpot R, Anderson MW, Wiseman RW. Characterization of p53 mutations in methylene chloride-induced lung tumors from B6C3F1 mice. Carcinogenesis. 1993;14:803–810. doi: 10.1093/carcin/14.5.803. [DOI] [PubMed] [Google Scholar]
- 12.Hong HL, Devereux TR, Melnick RL, Boorman GA, Sills RC. Mutations of ras protooncogenes and p53 tumor suppressor gene in cardiac hemangiosarcomas from B6C3F1 mice exposed to 1,3-butadiene for 2 years. Toxicol Pathol. 2000;28:529–534. doi: 10.1177/019262330002800404. [DOI] [PubMed] [Google Scholar]
- 13.Hong HL, Ton tv, Devereux TR, Moomaw C, Clayton N, Chan P, Dunnick JK, Sills RC. Chemical-specific alterations in ras, p53, and β-catenin Genes in hemangiosarcomas from B6C3F1 mice exposed to o-nitrotoluene or riddelliine for 2 years. Toxicol Appl Pharmacol. 2003;191:227–234. doi: 10.1016/s0041-008x(03)00165-0. [DOI] [PubMed] [Google Scholar]
- 14.Hong HL, Dunnick J, Herbert R, Devereux TR, Kim Y, Sills RC. Genetic alterations in K-ras and p53 cancer genes in lung neoplasms from Swiss (CD-1) male mice exposed transplacentally to AZT. Environ Mol Mutagen. 2007;48:299–306. doi: 10.1002/em.20197. [DOI] [PubMed] [Google Scholar]
- 15.Hsu SM, Raine l, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 16.Jackson L. Test Rule Support Document Cumene. Syracuse Research Corporation; Syracuse, NY: 1985. p. 170. SRC-TR-85–098. [Google Scholar]
- 17.Janssen YMW, Van Houten B, Borm PJA, Mossman BT. Cell and tissue responses to oxidative damage. Lab Invest. 1993;69:261–274. [PubMed] [Google Scholar]
- 18.Kino K, Sugiyama H. UVR-induced G-C to C-G transversions from oxidative DNA damage. Mutation Res. 2005;571:33–42. doi: 10.1016/j.mrfmmm.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Kratzke RA, Greatens TM, Rubins JB, Maddaus MA, Niewoehner DE, Niehans GA, Geradts J. Rb and p16INK4a expression in resected non- small cell lung tumors. Cancer Res. 1996;56:3415–3420. [PubMed] [Google Scholar]
- 20.Lambertini L, Surin k, Ton TT, Clayton N, Dunnick J, Kim Y, Hong hl, Devereux TR, Sills RC. Analysis of p53 tumor suppressor gene, H- ras protooncogene and profilerating cell nuclear antigen (PCNA) in squamous cell carcinomas of HRA/SKh mice following exposure to 8-methoxypsoralen (8-MOP) and UVA radiation (PUVA therapy) Toxicol Pathol. 2005;33:292–299. doi: 10.1080/019262390908380. [DOI] [PubMed] [Google Scholar]
- 21.Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Taniere P, Brennan P, Boffetta P, Zaridze DG, Hainaut P. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and currentsmokers. Cancer Res. 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 22.Lee EW. Cumene Section 1.5. In: Snyder R, editor. Ethel Browning’s Toxicity and Metabolism of Industrial Solvents. 2. I. Elsevier Science Publishers; New York, NY: 1987. Hydrocarbons. [Google Scholar]
- 23.Maronpot R, Anderson MW, Wiseman RW. Characterization of p53 mutations in methylene chloride-induced lung tumors from B6C3F1 mice. Carcinogenesis. 1993;14:803–810. doi: 10.1093/carcin/14.5.803. [DOI] [PubMed] [Google Scholar]
- 24.Maronpot RR, Fox T, Malarkey D, Goldsworthy T. Mutations in the ras proto-oncogene: clues to etiology and molecular pathogenesis of mouse liver tumors. Toxicology. 1995;101:125–156. doi: 10.1016/0300-483x(95)03112-s. [DOI] [PubMed] [Google Scholar]
- 25.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes & Development. 2005;19:643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 26.National Toxicology Program (NTP) In Vivo Cytogenetics Testing Results for Cumene, Micronucleus Induction Results. NIEHS; Research Triangle Park, NC: 1996. (Case No. 98-82-8) NTP TR 542. [Google Scholar]
- 27.Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, Fraire AE, Gabrielson EW, Gunning WT, Haines DC, Kaufman MH, Linnoila RL, Maronpot RR, Rabson AS, Reddick RL, Rehm S, Rozengurt N, Schuller HM, Shmidt EN, Travis WD, Ward JM, Jacks T. Classification of proliferative pulmonary lesions of the mouse recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 28.Osada H, Takahashi T. Genetic alterations of multiple tumor suppressors and oncogenes in the carcinogenesis and progression of lung cancer. Oncogene. 2002;21:7421–7434. doi: 10.1038/sj.onc.1205802. [DOI] [PubMed] [Google Scholar]
- 29.Patel AC, Anna CH, Foley JF, Stockton PS, Tyson fl, Barrett JC, Devereux TR. Hypermethylation of the p16Ink4a promoter in B6C3F1 mouse primary lung adenocarcinomas and mouse lung cell lines. Carcinogenesis. 2000;21:1691–1700. doi: 10.1093/carcin/21.9.1691. [DOI] [PubMed] [Google Scholar]
- 30.Research Triangle Institute (RTI) Metabolism, Disposition and Pharmacokinetics of Cumene in F 344 Rats Following Oral, IV Administration or Nose-Only Inhalation Exposure. RTI; Research Triangle Park, NC: 1989. RTI/4353-01F. [Google Scholar]
- 31.Reynolds SH, Stowers SJ, Maronpot RR, Aaronson SA, Anderson MW. Science. 1987;237:1309–1317. doi: 10.1126/science.3629242. [DOI] [PubMed] [Google Scholar]
- 32.Rodenhuis S, Van De Watering ML, Mooi WJ, Evers SG, Van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene – a possible pathogenetic favtor in adenocarcinoma of the lung. New Engl J Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 33.Rodin SN, Rodin AS. Origins and selection ofp53 mutations in lung carcinogenesis. Semin Cancer Biol. 2005;15:103–112. doi: 10.1016/j.semcancer.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Santillo M, Mondola P, Seru R, Annella T, Cassano S, Ciullo I, Tecce MF, Iacomino G, Damiano S, Cuda G, Paterno R, Martignetti V, Mele E, Feliciello A, Avvedimento EV. Opposing functions of Ki- and Ha-ras genes in the regulation of redox signals. Curr Biol. 2001;11:614–619. doi: 10.1016/s0960-9822(01)00159-2. [DOI] [PubMed] [Google Scholar]
- 35.Sato A, Nakajima T. Pharmacokinetics of organic solvent vapors in relation to their toxicity. Scand J Work Environ Health. 1987;13:81–93. doi: 10.5271/sjweh.2075. [DOI] [PubMed] [Google Scholar]
- 36.Shigenaga MK, Ames BN. Assays for 8-hydroxy-2′-deoxyguanosine: a biomarker of in vivo oxidative DNA damage. Free Radical Biol Med. 1991;10:211–216. doi: 10.1016/0891-5849(91)90078-h. [DOI] [PubMed] [Google Scholar]
- 37.Sills RC, Hong HL, Greenwell A, Herbert RA, Boorman GA, Devereux TR. Increased frequency of K-ras mutations in lung neoplasms from female B6C3F1 mice exposed to ozone for 24 or 30 months. Carcinogenesis. 1995;16:1623–1628. doi: 10.1093/carcin/16.7.1623. [DOI] [PubMed] [Google Scholar]
- 38.Sills RC, Hong HL, Melnick RL, Boorman GA, Devereux TR. High frequency of codon 61 K-ras A to T transversions in lung and Harderian gland neoplasms of B6C3F1 mice exposed to chloroprene (2-chloro-1,3-butadiene) for 2 years, and comparisons with the structurally related chemicals isoprene and 1,3- butadiene. Carcinogenesis. 1999;20:657–662. doi: 10.1093/carcin/20.4.657. [DOI] [PubMed] [Google Scholar]
- 39.Sills RC, Hong HL, Flake G, Moomaw C, Clayton N, Boorman GA, Dunnick J, Devereux TR. O-Nitrotolune-induced large intestinal tumors in B6C3F1 mice model human colon cancer in their molecular pathogenesis. Carcinogenesis. 2004;25:605–612. doi: 10.1093/carcin/bgh044. [DOI] [PubMed] [Google Scholar]
- 40.Snyder R, Hedli CC. An overview of benzene metabolism. Environ Health Perspect. 1996;104(Suppl 6):1165–1171. doi: 10.1289/ehp.961041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tam AS, Foley JF, Devereux TR, Maronpot RR, Massey TE. High frequency and heterogenous distribution of p53 mutations in aflatoxin B1-induced mouse lung tumors. Cancer Res. 1999;59:3634–3640. [PubMed] [Google Scholar]
- 42.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollamn AP, Nishimura S. 8-Oxogyanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci, USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.US Environmental Protection Agency (EPA) Toxicological Review of Cumene, EPA’s Summary Information on the Integrated Risk Information System. EPA; Washington, DC: 1997. [Google Scholar]
- 44.Valentine JL, Lee SS, Seaton MJ, Asgharian B, Farris G, Corton JC, Gonzalez FJ, Medinsky MA. Reduction of benzene metabolism and toxicity in mice that lack CYP2E1 expression. Toxicol Appl Pharmacol. 1996;141:205–213. doi: 10.1006/taap.1996.0277. [DOI] [PubMed] [Google Scholar]
- 45.Wakamatsu N, Collins BC, Parker JS, Tessama M, Clayton ND, Ton TT, Hong HL, Belinsky S, Devereus TR, Sills RC, Lahousse SA. Gene expression studies demonstrate that the K-ras/Erk MAP kinase signal transduction pathway and modification of histone, contribute to the pathogenesis of cumene- induced lung tumors. Toxicol Pathol. 2008 doi: 10.1177/0192623308320801. Submitted. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Wang Y, Vikis HG, Johnson L, Liu J, Anderson MW, Sills RC, Hong HL, Devereux TR, Jacks T, Guan KL, You M. Wild-type Kras 2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]