Abstract
Background
Although rheumatoid arthritis has been shown to have moderately strong genetic component, both linked loci identified in linkage analyses and susceptibility variants from association studies are short of adequately accounting for a comprehensive catalogue of the molecular factors underlying this complex disease. The objective of this study was to use supplementary phenotype based on cumulative hazard of rheumatoid arthritis to identify linkage evidence for new and additional rheumatoid arthritis loci in a genome-wide linkage analysis of 342 affected sibling pair families from the United Kingdom.
Methods
Using proportional hazards model, we estimated cumulative hazard of rheumatoid arthritis and then used it as a quantitative trait in a non-parametric multipoint variance component linkage analysis with 353 microsatellite markers distributed across the 22 autosomal chromosomes.
Results
We identified 3 new loci with genome-wide suggestive linkage evidence for rheumatoid arthritis on 9q21.13, 15p11.1 and 20q13.33. Our results also confirmed previously reported linkage evidence in the HLA-DRB1 region on chromosome 6 and on locus 1q32.1.
Conclusion
This study demonstrates the potential for information gain through the use of supplementary phenotypes in genetic study of complex diseases to identify new and additional potential linked loci that are not detected by linkage analysis of traditional phenotypes; and our results provide further evidence of the involvement of multiple loci in the genetic aetiology of rheumatoid arthritis.
Background
Rheumatoid arthritis (RA) is a chronic inflammatory condition that belongs to the family of diseases referred to as autoimmune. The population prevalence of RA is about 0.01 [1,2], and the relative risk for siblings of affected persons ranges between 2 and 17 [2-5]. In addition to evidence from familial aggregation, results from twin studies [6-8], segregation analysis [9] and genome-wide association studies [10-12] all indicate evidence of genetic influence in the aetiology of RA. This evidence is further strengthened by the relatively high heritability of RA, estimated to be about 65% [13] from twin studies; and also by reported linkage evidence for regions on chromosomes 2 and 11 [14], and 6 and 16 [15]. Although consistently strong evidence across studies points to association with the HLA locus on chromosome 6, it has however been observed that the RA-HLA region association accounts for only about one-third of the total genetic determinants of susceptibility to RA [16,17]. On the other hand, none of the other RA-linked loci reported to date has been consistently reported across studies [1,3,14,18,19]. The implication is that it is likely that a number of other loci involved in the genetic susceptibility to RA exist and are yet to be identified and their individual roles quantified; hence the complete role of genetic factor in the aetiology of this complex disease remains to be clearly understood.
As with many other complex human diseases, the difficulty in achieving a clear understanding of the role of genetics in disease can partly be attributed to limitations in phenotype definition and characterization. RA is a complex disease condition with variation in onset age, severity, and accompanying presence or absence of clinical features such as nodules, erosions, and rheumatoid factor. In addition, phenotypic, clinical and genetic heterogeneity exists between studies. To address the problem of heterogeneity associated with RA phenotype, John et al. [15] and Eyre et al. [20] used stratified analyses based on subgroups defined by age at disease onset, sex and clinical features such as rheumatoid factor (RF) status, erosion and shared epitopes. In addition to the affection status variable, age at RA onset provides the opportunity to investigate genetic influence based on the cumulative hazard of RA by using survival analysis tool in combination with linkage analysis. The premise is that detecting evidence of linkage in regions not previously identified in analysis of affection-based phenotype may be possible in analysis of supplementary phenotype of the same disease. The objective of this study was to use cumulative hazard estimates from proportional hazards model in genome-wide scan for linkage evidence for new and additional genetic loci linked to RA using affected sibling pairs data. We present here the results from the genome-wide linkage analysis of the cumulative hazard to RA in a sample of 728 relatives from 342 affected sibling pair families from the UK.
Methods
The RA affected sibling pair families used in this study were part of the UK National Repository of Multicase Rheumatoid Arthritis Families maintained by the Arthritis Research Campaign (http://www.medicine.manchester.ac.uk/epidemiology/research/arc/). Details of family recruitment and assessment protocols have been described elsewhere [1,20]. Briefly, subjects were examined by trained metrologists using standardized questionnaires and protocols. Radiographs of each subject's hands were reviewed and rheumatoid factor status of each subject was ascertained using a particle agglutination test. All RA affected subjects satisfied the American College of Rheumatology criteria [21,22]. Access to the UK RA data was requested and approved through the Genetic Analysis Workshop 15 (GAW15). The data used in the present study were from cohorts 1 and 2 families as made available through GAW15 [23]. Cohort 1 comprised of RA affected sibling pairs from the 182 families as described by Mackay et al [1] and cohort 2 was made up of RA affected sibling pairs from the additional 195 families which have been described elsewhere [20]. Available phenotypic and clinical information included RA affection status, age at onset of RA symptoms, erosion status, and RF status. Genotype data on 353 informative microsatellite markers from the ABI Prism Linkage Mapping set, spanning the 22 autosomes at an average inter-marker distance of 10 cM were available on subjects and used in the present study.
Estimation of cumulative hazard
We restricted all analyses to RA affection status, age at onset and sex since other measures such as severity of RA, erosion status and RF status are measures of post-RA affection symptoms. Our objective was to study only those variables that influence an individual's hazard to RA. Survival analysis was performed using RA status, sex, and onset age data on 750 affected subjects from 364 families. Since first symptoms of RA can begin at any point in time, it is reasonable to suppose that observed ties in event times (age at onset) among subjects are likely results of approximation or convenience in measurement and that there is a true but unknown ordering for the tied event times. This supposition appears more plausible for event as onset of RA and as such tied event times were treated according to the method described by Kalbfleisch and Prentice [24], and DeLong and colleagues [25]. Briefly, in the fitted proportional hazards model, exact conditional probability was computed for each event under the assumption that all tied event times occur before censored times of the same value or before larger values. The proportional hazards model was based on the assumption that the hazard to RA for any subject is a fixed proportion of the hazard to RA for any other subject in the study sample and the ratios of hazards are independent of time. The use of Cox regression to fit the proportional hazards model in this study allowed us not to specify any particular distribution for the underlying hazard function and also to test the hypothesis of proportionality of hazards. We included sex as a covariate and then modelled the survival function from which the cumulative hazard for each subject was estimated as the negative of the logarithm of the survival function as -logS(t); where S(t) is the survival function at time 't' estimated from the fitted proportional hazards model.
Linkage analysis
For the linkage analysis, 22 families with singletons were excluded because these were uninformative for linkage analysis. The linkage analysis was therefore based on 342 families with a total of 728 affected siblings. There were 353 microsatellite markers across the 22 autosomal chromosomes. For subjects not genotyped for some markers, their genotypes were set to missing for such markers. Allele frequencies for each marker were calculated using maximum likelihood estimation method. The marker-specific identity by descent (IBD) for sibling pairs within each family, and the multipoint IBDs were computed using the software SOLAR [26]. Since the estimated cumulative hazard was on continuous scale, we therefore used it as a quantitative trait in the linkage analysis as opposed to a dichotomous trait that is used when RA is analysed as affection status. The computed multipoint IBDs were used in a nonparametric multipoint linkage analysis of the cumulative hazard of RA using the software SOLAR. The use of nonparametric linkage method enabled us not to specify any inheritance model since the aetiology of RA is complex. Linkage evidence was expressed as logarithm of odds (LOD) scores for linkage at each chromosomal position. LOD scores which were defined as the logarithm to base 10 of likelihood ratio of the data assuming linkage versus the analysis model assuming no linkage, were used to summarize the evidence in favour of linkage and these plotted by chromosome against the genetic marker positions.
Results
The distributions of pairs of RA affected siblings and sexes in the total sample of 342 families included in the linkage analysis are shown in Table 1. There were 306 families with 2 affected siblings each, 31 families with 3 affected siblings each, and 5 families with more than 3 affected siblings each. The 728 subjects comprised of 537 females and 191 males. Mean age in years at onset of RA is 41.6 for the sample, 41.4 for females and 42.2 for males (P = 0.8360) (Table 1). Influence of gender on RA hazard was assessed by including sex as a covariate in the proportional hazards model and then testing the null hypothesis that its coefficient is not different from zero. We also tested the hypothesis of proportionality of hazards in our study sample. For both tests, we did not observe any statistical evidence against the null hypotheses of proportionality of hazards and that of equality between zero and coefficient of sex. The p-value for the coefficient of sex was 0.5109 and the p-value for the Kolmogorov-type supremum test for proportional hazards assumption was 0.9640 based on 1000 bootstrap samples.
Table 1.
Distributions of affected sibling pair sizes
| Size of sibling pair | No. of families | No. of subjects | Mean onset age (years ± standard error) |
|---|---|---|---|
| 2 | 306 | 612 | - |
| 3 | 31 | 93 | - |
| 4 | 3 | 12 | - |
| 5 | 1 | 5 | - |
| 6 | 1 | 6 | - |
| All | 342 | 728 | 41.63 ± 0.51 |
| Females | - | 537 | 41.44 ± 0.61 |
| Males | - | 191 | 42.17 ± 0.89 |
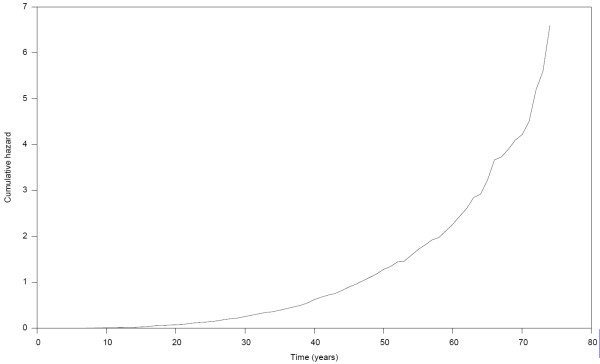
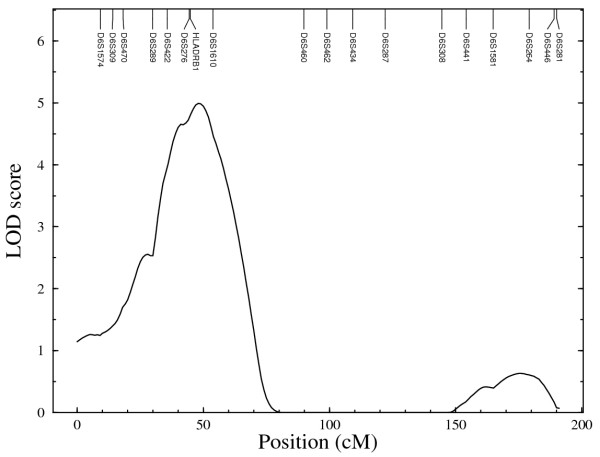
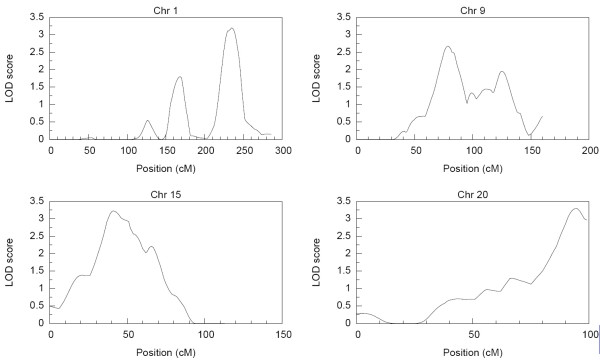
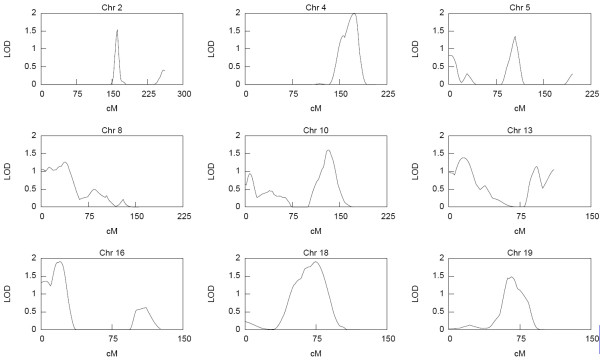
The distribution of the estimated cumulative hazard function is presented in Figure 1. The result shows a rather slow rise in cumulative hazard from the early years up till about age 42 years, which is also about the median survival time, and thereafter the cumulative hazard began to accelerate more rapidly with increasing years. The strongest linkage evidence observed on each chromosome from the results of the nonparametric multipoint linkage analysis using the 353 microsatellites markers are presented in Table 2. Based on generally accepted thresholds of LOD scores for genome-wide linkage studies [27], we observed genome-wide significant evidence (LOD score ≥ 3.6; P ≤ 2 × 10-5) for linkage on chromosome 6. The linkage signal spanned the HLA-DRB1 region (Figure 2) which has been reported in other studies [1-3,14,18,28,29] to contain potential RA loci. Other chromosomal regions with genome-wide suggestive evidence for linkage (LOD score ≥ 2.2; P ≤ 7.4 × 10-4) were identified on chromosomes 1, 9, 15 and 20 and are displayed in Figure 3. The LOD score of 3.3 on chromosome 20 at about 94 cM near the D20S173 was the second highest followed by the LOD score of 3.2 on chromosome 15 near the D15S994 marker at about 41 cM. The signal on chromosome 1 was observed in the region previously identified in other reports [1,18,20,28-30], whereas those on chromosomes 9, 15 and 20 have not previously been reported and thus represent newly identified potential RA loci. The lengths of the corresponding 1-LOD support intervals for the linkage evidence on chromosomes 1, 6 and 15 were 22 cM, 31 cM and 26 cM, respectively. The location of the highest linkage signal on chromosome 20 was near the end of the chromosome thus making a reliable estimation of its support interval difficult. Genome-wide nominal linkage evidence (LOD ≥ 0.8; P < 0.05) was identified on eleven other chromosomal regions (Figure 4), some of these corresponded to previously reported regions with similar level of linkage signal for RA [1,20,30].
Figure 1.
Distribution of cumulative hazard of rheumatoid arthritis.
Table 2.
Regions with highest LOD scores on each chromosome
| Chromosome | Locus | LOD score | Position (cM)‡ | Nearest marker |
|---|---|---|---|---|
| 1 | 1q32.1 | 3.2 | 235 | D1S425 |
| 2 | 2q24.1 | 1.5 | 161 | D2S142 |
| 3 | 3p26.3 | 1.0 | 8 | D3S1297 |
| 4 | 4q34.1 | 2.0 | 173 | D4S1539 |
| 5 | 5q15 | 1.3 | 105 | D5S644 |
| 6 | 6p21.3 | 5.0 | 48 | HLA-DRB1 |
| 7 | 7q36.2 | 0.5 | 181 | D7S2465 |
| 8 | 8q21.3 | 1.3 | 38 | D8S258 |
| 9 | 9q21.13 | 2.7 | 78 | D9S167 |
| 10 | 10q25.1 | 1.6 | 132 | D10S597 |
| 11 | 11q | 0.1 | 86 | D11S937 |
| 12 | 12q24.33 | 1.0 | 165 | D12S1723 |
| 13 | 13q12.3 | 1.4 | 15 | D13S217 |
| 14 | 14q13 | 0.6 | 57 | D14S276 |
| 15 | 15p11.1 | 3.2 | 41 | D15S994 |
| 16 | 16q13.13 | 1.9 | 20 | D16S404 |
| 17 | - | 0.8 | 104 | D17S1790 |
| 18 | 18q21.1 | 1.9 | 74 | D18S474 |
| 19 | 19q13.2 | 1.5 | 66 | D19S420 |
| 20 | 20q13.33 | 3.3 | 94 | D20S173 |
| 21 | - | 0.0 | - | - |
| 22 | 22q13.2 | 0.2 | 52 | D22S274 |
‡cM: centiMorgan on Marshfield map
Figure 2.
Region of significant genome-wide linkage evidence on chromosome 6.
Figure 3.
Chromosomal regions with suggestive linkage evidence.
Figure 4.
Selected chromosomal regions with nominal linkage evidence.
Discussion
We report here genome-wide linkage evidence for RA based on analysis of cumulative hazard estimated from family data on RA affected sibling pairs. The objective of the study was to identify additional evidence of linkage for RA by using supplementary RA phenotype that we envisioned would capture linkage information otherwise undetected by the traditional affection-status phenotype definition used in previous linkage studies of RA. The supplementary phenotype approach was based on application of proportional hazards modelling to estimate cumulative hazard of RA based on affection status and RA onset age for each study subject. 750 affected subjects from 364 families were included in the estimation of cumulative hazard using proportional hazards model. The estimated cumulative hazard was then used as quantitative trait in linkage analysis of genotypes on 353 microsatellite markers spanning the 22 autosomes at an average inter-marker distance of 10 cM.
In this study we have identified three new potential RA loci which have not been reported or detected in previous linkage analyses that used the traditional RA affection status. The new potential RA loci with strong genome-wide suggestive linkage evidence are 9q21.13 with LOD score of 2.7, 15p11.1 with LOD score of 3.2 and 20q13.33 with LOD score of 3.3. In addition to the identified new potential RA loci, we also obtained linkage evidence in chromosomal regions that are consistent with previous findings. The most significant genome-wide linkage evidence was detected on chromosome 6 in the HLA-DRB1 region which is consistent with previous reports [1-3,14,18,28,29]. The genome-wide suggestive linkage evidence detected in 1q32.1 is also similar to reported level of linkage evidence in previous studies for the region [30]. Among the candidate genes located in locus 1q32.1 are CHI3L1 (chitinase 3-like 1 (cartilage glycoprotein-39)) gene which has been implicated in local autoimmune response that leads to chronic inflammation and joint destruction in RA patients [31] and the IKBKE (inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon) gene which has been reported to play a role in synovial inflammation, extracellular matrix destruction, and activation of the viral program and innate immune response in RA [32]. Also, the HRH3 (histamine receptor H3) gene located in locus 20q13.33, has a role in neurogenic inflammation [33].
We note that in a previous linkage analysis of a subset of these data (cohort 1 families), Mackay et al [1] reported significant genome-wide evidence on chromosome 6 in the same HLA-DRB1 region on which we have also detected significant evidence, and also identified a region on same chromosome with suggestive evidence plus 9 regions of nominal evidence on other chromosomes. Likewise, in a slightly larger sample from the same UK cohort 1 families, John et al [15] confirmed the linkage evidence on chromosome 6 previously reported by Mackay et al [1], but did not detect any suggestive linkage evidence outside of chromosome 6. However, in another linkage study involving both cohorts 1 and 2 families, Eyre et al [20] failed to detect any significant or suggestive linkage evidence. The fact that previously reported potential RA loci are replicated in the present study underscore the validity of the supplementary RA phenotype approach used here. The identification of new potential RA loci in this study is a demonstration of the importance of the use of supplementary phenotypes for genetic mapping of complex diseases such as RA.
It is worth noting that in this study we did not include clinical covariates such as rheumatoid factor status and erosion status that were included in some of the previous studies because these covariates measure symptoms resulting from RA affection. It is therefore not illogical to assume that an individual's cumulative hazard to RA could not be influenced by such post-RA onset phenotypes or clinical symptoms. We therefore chose not to include in the model variables that we think do not influence an individual's hazard of being affected by RA. Whether all the identified linkage regions in the present study are true positives requires further studies. We declare that the reproduction of linkage evidence at previously reported potential RA loci provides support for the validity of the definition of supplementary RA phenotype and modelling that we have used in the present study. We recognise that supplementary phenotype as defined and treated as quantitative trait in the present study may not be practicable for every complex human disease phenotype. However, for complex diseases for which this approach is applicable, this can lead to identification of new additional genetic loci that could explain part of the missing heritability [34] observed in susceptibility variants identified in association studies of complex diseases. This supplementary phenotype approach can be used to identify new candidate loci or narrow candidate regions in linkage and association studies. It is envisioned that results from both analyses of supplementary disease phenotypes and their corresponding traditional disease phenotype definitions would enhance the efficiency of the "genetic dragnet" at capturing important genetic variants contributing to complex diseases.
Conclusion
This present study demonstrates the potential information gain through the use of supplementary phenotypes in genetic study of complex diseases such as RA to detect new and additional linked loci that are undetected in linkage analysis of the traditional phenotypes. Also, our results provide further evidence of the involvement of multiple loci in the genetic aetiology of RA.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BOT conceived of the study, performed all statistical analyses, interpreted results and wrote the manuscript. YL participated in the design, statistical analyses, and preparation of manuscript. AK, AM, MT and RSC participated in the design and preparation of manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Bamidele O Tayo, Email: btayo@lumc.edu.
Yulan Liang, Email: ylian001@umaryland.edu.
Arpad Kelemen, Email: akele001@umaryland.edu.
Austin Miller, Email: am65@buffalo.edu.
Maurizio Trevisan, Email: maurizio_trevisan@nshe.nevada.edu.
Richard S Cooper, Email: rcooper@lumc.edu.
Acknowledgements
This work is based on data that was gathered with the support of grants from the National Institutes of Health (NO1-AR-2-2263 and RO1-AR-44422), and the National Arthritis Foundation; and made available through Genetic Analysis Workshop (GAW) supported by grant R01 GM031575. This work was supported in part by the National Science Foundation grant DMS-06-04639 and by the National Heart, Lung, and Blood Institute, NIH grant R01 HL053353.
References
- MacKay K, Eyre S, Myerscough A, Milicic A, Barton A, Laval S, Barrett J, Lee D, White S, John S, Brown MA, Bell J, Silman A, Ollier W, Wordsworth P, Worthington J. Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis and rheumatism. 2002;46(3):632–639. doi: 10.1002/art.10147. [DOI] [PubMed] [Google Scholar]
- Seldin MF, Amos CI, Ward R, Gregersen PK. The genetics revolution and the assault on rheumatoid arthritis. Arthritis and rheumatism. 1999;42(6):1071–1079. doi: 10.1002/1529-0131(199906)42:6<1071::AID-ANR1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, Kern M, Criswell LA, Albani S, Nelson JL, Clegg DO, Pope R, Schroeder HW Jr, Bridges SL Jr, Pisetsky DS, Ward R, Kastner DL, Wilder RL, Pincus T, Callahan LF, Flemming D, Wener MH, Gregersen PK. A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. American journal of human genetics. 2001;68(4):927–936. doi: 10.1086/319518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Junco D, Luthra HS, Annegers JF, Worthington JW, Kurland LT. The familial aggregation of rheumatoid arthritis and its relationship to the HLA-DR4 association. American journal of epidemiology. 1984;119(5):813–829. doi: 10.1093/oxfordjournals.aje.a113802. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Young A, Gorsuch AN, Bottazzo GF, Cudworth AG. Evidence for an association between rheumatoid arthritis and autoimmune endocrine disease. Annals of the rheumatic diseases. 1983;42(3):297–300. doi: 10.1136/ard.42.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho K, Koskenvuo M, Tuominen J, Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. The Journal of rheumatology. 1986;13(5):899–902. [PubMed] [Google Scholar]
- Lawrence JS, Valkenburg HA, Bremner JM, Ball J. Rheumatoid factors in families. Annals of the rheumatic diseases. 1970;29(3):269–274. doi: 10.1136/ard.29.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A, Ollier WE. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. British journal of rheumatology. 1993;32(10):903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- Lynn AH, Kwoh CK, Venglish CM, Aston CE, Chakravarti A. Genetic epidemiology of rheumatoid arthritis. American journal of human genetics. 1995;57(1):150–159. [PMC free article] [PubMed] [Google Scholar]
- Julia A, Ballina J, Canete JD, Balsa A, Tornero-Molina J, Naranjo A, Alperi-Lopez M, Erra A, Pascual-Salcedo D, Barcelo P, Camps J, Marsal S. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis and rheumatism. 2008;58(8):2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- Hinks A, Barton A, Shephard N, Eyre S, Bowes J, Cargill M, Wang E, Ke X, Kennedy GC, John S, Worthington J, Thomson W. Identification of a novel susceptibility locus for juvenile idiopathic arthritis by genome-wide association analysis. Arthritis and rheumatism. 2009;60(1):258–263. doi: 10.1002/art.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Taylor KE, Destefano AL, Criswell LA, Izmailova ES, Parker A, Roubenoff R, Plenge RM, Weinblatt ME, Shadick NA, Karlson EW. Genome-wide association study of determinants of anti-cyclic citrullinated peptide antibody titer in adults with rheumatoid arthritis. Mol Med. 2009;15(5-6):136–143. doi: 10.2119/molmed.2009.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, Silman AJ. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis and rheumatism. 2000;43(1):30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Amos CI, Chen WV, Lee A, Li W, Kern M, Lundsten R, Batliwalla F, Wener M, Remmers E, Kastner DA, Criswell LA, Seldin MF, Gregersen PK. High-density SNP analysis of 642 Caucasian families with rheumatoid arthritis identifies two new linkage regions on 11p12 and 2q33. Genes and immunity. 2006;7(4):277–286. doi: 10.1038/sj.gene.6364295. [DOI] [PubMed] [Google Scholar]
- John S, Amos C, Shephard N, Chen W, Butterworth A, Etzel C, Jawaheer D, Seldin M, Silman A, Gregersen P, Worthington J. Linkage analysis of rheumatoid arthritis in US and UK families reveals interactions between HLA-DRB1 and loci on chromosomes 6q and 16p. Arthritis and rheumatism. 2006;54(5):1482–1490. doi: 10.1002/art.21794. [DOI] [PubMed] [Google Scholar]
- Deighton CM, Walker DJ, Griffiths ID, Roberts DF. The contribution of HLA to rheumatoid arthritis. Clinical genetics. 1989;36(3):178–182. doi: 10.1111/j.1399-0004.1989.tb03185.x. [DOI] [PubMed] [Google Scholar]
- Wordsworth BP. HLA class II antigens in susceptibility to rheumatoid arthritis. British journal of rheumatology. 1991;30(2):151–152. doi: 10.1093/rheumatology/30.2.151. [DOI] [PubMed] [Google Scholar]
- Cornelis F, Faure S, Martinez M, Prud'homme JF, Fritz P, Dib C, Alves H, Barrera P, de Vries N, Balsa A, Pascual-Salcedo D, Maenaut K, Westhovens R, Migliorini P, Tran TH, Delaye A, Prince N, Lefevre C, Thomas G, Poirier M, Soubigou S, Alibert O, Lasbleiz S, Fouix S, Bouchier C, Liote F, Loste MN, Lepage V, Charron D, Gyapay G, Lopes-Vaz A, Kuntz D, Bardin T, Weissenbach J. New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(18):10746–10750. doi: 10.1073/pnas.95.18.10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa S, Hayashi S, Tsukamoto Y, Goko H, Kawasaki H, Wada T, Shimizu K, Yasuda N, Kamatani N, Takasugi K, Tanaka Y, Shiozawa K, Imura S. Identification of the gene loci that predispose to rheumatoid arthritis. International immunology. 1998;10(12):1891–1895. doi: 10.1093/intimm/10.12.1891. [DOI] [PubMed] [Google Scholar]
- Eyre S, Barton A, Shephard N, Hinks A, Brintnell W, MacKay K, Silman A, Ollier W, Wordsworth P, John S, Worthington J. Investigation of susceptibility loci identified in the UK rheumatoid arthritis whole-genome scan in a further series of 217 UK affected sibling pairs. Arthritis and rheumatism. 2004;50(3):729–735. doi: 10.1002/art.20039. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis and rheumatism. 1988;31(3):315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- MacGregor AJ, Bamber S, Silman AJ. A comparison of the performance of different methods of disease classification for rheumatoid arthritis. Results of an analysis from a nationwide twin study. The Journal of rheumatology. 1994;21(8):1420–1426. [PubMed] [Google Scholar]
- Amos CI, Chen WV, Remmers E, Siminovitch KA, Seldin MF, Criswell LA, Lee AT, John S, Shephard ND, Worthington J, Cornelis F, Plenge RM, Begovich AB, Dyer TD, Kastner DL, Gregersen PK. Data for Genetic Analysis Workshop (GAW) 15 Problem 2, genetic causes of rheumatoid arthritis and associated traits. BMC Proc. 2007;1(Suppl 1):S3. doi: 10.1186/1753-6561-1-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. New York: John Wiley & Sons, Inc; 1980. [Google Scholar]
- DeLong DM, Guirguis GH, So YC. Efficient Computation of Subset Selection Probabilities with Application to Cox Regression. Biometrika. 1994;81:607–611. doi: 10.1093/biomet/81.3.607. [DOI] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature genetics. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Osorio YFJ, Bukulmez H, Petit-Teixeira E, Michou L, Pierlot C, Cailleau-Moindrault S, Lemaire I, Lasbleiz S, Alibert O, Quillet P, Bardin T, Prum B, Olson JM, Cornelis F. Dense genome-wide linkage analysis of rheumatoid arthritis, including covariates. Arthritis and rheumatism. 2004;50(9):2757–2765. doi: 10.1002/art.20458. [DOI] [PubMed] [Google Scholar]
- John S, Shephard N, Liu G, Zeggini E, Cao M, Chen W, Vasavda N, Mills T, Barton A, Hinks A, Eyre S, Jones KW, Ollier W, Silman A, Gibson N, Worthington J, Kennedy GC. Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. American journal of human genetics. 2004;75(1):54–64. doi: 10.1086/422195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzel CJ, Chen WV, Shepard N, Jawaheer D, Cornelis F, Seldin MF, Gregersen PK, Amos CI. Genome-wide meta-analysis for rheumatoid arthritis. Hum Genet. 2006;119(6):634–641. doi: 10.1007/s00439-006-0171-8. [DOI] [PubMed] [Google Scholar]
- Steenbakkers PG, Baeten D, Rovers E, Veys EM, Rijnders AW, Meijerink J, De Keyser F, Boots AM. Localization of MHC class II/human cartilage glycoprotein-39 complexes in synovia of rheumatoid arthritis patients using complex-specific monoclonal antibodies. J Immunol. 2003;170(11):5719–5727. doi: 10.4049/jimmunol.170.11.5719. [DOI] [PubMed] [Google Scholar]
- Sweeney SE, Hammaker D, Boyle DL, Firestein GS. Regulation of c-Jun phosphorylation by the I kappa B kinase-epsilon complex in fibroblast-like synoviocytes. J Immunol. 2005;174(10):6424–6430. doi: 10.4049/jimmunol.174.10.6424. [DOI] [PubMed] [Google Scholar]
- Akdis CA, Jutel M, Akdis M. Regulatory effects of histamine and histamine receptor expression in human allergic immune responses. Chem Immunol Allergy. 2008;94:67–82. doi: 10.1159/000154858. full_text. [DOI] [PubMed] [Google Scholar]
- Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456(7218):18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]