Abstract
Vibrio alginolyticus ZJ-51 displays phase variation between opaque/rugose colonies (Op) and translucent/smooth colonies (Tr). These colony variants show great differences in biofilm formation and motility. In this study, a gene encoding for an rpoS-like sigma factor, rpoX, has been cloned and characterized. The absence of rpoX did not affect colony switching rate but did decrease biofilm formation in both the Op and the Tr variants. When challenged with hydrogen peroxide, the ΔrpoX in the Op background showed a slightly higher survival rate compared with the wild type, whereas survival was decreased in the Tr background. Deletion of rpoX in the Tr background resulted in a higher ability to resist ethanol challenges and to survive hyperosmolarity challenges, and in the Op background the opposite phenotype was observed. This indicates that the rpoX gene is involved in biofilm formation and stress response but the effects are controlled by colony phase variation in V. alginolyticus.
1. Introduction
Sigma factors are a class of proteins constituting essential dissociable subunits that confer sequence-specific DNA binding properties to RNA polymerase (RNAP). Within the RNAP holoenzyme, sigma factors provide promoter recognition specificity to the polymerase and contribute to DNA strand separation, then they dissociate from RNAP core enzyme and transcription initiation follows [1]. Besides the vegetative sigma factor, which recognizes the “standard” promoter, many bacteria possess alternative sigma factors that confer altered promoter specificity upon RNAP [2]. Alternative sigma factors are synthesized or activated in response to certain environmental conditions. They partially and/or temporarily replace the vegetative sigma factor and are thus crucial for many stress responses [2].
Sigma factors can be divided into two structurally unrelated families: the σ70 and the σ54 families. The σ70 family consists of primary sigma factors, as well as related alternative sigma factors [3, 4]. These alternative sigma factors are further classified by the physiological processes that they control, for example, stress response and flagella biosynthesis [3, 4]. The extracytoplasmic function factors (ECF) belong to the σ70 family of sigma factors, but are phylogenetically distinct. Many ECF members are responsible for sensing and reacting to signals from the extracytoplasmic environment [3–5]. The σ70 family proteins generally contain four regions, region 1 to region 4, but only region 2 and 4 are well conserved in all members [3, 4]. rpoS or σs, a member of the σ70 alternative sigma factors subfamily, is important for survival under various stress conditions including starvation, oxidative stress, osmotic shock, extreme temperature, low pH (acidic stress), and exposure to UV (DNA damage) [6–15].
V. alginolyticus is an opportunistic pathogen of marine fish, shellfish, as well as human [16]. On the coast of Southern China, V. alginolyticus is the dominant Vibrio species found in the seawater and in farmed marine animals [17]. V. alginolyticus ZJ-51, isolated from diseased grouper, has shown colony phase variation between opaque/rugose (Op, opaque variant) and translucent/smooth (Tr, translucent variant). The two variants exhibit different capability to form biofilms and to express genes involved in polysaccharide biosynthesis, flagellar synthesis, and the AI-2 quorum-sensing system [18]. Recently, genome sequences revealed that in addition to rpoS, an rpoS-like sigma factor was found in V. alginolyticus 12G01. The protein is composed of 279 amino acids. Amino acid 10 to 277 contains an rpoS-proteo region with three conserved sigma 70 regions. However, the function of this protein has not been determined yet. Based on this, we wanted to identify this protein in V. alginolyticus ZJ-51 and study whether it has a role in biofilm formation and stress response in the two colony phase variants.
2. Materials and Methods
2.1. Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids are described in Table 1. ZJ-O is the opaque/rugose variant of the wild type ZJ-51 and ZJ-T is the translucent/smooth variant derived from ZJ-O. Bacteria were routinely grown in Trypticase Soy Broth (TSB) (Becton, Dickinson and company), or 1.5% TSB agar plate (TSA) at 30°C, with the following antibiotics: 100 μg/mL ampicillin (Ap) and 10 μg/mL chloramphenicol (Cm). When necessary, Luria-Bertani (LB) and 1.5% LB agar plate (LA) were used. Biofilm growth medium (BF medium) contains minimal 1 × M63 salts supplemented with 1% NaCl, 1.5% Casamino acids, 1% glucose, 1 mM MgSO4, and 10 μg Thiamine per liter. For selection of conjugants, Thiosulfate Citrate Bile Salts Sucrose Agar (TCBS) (HUANKAI MICROBIAL SCI.&TECH., GD, China) was used with Cm at 10 μg/mL. For sucrose selection, TSA and TSB containing 10% sucrose were used.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| SY327 | Δ(lac pro) argE(Am) rif malA recA56 λ pir lysogen | [19] |
| S17-1 | thi pro hsdR hsdM+ recA RP4-2-Tc::Mu-Km::Tn7 λ pir lysogen | [20] |
| V. alginolyticus | ||
| ZJ-O | Opaque variant of wild strain, isolated from diseased cultured fish in Guangdong province on the Southern China coast. Apr | [17] |
| ZJ-T | Translucent variant of wild strain; isolated from diseased cultured fish in Guangdong province on the Southern China coast. Apr | [17] |
| ΔrpoX-O | ZJ-O carrying an in-frame deletion of rpoX. Apr | This study |
| ΔrpoX-T | ZJ-T carrying an in-frame deletion of rpoX. Apr | This study |
| CrpoX-O | ΔrpoX-O complemented in trans with wild-type rpoX gene. Apr and Cmr | This study |
| CrpoX-T | ΔrpoX-T complemented in trans with wild-type rpoX gene. Apr and Cmr | This study |
| Plasmids | ||
| pDM4 | Cmr, suicide vector with an R6K origin(pir requiring) and sacBR of Bacillus subtilis | [21] |
| pDMrpoX | Cmr, pDM4 containing a mutant allele of rpoX | This study |
| pDMCrpoX | Cmr, pDM4 containing the wild-type allele of rpoX | This study |
aCmr, chloramphenicol resistant
Apr, ampicillin resistant.
2.2. Cloning the rpoX Gene from V. alginolyticus
To clone rpoX from V. alginolyticus ZJ-51, a pair of primers derived from the rpoS-like sigma factor gene (NZ_AAPS01000024) in V. alginolyticus 12G01 were designed as follows: rpoX-I (5′-CCCCACCTGCTCACG-3′) and rpoX-II (5′-GAGACACAAGCGCAA-3′). PCR was performed at 95°C for 5 min, 35 cycles at 94°C for 30 sec, 30 sec at 55°C, 1 min at 72°C, followed by incubation at 72°C for 10 min. A 1.199-bp DNA fragment from the V. alginolyticus ZJ-51 genomic DNA was amplified and sequenced. Nucleotide and deduced protein sequences were analyzed with BLAST and CLUSTAL (version 1.81).
2.3. Mutant and Complementation Strains
To create null mutants of rpoX in both Op and Tr backgrounds, an in-frame deletion was made by allelic exchanges as described previously [21]. Briefly, the allele was created by two rounds of PCR. The first round of PCR was carried out to generate two fragments flanking the coding region of rpoX. The first fragment was amplified using primers rpoX-A (ATAGATCTCCCCACCTGCTCACG), which contains a SalI site at the 5′-end and rpoX-B (GGGCTAAAAATCTTATGACAGTTCATCGT); rpoX-D (ATGTCGACTCGAGACACAAGCGCAA), which contains a BglII site at the 5′-end and rpoX-C (ACGGATGAACTGTCATAAGATTTTTAGCCC) were used to amplify the second fragment. Both fragments contain a 10-bp overlapping sequence and were used as templates for the second PCR using the primers rpoX-A and rpoX-D. The resulting fragment was ligated into the suicide vector pDM4 that had been cut with SalI and BglII to generate the plasmid pDMrpoX. The recombinant suicide plasmid was then conjugated into V. alginolyticus ZJ-O and ZJ-T. Conjugants were selected on the TCBS plates containing 10 μg/mL Cm, followed by sucrose selection. The in-frame deletions, where the wild-type coding sequence of rpoX was completely removed by allelic exchange, were confirmed by sequencing.
To construct complemented strains, the open reading frame (ORF) of rpoX and flanking DNA were amplified using the primers rpoX-A and rpoX-D. The fragment was ligated into pDM4, resulting in pDMCrpoX. The plasmid was introduced into the rpoX null mutants by conjugation. Cm- and Ap-resistant transconjugants were selected, and the presence of the plasmid was confirmed by PCR analysis and sequencing.
2.4. Growth Rate Assay
Overnight cultures of the bacteria were inoculated in TSB with the initial OD600 of 0.01. The optical density was measured every 2 hours.
2.5. Colony Phase Variation Assay
Strains of the Op background were incubated statically in LB at 30°C for 24 hours as described before [18]. Then the cultures were serially diluted and plated on LA. Colony phase was observed and the variation rate was calculated as percentage of Tr cells in the total number of cells. The experiment was repeated three times.
2.6. Biofilm Assay
Biofilm formation was quantitatively analyzed with crystal violet staining as described previously [18]. Biofilm structures were observed with scanning electron microscope (SEM) as described previously [22]. Briefly, biofilms formed on sterilized glass coverslips that were placed in 50 mL centrifuge tubes containing 10 mL BF medium. After 6-hour statical incubation, the coverslips were removed and biofilm structures on the coverslip were observed with SEM S-3400N.
2.7. Stress Response Assays
Bacterial viability was quantitatively determined using the LIVE/DEAD Baclight Bacterial Viability Kit for microscopy and quantitative assay (Molecular Probes, OR) according to the manufacture instruction. Fluorescence output was measured with a Victor 3 multiple label microplate reader (PE, USA) with the excitation and emission wavelengths of 485 nm and 535 nm, respectively.
Before exposure to various stresses, overnight cultures were centrifuged and washed twice with 0.9% NaCl. Then the equal cell numbers, corresponding to an OD600 of 0.5/mL, were diluted in 0.9% NaCl containing either 18% ethanol (Ethanol challenge) or 30 mM H2O2 (oxidative challenge) and 2.5 M NaCl (osmotic challenge). Incubations were carried out at 30°C. The heat challenge was performed in pre-warmed (45°C) 0.9% NaCl. At selected time points, samples were withdrawn, centrifuged, and resuspended in the same volume of 0.9% NaCl, then fluorescence was measured. All experiments were done three times (Gene Accesion Number FJ358498).
3. Results
3.1. Identification of rpoX of V. alginolyticus ZJ-51
A fragment of 1.199 bp in length was amplified from chromosomal DNA of V. alginolyticus ZJ-51 using a pair of primers derived from the sequence of the V. alginolyticus 12G01 rpoS-like gene. The sequence contained the ORF of the rpoX gene (840 bp) and the flanking regions. BlastN analysis showed a 73% identity of the DNA fragment to the rpoS-like gene of Vibrio splendidus LGP329 (FM954973), in addition to a 98% identity to that of V. alginolyticus 12G01 (NZ_AAPS01000024). The sequence of the deduced protein was the same as the rpoS-like sigma factor of V. alginolyticus 12G01 and showed 80% identity to the rpoS-like sigma factor of Vibrio sp. MED222 and 79% identity to the rpoS-like sigma factor of V. splendidus LGP32. Interestingly, RpoQ of V. fischeri ES114 and rpoS of V. alginolyticus VIB283 were also picked up in the search with 40% and 36% identity, respectively (Figure 1).
Figure 1.
Alignment of the amino acid sequences of rpoX from V. alginolyticus ZJ-51 (this study, the rpoX gene gb: FJ358498), rpoS-like sigma factor from V. alginolyticus 12G01(ZP_01261551), Vibrio sp. MED222 (ZP_01063551) and V. splendidus LGP32 (YP_002395435), RpoQ from V. fischeri ES114 (YP_206973), and rpoS from V. alginolyticus (ABW86844) with the accession number of the databases indicated in parentheses. Identical amino acid residues present in three or more proteins are boxed in black. Similar residues are highlighted in gray. The V. alginolyticus rpoX translated sequence overall identity ranges from 36% to 100% for the other sequences shown.
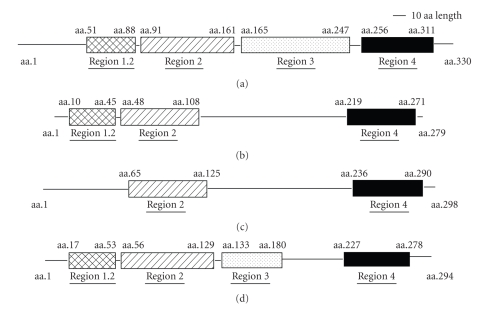
The predicted protein sequence of rpoX contains three conserved domains: (i) sigma 70 region 1.2 (amino acid 10 to 45), (ii) sigma 70 region 2 (amino acid 48 to 108), and (iii) sigma region 4 (amino acid 219 to 271) (Figure 2). Protein sequence alignment revealed that the reason for a 40% identity to the rpoS of V. alginolyticus was due to the second conserved region, which belongs to the sigma 70 region 2 superfamily. In contrast to rpoS and RpoQ, rpoX does not contain the sigma 70 region 3, but it possesses sigma 70 region 1.2, which is absent in the rpoS-like sigma factors of Vibrio sp. MED222 and V. splendidus LGP32 (Figure 2).
Figure 2.
Comparison of the conservative domains among rpoS, rpoX, and the other rpoS-like sigma factors. (a) Conserved rpoS domains in V. alginolyticus, (b) rpoX conserved domains in V. alginolyticus 12G01 and ZJ-51, (c) conserved domains in rpoS-like sigma factors in Vibrio sp. MED222 and V. splendidus LGP32, and (d) RpoQ conserved domains in V. fischeri ES114. Compared with rpoS and RpoQ, rpoX does not have the sigma 70 region 3, but it contains sigma 70 region 1.2, a region that is absent in the rpoS-like sigma factors of Vibrio sp. MED222 and V. splendidus LGP32.
3.2. Growth Ability
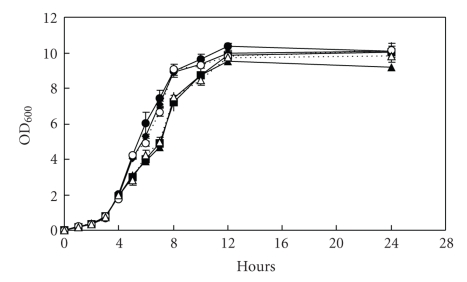
Bacterial growth was investigated for wild types, the rpoX mutants, and the complemented strains. All the mutants showed the same growth rate as the wild types in both Op and Tr backgrounds (Figure 3). Deletion of rpoX does not interfere with the cell growth.
Figure 3.
Kinetics of cell growth of both mutant and wild type of V. alginolyticus ZJ-51, wild type (Op: closed circle; Tr: closed triangle), rpoX null mutant (Op: closed diamond; Tr: closed square), and complemented strains (Op: open circle; Tr: open triangle). Bacteria were grown in TSB medium in Erlenmeyer flasks, which were incubated at 30°C with 240 rpm. Samples were taken at 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, and 24 hours. For every sample, the OD at 600 nm was measured.
3.3. Colony Phase Variation Ability
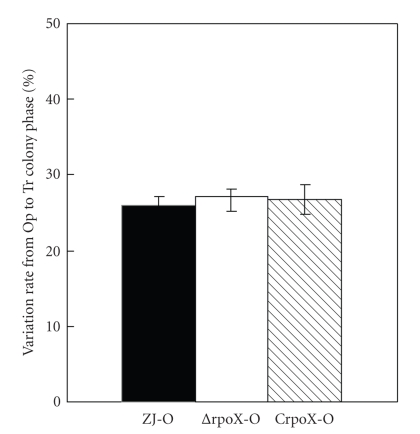
In the previous study [18], transition from Tr to Op has never been observed, thus variation from Op to Tr colony was examined only in the Op background (ZJ-O, ΔrpoX-O, and CrpoX-O). After incubation in LB without shaking for 24 hours, the switching rates from Op to Tr were 26.00% (ZJ-O), 27.12% (ΔrpoX-O), and 26.78% (CrpoX-O), respectively (Figure 4). It suggested that rpoX has no effect on colony phase variation rate.
Figure 4.
Variation rate from the opaque to translucent colony phase was examined in ZJ-O, ΔrpoX-O, and CrpoX-O of V. alginolyticus ZJ-51. After static incubation in LB for 24 hours, percentages of switched colonies were 26.00%, 27.12%, and 26.78% in the wild, mutant, and complemented strains, respectively.
3.4. Biofilm Assay
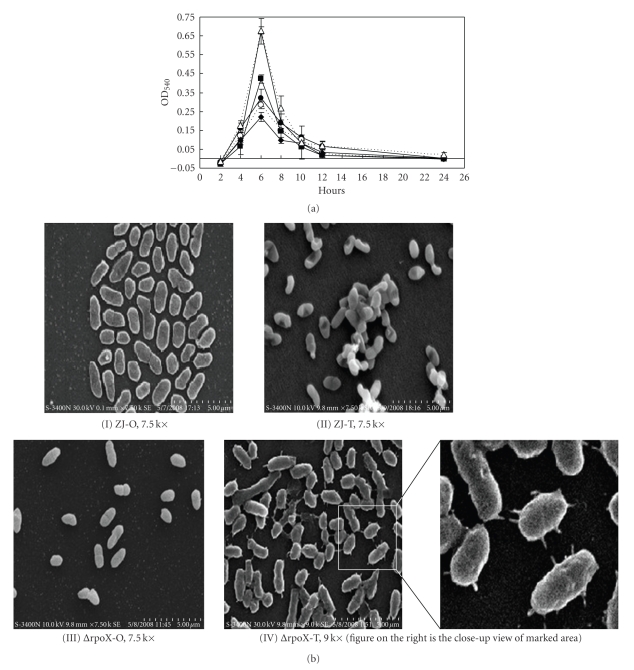
Biofilm formation was investigated for wild types, the rpoX mutants, and the complemented strains. As shown in Figure 5(a), all strains exhibited similar biofilm formation kinetics. The amount of attached cells peaked when the bacteria were in late exponential growth phase (6 hours). The peak was followed by a dramatic decrease to a basal level of attached cells. However, compared to their parental strains, rpoX mutants in both backgrounds were reduced in their maximal attached biomass. The structure of the biofilms was observed using SEM. As shown in Figure 5(b), ZJ-O and ΔrpoX-O attached to the surface with single cells, but cells of ZJ-T and ΔrpoX-T were stacked together and formed structures resembling microcolonies. Surprisingly, the ΔrpoX-T cell had a pilus-like structure on the cell surface, which is absent in the other three strains.
Figure 5.
(a) Kinetics of biofilm formation of the V. alginolyticus ZJ-51, wild type (Op: closed circle; Tr: closed triangle), rpoX null mutant (Op: closed diamond; Tr: closed square), and complemented strains (Op: open circle; Tr: open triangle). Bacteria were grown in BF medium in a 24-well polystyrene plate that was incubated statically at 30°C for 2, 4, 6, 8, 10, 12, and 24 hours. Cells were processed as mentioned in Materials and Methods section. Biofilms were quantified by measuring the OD at 540 nm. (b) Observation of biofilm structures by SEM S-3400N: (I) ZJ-O, 7.5 k×, (II) ZJ-T, 7.5 k×, (III) ΔrpoX-O, 7.5 k×, (IV) ΔrpoX-T, 9.0 k×.
3.5. Stress Response
The rpoS mutants in Vibrio species have been shown to be more susceptible to environmental stresses than the wild type [9, 12–15]. Therefore we examined the effect of the rpoX deletion in V. alginolyticus ZJ-O and ZJ-T on the survival of these cells under various stress conditions.
As shown in Figure 6(a), when challenged with heat (45°C) for 90 minutes, more than 60% of the cells had died in all strains. The survival rate for all strains was similar. This suggests that rpoX is not involved in the heat shock response in V. alginolyticus.
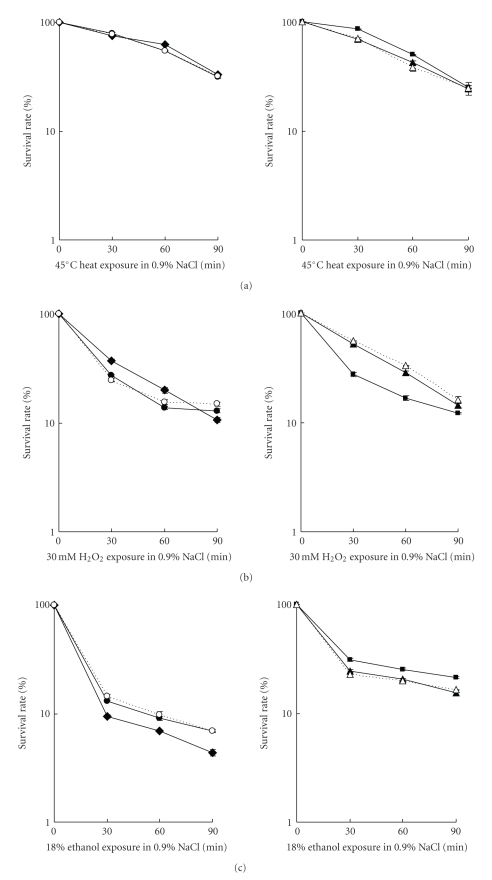
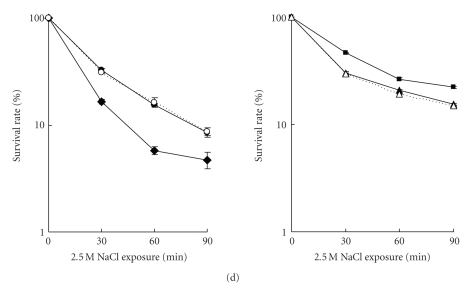
Figure 6.
Stress survival of stationary-phase V. alginolyticus cells. Wild type (Op: closed circle; Tr: closed triangle), rpoX null mutant (Op: closed diamond; Tr: closed square), and complemented strains (Op: open circle; Tr: open triangle) were exposed to various stress conditions and the survival rate was determined as percentage of viable cells at the indicated time points. One hundred percent survival corresponds to number of viable cells prior to exposure to the indicated stress: (a) 45°C heat shock, (b) 30 mM H2O2, (c) 18% ethanol (d) and 2.5 M NaCl.
When exposed to hydrogen peroxide (30 mM H2O2), the rpoX mutant in the Op background was slightly better than the wild type to survive the first 60 minutes, but after 90 minutes, no differences could be observed. ΔrpoX in the Tr background resulted in cells more sensitive to hydrogen peroxide, compared to wild type. This phenotype could be complemented (Figure 6(b)).
Interestingly, when exposed to 18% ethanol and/or to 2.5 M NaCl (hyperosmolarity), the ΔrpoX in the Op background showed decreased survival rates than the wild type and complemented strain. However, deletion of rpoX in the Tr background resulted in higher survival ability to ethanol and NaCl than the wild and complemented cells (Figures 6(c) and 6(d)). This suggests that the function of rpoX is connected to the colony morphotype: in the Op background, rpoX is required for resistance towards ethanol and hyperosmolarity, whereas in the Tr background, rpoX may be detrimental, as its loss increases resistance.
4. Discussion
In this study, the gene rpoX, an rpoS-like sigma factor from V. alginolyticus ZJ-51, was identified and sequenced. The nucleotide sequence showed no similarity with other genes, except for rpoS of V. splendidus. In contrast the deduced protein exhibited a certain level of identity to rpoS-like sigma factors in a few Vibrio species. rpoX contains three conserved domains: (i) sigma 70 region 1.2, (ii) region 2, and (iii) region 4. Sequence alignments revealed that the similarity to rpoS originated from region 2. All members of the σ70 family contain regions 2 and 4 [3, 4]. Region 2 is the most conserved region in the family since it contains both the −10 promoter recognition residues and the primary core RNAP binding domain, which is essential for the specificity and initiation of transcription [4, 23]. Compared to rpoS and RpoQ, rpoX does not have the sigma 70 region 3. Instead a sigma 70 region 1.2 is present, which is absent in the rpoS-like sigma factors of Vibrio sp. MED222 and V. splendidus LGP32. The σ70 region 1.2 is a subregion of region 1 and is found in the σ70 family except for the Sigma B of Myxococcus xanthus and the FliA protein of Salmonella typhimurium. Region 3 forms a discrete compact three-helix domain within the sigma factor. This region is usually not involved in the recognition of promoter DNA. But region 3 plays an important role in binding the core RNA polymerase in the holoenzyme for some specific bacterial promoters that contain an extended −10 promoter element [4, 23]. This region is weakly conserved or virtually absent from most of the sigma factors including the ECF subfamily [4]. The difference in the conserved domains suggests that rpoX may have functions other than those exerted by rpoS.
In a previous study we have shown that the Tr variant produced copious biofilms with mature three-dimensional structures in contrast to the basic level of biofilm with mostly individual cells attached on the glass that is formed by the Op variant [18]. A deletion of rpoX did not affect the kinetics of biofilm formation. However, this gene could be involved in the attachment and not in the maturation of the biofilm, most likely through a change in cell surface properties. This theory is supported by accessory filamentous-like structures observed on the attached cells of ΔrpoX-T, though we do not yet know the role of these structures in the biofilm formation of V. alginolyticus. In comparison, the rpoS gene was reported to be important for the maturation of a biofilm via regulation of the energy metabolism and motility [24]. Deletion of the rpoS gene decreased the attachment of E. coli to the surface and the attached cells maintained as microcolonies and did not develop a mature biofilm [25].
Although the rpoX gene is important for survival of V. alginolyticus in high concentrations of hydrogen peroxide, the effect is not as strong as in an rpoS deletion [6, 9, 15, 26]. This suggests that rpoX might assist rpoS to ensure survival in the presence of hydrogen peroxide. However, the rpoX was not required for heat shock, although rpoS is important for that [15]. In the present study, rpoX had no effect on the rate of colony phase variation, but interestingly, when the cells were treated with ethanol and or sodium chloride, the effect of the rpoX gene on survival ability depended upon the strain background (translucent versus opaque). In our study on ValR, the master regulator of the quorum sensing signaling pathway, we observed that ValR regulated flagella and exopolysaccharide biosynthesis genes in a colony phase dependent way (submitted manuscript). Sequence alignment revealed that rpoX shared 43% identity with the rpoS-like sigma factor (RpoQ) of Vibrio fischeri ES114. This sigma factor was shown to be regulated by quorum sensing system [27]. Therefore, we suggest that the rpoX gene is regulated by the quorum sensing system.
In conclusion, the rpoX sigma factor in V. alginolyticus has functions that are different from those of the rpoS sigma factor. Although both sigma factors are involved in biofilm formation and stress response, the regulatory mechanism of rpoX and the relationship between rpoX and rpoS require further investigation. Moreover, it is important to elucidate the effect of colony phase variation on the regulation of rpoX, which may help this pathogen to survive changes in the environment.
Acknowledgments
This work was supported by National Basic Research Program of China (no. 2006CB101803), the Program for Young Scholars of SCSIO (2007), the Open Research Fund Program of the Key Laboratory of Aquaculture Resources and Technology (Chinese Academy of Fishery Sciences), and the Open Research Fund Program of LMB (2007A01) which are gratefully acknowledged. The first and the second authors contributed equally to the paper.
References
- 1.Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Current Opinion in Microbiology. 2003;6(2):93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 2.Hengge-Aronis R. A role for the σS subunit of RNA polymerase in the regulation of bacterial virulence. Advances in Experimental Medicine and Biology. 2000;485:85–93. doi: 10.1007/0-306-46840-9_11. [DOI] [PubMed] [Google Scholar]
- 3.Paget MSB, Helmann JD. The σ70 family of sigma factors. Genome Biology. 2003;4(1):p. 203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lonetto M, Gribskov M, Gross CA. The σ70 family: sequence conservation and evolutionary relationships. Journal of Bacteriology. 1992;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Advances in Microbial Physiology. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 6.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Molecular Microbiology. 1991;5(1):49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 7.Fang FC, Libby SJ, Buchmeier NA, et al. The alternative σ factor katF (rpoS) regulates Salmonella virulence. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(24):11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badger JL, Miller VL. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. Journal of Bacteriology. 1995;177(18):5370–5373. doi: 10.1128/jb.177.18.5370-5373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildiz FH, Schoolnik GK. Role of rpoS in stress survival and virulence of Vibrio cholerae. Journal of Bacteriology. 1998;180(4):773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos-Gonzalez MI, Molin S. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. Journal of Bacteriology. 1998;180(13):3421–3431. doi: 10.1128/jb.180.13.3421-3431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SEH, Ohman DE. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. Journal of Bacteriology. 1999;181(13):3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YH, Miyamoto C, Meighen EA. Cloning, sequencing, and functional studies of the rpoS gene from Vibrio harveyi. Biochemical and Biophysical Research Communications. 2002;293(1):456–462. doi: 10.1016/S0006-291X(02)00245-0. [DOI] [PubMed] [Google Scholar]
- 13.Rosche TM, Smith DJ, Parker EE, Oliver JD. RpoS involvement and requirement for exogenous nutrient for osmotically induced cross protection in Vibrio vulnificus. FEMS Microbiology Ecology. 2005;53(3):455–462. doi: 10.1016/j.femsec.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Weber B, Croxatto A, Chen C, Milton DL. RpoS induces expression of the Vibrio anguillarum quorum-sensing regulator VanT. Microbiology. 2008;154(3):767–780. doi: 10.1099/mic.0.2007/014167-0. [DOI] [PubMed] [Google Scholar]
- 15.Tian Y, Wang QY, Liu Q, Ma Y, Cao XD, Zhang Y. Role of RpoS in stress survival, synthesis of extracellular autoinducer 2, and virulence in Vibrio alginolyticus. Archives of Microbiology. 2008;190(5):585–594. doi: 10.1007/s00203-008-0410-6. [DOI] [PubMed] [Google Scholar]
- 16.Balebona MC, Andreu MJ, Bordas MA, Zorilla I, Morinigo MA, Borrego JJ. Pathogenicity of Vibrio alginolyticus for cultured gilt-head sea bream (Sparus aurata L.) Applied and Environmental Microbiology. 1998;64(11):4269–4275. doi: 10.1128/aem.64.11.4269-4275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie ZY, Hu CQ, Chen C, Zhang LP, Ren CH. Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Letters in Applied Microbiology. 2005;41(2):202–207. doi: 10.1111/j.1472-765X.2005.01688.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Xie J, Hu CQ. Phenotypic and genetic differences between opaque and translucent colonies of Vibrio alginolyticus. Biofouling. 2009;25(6):525–531. doi: 10.1080/08927010902964578. [DOI] [PubMed] [Google Scholar]
- 19.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. Journal of Bacteriology. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio-Technology. 1983;1(9):787–796. [Google Scholar]
- 21.Milton DL, O'Toole R, Hörstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. Journal of Bacteriology. 1996;178(5):1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passey S, Pellegrin S, Mellor H. Scanning electron microscopy of cell surface morphology. Current Protocols in Cell Biology. 2007;37, chapter 4, unit 4.17:1–13. doi: 10.1002/0471143030.cb0417s37. [DOI] [PubMed] [Google Scholar]
- 23.Campbell EA, Muzzin O, Chlenov M, et al. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Molecular Cell. 2002;9(3):527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 24.Ito A, May T, Kawata K, Okabe S. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnology and Bioengineering. 2008;99(6):1462–1471. doi: 10.1002/bit.21695. [DOI] [PubMed] [Google Scholar]
- 25.Adams JL, McLean RJC. Impact of rpoS deletion on Escherichia coli biofilms. Applied and Environmental Microbiology. 1999;65(9):4285–4287. doi: 10.1128/aem.65.9.4285-4287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson M, Pollitt CE, Roberts IS, Eastgate JA. Identification and characterization of the Erwinia amylovora rpoS gene: rpoS is not involved in induction of fireblight disease symptoms. Journal of Bacteriology. 1998;180(24):6789–6792. doi: 10.1128/jb.180.24.6789-6792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandel MJ, Stabb EV, Ruby EG. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics. 2008;9, article 138 doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]