Abstract
Bacterial and archaeal aerobic communities were recovered from sediments from the shallow El-Djerid salt lake in Tunisia, and their salinity gradient distribution was established. Six samples for physicochemical and microbiological analyses were obtained from 6 saline sites in the lake for physico-chemical and microbiological analyses. All samples studied were considered hypersaline with NaCl concentration ranging from 150 to 260 g/L. A specific halophilic microbial community was recovered from each site, and characterization of isolated microorganisms was performed via both phenotypic and phylogenetic approaches. Only one extreme halophilic organism, domain Archaea, was isolated from site 4 only, whereas organisms in the domain Bacteria were recovered from the five remaining sampling sites that contained up to 250 g/L NaCl. Members of the domain Bacteria belonged to genera Salicola, Pontibacillus, Halomonas, Marinococcus, and Halobacillus, whereas the only member of domain Archaea isolated belonged to the genus Halorubrum. The results of this study are discussed in terms of the ecological significance of these microorganisms in the breakdown of organic matter in Lake El-Djerid and their potential for industry applications.
1. Introduction
Hypersaline environments are found in a wide variety of aquatic and terrestrial ecosystems. They are inhabited by halotolerant microorganisms but also halophilic microorganisms ranging from moderate halophiles with higher growth rates in media containing between 0.5 M and 2.5 M NaCl to extreme halophiles with higher growth rates in media containing over 2.5 M NaCl [1]. Aerobic, anaerobic, and facultative anaerobic microbes belonging to domains Archaea and Bacteria have been recovered from these extreme ecosystems, where they participate in overall organic matter oxidation [2–6].
Moderate and extreme halophiles have been isolated not only from hypersaline ecosystems (salt lakes, marine salterns and saline soils) but also from alkaline ecosystems (alkaline lakes). The most widely studied ecosystems are the Great Salt Lake (Utah, USA), the Dead Sea (Israël), the alkaline brines of Wadi Natrun (Egypt), and Lake Magadi (Kenya) [7–9]. It is noteworthy that low taxonomic biodiversity is observed in all these saline environments [10, 11], most probably due to the highly salt concentrations measured in these environments.
To adapt to high saline conditions, halophilic microorganisms have developed various biochemical strategies, including compatible solute synthesis to maintain cell structure and function [12–14]. These solutes (e.g., ectoïne) plus other compounds (bacteriorhodopsins, exopolysaccharides, hydrolases, biosurfactants) produced by halophilic microbes are clearly of industrial interest. Besides these metabolical and physiological features, halophilic microorganisms are known to play important roles in fermenting fish sauces and in transforming and degrading waste and organic pollutants in saline wastewaters [15–17].
Southern Tunisia features numerous ecosystems including extreme (hypersaline) environments in which microbial diversity has been poorly studied. El-Djerid Sebkha, the largest saline lake (5000 km2) in southern Tunisia, is an important source of salt for food, but its microbial diversity has never yet been studied. Given its economic value for the region as a salt source, we conducted a microbial survey to gain better knowledge of the microbial diversity thriving in this extreme ecosystem. The purpose of this research was to chemically analyse salt and brine samples collected from the lake, isolate any novel extremely halophilic aerobic or facultative anaerobic microorganisms, and examine their phenotypic features and physiological and biochemical characteristics with a view to screening for metabolites of industrial interest produced by the novel halophilic isolates.
2. Material and Methods
2.1. Sample Collections
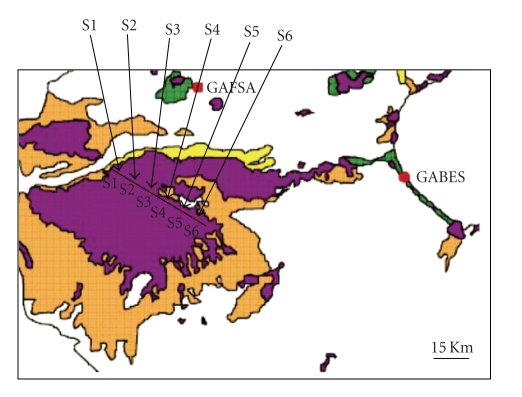
The studied strains were isolated from water and sediments of the El-Djerid Sebkha, a shallow lake located in southern Tunisia. According to in situ physico-chemical conditions and level of wastewater pollutants, the Sebkha was divided into six experimental sites (Figure 1). The samples were collected in February 2006. Water and sediment samples were collected at the surface and at various depths (0.1, 0.2, 0.3 m) in each site. All samples were collected into sterile bottles and stored in ice boxes in the laboratory.
Figure 1.
Site map of the El-Djerid lake (Tunisia) and sampling points.
2.2. Physicochemical Analysis of the Samples
pH, moisture content, and Na+, K+, Ca2+, Mg2+, and Cl− content of the salt and sediment samples were measured according to standard methods of Trussel et al. [18]; Cl− was quantified by titration with AgNO3, Mg2+ was quantified by atomic absorption spectrophotometry, Na+ was quantified by flame spectrophotometry, and Ca2+ was quantified by complexometry using EDTA. Temperature and pH were measured in situ.
2.3. Enrichment and Isolation
Enrichment cultures and isolation procedures to recover aerobic or facultatively anaerobic moderately to extremely halophilic microorganisms were performed in medium containing (per liter): NaCl, 250 g; MgCl2 6H2O, 13 g; MgSO4 7H2O, 20 g; KCl, 4 g; CaCl2 2H2O, 1 g; NaBr, 0.5 g; NaHCO3, 0.2 g; yeast extract, 5 g; tryptone, 8 g; and glucose, 1 g. pH was adjusted to 7.2 with 10 M NaOH before autoclaving. Enrichment cultures were subcultured several times under the same conditions. Strains were grown in 100 mL of medium in 250-mL Erlenmeyer flasks in a rotary shaker at 37°C under agitation at 150 rpm. Aliquots (100 μl) of 10−1–10−4 dilutions were plated onto agar medium. After two weeks of incubation at 37°C, there were red, orange-red, pale-pink, yellowish, cream, white, and transparent colonies. Different colonies were picked and restreaked several times to obtain pure cultures. Microbial cultures were stored at −80°C in the isolation medium supplemented with 50% glycerol.
2.4. Characterization and Identification of Isolates
Among the 130 strains isolated, only 36 showed different phenotypic characteristics and phylogenetic signatures (ARDRA, 16S rRNA gene sequences). These were chosen for further characterization. Isolates were examined for colony and cell morphology and motility. Colonial morphologies were described using standard microbiological criteria, with special emphasis on pigmentation, diameter, colonial elevation, consistency, and opacity [19]. These characteristics were described for cultures grown at optimum temperature, pH, and salt concentration.
For biochemical tests, the strains were grown in flasks and cultures were incubated at 37°C. The optimal ionic content (per liter: 4 g of KCl, 13 g of MgCl2 6H2O, 1 g of CaCl2 2H2O, 20 g of MgSO4 7H2O, 0.5 g of NaBr, 0.2 g of NaHCO3, 250 g of NaCl) was used in all the biochemical test media. Oxidase reaction was performed according to Kovacs (1956) [20]. Catalase was determined by adding 10 volumes of H2O2 to each strain culture (after 18 hour incubation at 37°C) on solid medium. Gelatinase, β-galactosidase, urease, indol production, and Voges-Proskauer tests were performed using standard procedures. Other phenotypic characteristics were determined using API 20E and API 20NE kits (BioMérieux, Marcy l'Etoile, France) according to Logan and Berkeley (1984) [21].
2.5. PCR Amplification of 16S rDNA
The DNA from bacterial cultures was extracted using a Wizard Genomic DNA Purification Kit. The 16S rRNA gene of the isolate strain was amplified by adding 1 μL of cell culture to a thermocycler microtube containing 5 μL of 10 × taq buffer, 0.5 μL of each 50 nM Fd1 and Rd1 primers, 5 μL of 25 mM MgCl2 6H2O, 0.5 μL of 25 mM dNTPs, 0.5 μL of Taq polymerase (5U μL−1), and 38 μL of sterilized distilled water. Universal primers Fd1 and Rd1 (Fd1, 5′-AGAGTTTGATCCTGGCTCAG-3′ and Rd1, 5′-AAGGAGGTGATCCAGCC-3′) were used to obtain a PCR product of ~1.5 kb corresponding to base positions 8-1542 based on Escherichia coli numbering of the 16S rRNA gene [22]. The sample was placed in a hybrid thermal reactor thermocycler (BIOMetra, Leusden, The Netherlands), denatured for 1 minute at 95°C and subjected to 30 cycles for 20 seconds at 95°C, 30 seconds at 55°C, and 1 minute and 30 seconds at 72°C. This was followed by a final elongation step for 5 minutes at 72°C. The PCR products were analysed on 1% (w/v) agarose gels and sent to GATC (Germany) for sequencing. Sequence data were imported into the BioEdit version 5.0.9 sequence editor [23]; base-calling was examined, and a contiguous sequence was obtained. The full sequence was aligned using the RDP Sequence Aligner program [24]. The consensus sequence was manually adjusted to conform to the 16S rRNA gene secondary structure model [22]. A nonredundant BLAST search [25] identified its closest relatives. Sequences used in the phylogenetic analysis were obtained from the RDP [24] and GenBank databases [26]. Sequence positions and alignment ambiguities were eliminated and pairwise evolutionary distances were calculated using the method of Jukes and Cantor (1969) [27]. A dendrogram was constructed using the neighbour-joining method [28]. Confidence in tree topology was determined using 100-bootstrapped trees [29].
2.6. Restriction Endonuclease Digestions
Enzymatic digestions were performed by incubating 5 μL of the PCR products with 10 U of each endonuclease and the corresponding enzyme buffer. Digestions were continued for one hour at 37°C for AluI, HaeIII, and RsaI. Digested products were analysed on 2% (w/v) agarose gels.
3. Results
3.1. Physicochemical Analyses
Temperature at the sampling sites was 15°C at 6 A.M. The physico-chemical characteristics of the sediment samples are shown in Table 1. The pH of sediment samples was between 7.8 and 8.8. The highest moisture content values were found in the S4 sample. Na+ content was the highest in the S5 sample, and Ca2+ content was the highest in the S3 sample, whereas K+ concentration was the highest in the S2 sample (Table 1). Total salt composition was higher at the S4 sampling site (335 mg/g) than the other sampling sites (Table 1). All sediment samples from the studied lake were dominated by Cl− and high levels of Ca2+. Total ionic composition of the lake differed depending on the area sampled. Given the mineral composition of the lake and its concentration in Na+, K+, Ca2+, Mg2+, and Cl−, it should clearly be inhabited by halophilic microorganisms, thus justifying the microbial survey.
Table 1.
Physico-chemical characteristics of the sediment samples.
| Sampling site | Colour of sampling site | pH | Hardness (%) | Ca2+ (mg/g) | Mg2+ (mg/g) | Cl− (mg/g) | Na+ (mg/g) | K+ (mg/g) | Total |
|---|---|---|---|---|---|---|---|---|---|
| S1 | Dark-cream | 7.9 | 7.54 | 80.52 | 19.42 | 131.50 | 26.82 | 8.55 | 266.83 |
| S2 | Cream | 7.5 | 4.22 | 68.28 | 21.37 | 177.96 | 44.62 | 14.40 | 326.65 |
| S3 | Cream | 7.9 | 6.92 | 82.43 | 15.84 | 113.66 | 69.22 | 7.10 | 288.27 |
| S4 | Cream | 7.3 | 7.78 | 57.94 | 12.26 | 190.50 | 67.14 | 7.48 | 335.34 |
| S5 | Dark-cream | 8.3 | 6.60 | 64.36 | 14.94 | 110.93 | 76.47 | 10.42 | 277.14 |
| S6 | Brown-black | 8.2 | 7.29 | 75.57 | 20.48 | 141.60 | 59.41 | 10.45 | 307.54 |
|
| |||||||||
| Average | 7.87 | 6.72 | 71.52 | 17.39 | 144.36 | 57.28 | 9.73 | 300.29 | |
3.2. Microbiological Analyses
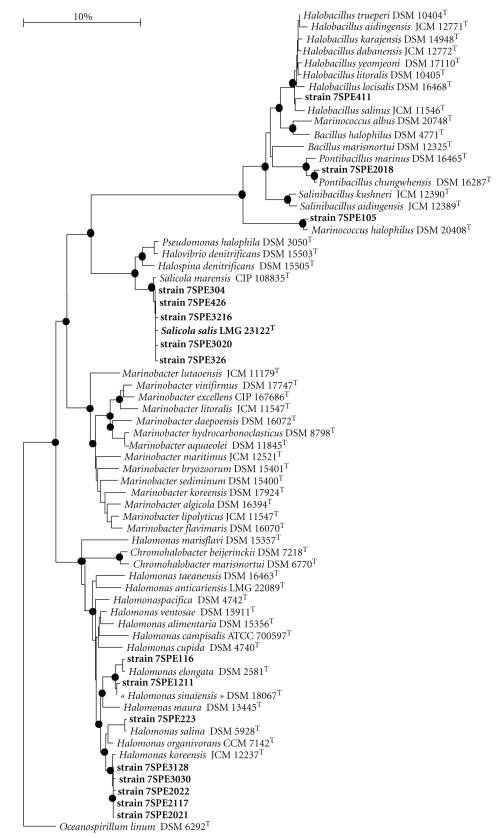
After several dilutions and subculturing in the same liquid medium under aerobic conditions, colonies were isolated in the agar medium containing 25% NaCl. A total of 130 extremely halophilic strains were isolated under aerobic conditions from the six samples. However, on the basis of phenotypic characteristics (macro and microscopic analysis), physiological analyses (NaCl, pH), biochemical tests (API 20E, API 20NE), and molecular approaches [PCR 16S, ARDRA (digestion by three enzymes AluI, HaeIII and RsaI)], only 36 isolates were selected and examined in greater detail. These strains were identified by analyzing sequences of genes encoding for 16S rRNA (Figure 2). The highest total bacterial number (1 × 104 cfu/g) growing under aerobic conditions was found in the S1 sampling site. Some colonies were white or transparent whereas others showed various pigmentations, that is, red, orange-red, bright-pink, or yellowish-cream. Cream-coloured colonies were found to be the most numerous in the lake (Table 3).
Figure 2.
16S rRNA gene-based phylogenetic tree of the bacterial domain, including 16S rDNA sequences from sediment samples from El-Djerid lake. Topologies of the phylogenetic tree built using maximum-likelihood and maximum-parsimony algorithms were similar to those of the tree constructed by neighbour-joining analysis. Solid circles indicate nodes with a bootstrap value higher than 80%.
Table 3.
Phenotypic features of the 36 strains studied.
| Strains (7SPE) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sampling sites (S)/depth (cm)/ characteristics | 2021 S2/0 | 2117 S2/10 | 2022 S2/0 | 3′030S3/0 | 3′128S3/10 | 223 S2/20 | 1′211S1/20 | 116 S1/10 |
|
| ||||||||
| Taxonomical status | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. |
| Colonial morphology | Circular | Circular | Circular | Circular | Circular | Circular | irregular and spreading | irregular and spreading |
| Colony size | 1 mm | 1-2 mm | 1 mm | 1-2 mm | 1 mm | 1-2 mm | 2 mm | 1-2 mm |
| Colony | convex | convex | convex | slightly raised | flat | convex | convex | convex |
| Colony density | opaque matt | opaque matt | opaque matt | translucent glistening | opaque matt | opaque matt | translucent glistening | translucent glistening |
| Pigmentation | cream | white | cream | transparent-white | cream | cream | white | white |
| Cell shape | pleomorphic rods | pleomorphic rods | pleomorphic cells | pleomorphic cells | pleomorphic rods | pleomorphic cells | pleomorphic cells | pleomorphic rods |
| Cell arrangement | single and paired cells | single, paired cells and long chains | single and paired cells | single and paired cells | paired cells and long chains | single and paired cells | single and paired cells | paired chains |
| Chains | − | + | − | − | + | − | − | + |
| Motile | − | − | − | − | + | − | − | + |
| Cell size; length and width (μm) | 2–5 × 1 | 3–7.5 × 1 | 1–4 × 1 | 1–4 × 1 | 2–5 × 1 | 3–7.5 × 1 | 2–7.5 × 1.75 | 2–7.5 × 1 |
| Oxidase | + | − | + | − | + | − | − | − |
| Catalase | − | − | − | − | − | − | + | − |
|
| ||||||||
| Growth at 37°C, pH 7.2 | ||||||||
| 0% NaCl | − | − | − | − | − | − | − | − |
| 2% NaCl | + | − | − | − | + | − | + | + |
| 5% NaCl | + | − | − | − | + | − | + | + |
| 8% NaCl | + | + | − | − | + | + | + | + |
| 10% NaCl | + | + | + | + | + | + | + | + |
| 15% NaCl | + | + | + | + | + | + | + | + |
| 25% NaCl | + | + | + | + | + | + | + | + |
| 30% NaCl | − | − | − | − | − | − | − | − |
|
| ||||||||
| Growth at 37°C | ||||||||
| pH 4.5 | − | − | − | − | − | − | − | − |
| pH 6 | + | + | + | + | + | + | + | + |
| pH 7 | + | + | + | + | + | + | + | + |
| pH 7.5 | + | + | + | + | + | + | + | + |
| pH 8 | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20E: | ||||||||
| Hydrolysis of: | ||||||||
| ONPG | − | − | − | − | − | − | − | − |
| Arginine dihydrolase | + | + | + | + | + | + | + | + |
| Lysine decarboxylase | + | + | + | + | + | + | + | + |
| Ornithine decarboxylase | + | + | + | + | + | + | + | + |
| Citrate utilization | − | − | − | − | − | − | − | + |
| H2S production | − | − | − | − | − | − | − | − |
| Urease | + | + | + | + | + | + | + | + |
| Tryptophan deaminase | − | − | − | − | − | − | − | − |
| Indol production | − | − | − | − | − | − | − | − |
| Voges-Proskauer | + | + | + | − | − | − | − | − |
| Gelatinase activity | − | − | − | − | − | − | − | − |
| Fermentation/oxidation: | ||||||||
| D-glucose | − | − | − | − | − | − | − | − |
| D-mannitol | − | − | − | − | − | − | − | − |
| Inositol | − | − | − | − | − | − | − | − |
| D-sorbitol | − | − | − | − | − | − | − | − |
| L-rhamnose | − | − | − | − | − | − | − | − |
| D-sucrose | − | − | − | − | − | − | − | − |
| D-melibiose | − | − | − | − | − | − | − | − |
| Amygdalin | − | − | − | − | − | − | − | − |
| L-arabinose | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20NE: | ||||||||
| NO3 reduction | − | − | − | − | − | − | + | + |
| NO2 reduction | − | − | − | − | − | − | − | − |
| Hydrolysis of: | ||||||||
| Aesculin | − | − | − | − | − | − | − | − |
| PNPG | − | − | − | − | − | − | − | − |
| Assimilation of: | ||||||||
| D-mannose | − | − | − | − | − | − | − | − |
| N-acetyl-glucosamine | + | − | − | − | − | − | − | + |
| D-maltose | + | − | − | − | − | − | − | − |
| Potassium gluconate | − | − | − | − | − | − | − | − |
| Capric acid | − | − | − | − | − | − | − | − |
| Adipic acid | − | − | − | − | − | − | − | − |
| Malic acid | + | − | − | − | − | − | − | + |
| Phenylacetic acid | − | − | − | − | − | − | − | − |
|
| ||||||||
| Sampling sites (S)/depth (cm)/ characteristics | 604 S6/0 | 6′02S6/0 | 419 S4/10 | 402 S4/0 | 605 S6/0 | 2019 S2/0 | 214 S2/10 | 403 S4/0 |
|
| ||||||||
| Taxonomical status | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. |
| Colonial morphology | Circular | Circular | Circular | Circular | irregular and spreading | Circular | Circular | irregular and spreading |
| Colony size | 1 mm convex | 1-2 mm convex | 1-2 mm convex | 1-2 mm convex | 1-2 mm convex | 1 mm convex | 1 mm flat | 3 mm convex |
| Colony density | transparent glistening | opaque matt | opaque matt | opaque matt | translucent glistening | opaque matt | opaque matt | translucent glistening |
| Pigmentation | cream | white | cream | white | white | white | cream | white |
| Cell shape | pleomorphic cells | pleomorphic cells | pleomorphic rods | pleomorphic cells | pleomorphic cells | pleomorphic cells | pleomorphic cells | pleomorphic cells |
| Cell arrangement | single and paired cells | single and paired cells | long chains | single and paired cells | single and paired cells | single and paired cells | single and paired cells | single and paired cells |
| Chains | − | − | + | + | + | + | − | + |
| Motile | − | − | − | + | − | + | + | + |
| Cell size; length and width (μm) | 2–5 × 1 | 3–7.5 × 1.5 | 3–7.5 × 1 | 2–7.5 × 1 | 2–7.5 × 1 | 2–5 × 1 | 2–5 × 1 | 2–7.5 × 1 |
| Oxidase | − | − | − | − | − | − | − | − |
| Catalase | − | − | − | − | − | − | − | − |
|
| ||||||||
| Growth at 37°C, pH 7.2 | ||||||||
| 0% NaCl | − | − | − | − | − | − | − | − |
| 2% NaCl | − | − | − | + | + | + | + | + |
| 5% NaCl | − | − | − | + | + | + | + | + |
| 8% NaCl | + | + | + | + | + | + | + | + |
| 10% NaCl | + | + | + | + | + | + | + | + |
| 15% NaCl | + | + | + | + | + | + | + | + |
| 25% NaCl | + | + | + | + | + | + | + | + |
| 30% NaCl | − | − | − | − | − | − | − | − |
|
| ||||||||
| Growth at 37°C | ||||||||
| pH 4.5 | − | − | − | − | − | − | − | − |
| pH 6 | + | + | + | + | + | + | + | + |
| pH 7 | + | + | + | + | + | + | + | + |
| pH 7.5 | + | + | + | + | + | + | + | + |
| pH 8 | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20E: | ||||||||
| Hydrolysis of: | ||||||||
| ONPG | − | − | − | − | − | − | − | − |
| Arginine dihydrolase | + | + | + | + | + | + | + | + |
| Lysine decarboxylase | + | + | + | + | + | + | + | + |
| Ornithine decarboxylase | + | + | + | + | + | + | + | + |
| Citrate utilization | − | − | − | + | + | − | − | + |
| H2S production | − | − | − | − | − | − | − | − |
| Urease | + | + | + | + | + | + | + | + |
| Tryptophan deaminase | − | − | − | − | − | − | − | − |
| Indol production | − | − | − | − | − | − | − | − |
| Voges-Proskauer | − | − | − | − | − | − | − | − |
| Gelatinase activity | − | − | − | − | − | − | − | − |
| Fermentation/oxidation: | ||||||||
| D-glucose | − | − | − | − | − | − | − | + |
| D-mannitol | − | − | − | − | − | − | − | − |
| Inositol | − | − | − | − | − | − | − | − |
| D-sorbitol | − | − | − | − | − | − | − | − |
| L-rhamnose | − | − | − | − | − | − | − | − |
| D-sucrose | − | − | − | − | − | − | − | − |
| D-melibiose | − | − | − | − | − | − | − | − |
| Amygdalin | − | − | − | − | − | − | − | − |
| L-arabinose | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20NE: | ||||||||
| NO3 reduction | − | − | − | + | + | + | − | + |
| NO2 reduction | − | − | − | − | − | − | − | − |
| Hydrolysis of: | ||||||||
| Aesculin | − | − | − | − | − | − | − | − |
| PNPG | − | − | − | − | − | − | − | − |
| Assimilation of: | ||||||||
| D-mannose | − | − | − | − | − | − | − | − |
| N-acetyl-glucosamine | − | − | − | + | − | − | − | − |
| D-maltose | − | − | − | − | − | − | − | − |
| Potassium gluconate | − | − | − | − | − | − | − | − |
| Capric acid | − | − | − | − | − | − | − | − |
| Adipic acid | − | − | − | − | − | − | − | − |
| Malic acid | − | − | − | − | − | − | − | − |
| Phenylacetic acid | − | − | − | − | − | − | − | − |
|
| ||||||||
| Sampling sites (S)/depth (cm)/ characteristics | 139 S1/30 | 2015 S2/0 | 1′010S1/0 | 2230 S2/20 | 101 S1/0 | 1′115S1/10 | 1′213S1/20 | 108 S1/0 |
|
| ||||||||
| Taxonomical status | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. | Halomonas sp. |
| Colonial morphology | Irregular and spreading | Circular | Circular | Circular | Circular | irregular and spreading | Circular | irregular and spreading |
| Colony size | 1-2 mm convex | 1 mm convex | 1-2 mm convex | 1 mm convex | 1 mm convex | 1-2 mm convex | 0.3 mm slightly raised | 1-2 mm convex |
| Colony density | translucent glistening | opaque matt | translucent glistening | opaque matt | translucent glistening | translucent glistening | translucent matt | translucent glistening |
| Pigmentation | white | white | white | cream | white | white | white | white |
| Cell shape | pleomorphic cells | Pleomorphic rods | pleomorphic cells | pleomorphic cells | pleomorphic rods | pleomorphic rods | pleomorphic cells | pleomorphic cells |
| Cell arrangement | single and paired cells | long chains | single and paired cells | single and paired cells | paired cells | single and paired cells | single and paired cells | single and paired cells |
| Chains | − | + | − | + | + | + | − | + |
| Motile | − | + | − | + | + | + | − | − |
| Cell size; length and width (μm) | 2–7.5 × 1 | 2–5 × 1 | 2–7.5 × 1.75 | 2–5 × 1 | 2–7.5 × 1 | 2–7.5 × 1.5 | 2–7.5 × 1 | 2–7.5 × 1 |
| Oxidase | − | + | − | − | − | − | − | − |
| Catalase | − | − | − | − | − | − | + | − |
|
| ||||||||
| Growth at 37°C, pH 7.2 | ||||||||
| 0% NaCl | − | − | − | − | − | − | − | − |
| 2% NaCl | + | + | + | + | + | + | − | + |
| 5% NaCl | + | + | + | + | + | + | + | + |
| 8% NaCl | + | + | + | + | + | + | + | + |
| 10% NaCl | + | + | + | + | + | + | + | + |
| 15% NaCl | + | + | + | + | + | + | + | + |
| 25% NaCl | + | + | + | + | + | − | + | + |
| 30% NaCl | − | − | − | − | − | − | − | − |
|
| ||||||||
| Growth at 37°C | ||||||||
| pH 4.5 | − | − | − | − | − | − | − | − |
| pH 6 | + | + | + | + | + | + | + | + |
| pH 7 | + | + | + | + | + | + | + | + |
| pH 7.5 | + | + | + | + | + | + | + | + |
| pH 8 | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20E: | ||||||||
| Hydrolysis of: | ||||||||
| ONPG | − | − | − | − | − | − | − | − |
| Arginine dihydrolase | + | + | + | + | + | + | + | + |
| Lysine decarboxylase | + | + | + | + | + | + | + | + |
| Ornithine decarboxylase | + | + | + | + | + | + | + | + |
| Citrate utilization | − | − | − | − | + | − | − | − |
| H2S production | − | − | − | − | − | − | − | − |
| Urease | + | + | + | + | + | − | + | + |
| Tryptophan deaminase | − | − | − | − | − | − | − | − |
| Indol production | − | − | − | − | − | − | − | − |
| Voges-Proskauer | − | − | − | − | − | − | − | − |
| Gelatinase activity | − | − | − | − | − | − | − | − |
| Fermentation/oxidation: | ||||||||
| D-glucose | − | − | − | − | − | − | − | − |
| D-mannitol | − | − | − | − | − | − | − | − |
| Inositol | − | − | − | − | − | − | − | − |
| D-sorbitol | − | − | − | − | − | − | − | − |
| L-rhamnose | − | − | − | − | − | − | − | − |
| D-sucrose | − | − | − | − | − | − | − | − |
| D-melibiose | − | − | − | − | − | − | − | − |
| Amygdalin | − | − | − | − | − | − | − | − |
| L-arabinose | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20NE: | ||||||||
| NO3 reduction | + | − | + | − | + | + | + | + |
| NO2 reduction | − | + | − | − | − | − | − | − |
| Hydrolysis of: | ||||||||
| Aesculin | − | − | − | − | − | − | − | − |
| PNPG | − | − | − | − | − | − | − | − |
| Assimilation of: | ||||||||
| D-mannose | − | − | − | − | − | − | − | − |
| N-acetyl-glucosamine | − | − | − | − | − | − | − | − |
| D-maltose | − | − | − | − | − | − | − | − |
| Potassium gluconate | − | − | − | − | − | − | − | − |
| Capric acid | − | − | − | − | − | − | − | − |
| Adipic acid | − | − | − | − | − | − | − | − |
| Malic acid | − | − | − | − | − | − | − | − |
| Phenylacetic acid | − | − | − | − | − | − | − | − |
|
| ||||||||
| Sampling sites (S)/depth (cm)/ characteristics | 137 S1/30 | 103 S1/0 | 3′020S3/0 | 326 S3/20 | 304 S3/0 | 3′09S3/0 | 426 S4/20 | 3216 S3/20 |
|
| ||||||||
| Taxonomical status | Halomonas sp. | Halomonas sp. | Salicola sp. | Salicola sp. | Salicola sp. | Salicola sp. | Salicola sp. | Salicola sp. |
| Colonial morphology | irregular and spreading | Circular | Circular | Circular | Circular | Circular | irregular and spreading | Circular |
| Colony size | 1-2 mm convex | 0.5–1 mm convex | 0.3 mm flat | 0.3 mm convex | 0.3 mm flat | 0.2-0.3 mm flat | 1 mm flat | 0.2-0.3 mm slightly raised |
| Colony density | translucent glistening | translucent matt | opaque matt | translucent matt | opaque matt | transparent matt | opaque matt | translucent matt |
| Pigmentation | white | white | yellowish- cream | white | transparent white | transparent- white | transparent- white | transparent- white |
| Cell shape | pleomorphic cells | pleomorphic cells | pleomorphic rods | pleomorphic cells | pleomorphic rods | pleomorphic cells | pleomorphic cells | pleomorphic cells |
| Cell arrangement | single and paired cells | single and paired cells | single and paired cells | single and paired cells | single and paired cells | single and paired cells | single and paired cells | single and paired cells |
| Chains | − | + | − | + | + | − | − | − |
| Motile | − | + | − | − | − | − | + | − |
| Cell size; length and width (μm) | 2–7.5 × 1 | 2–7.5 × 1.5 | 1–3 × 0.5 | 1–4 × 0.5 | 1–3 × 0.5 | 1–3 × 0.5 | 1–4 × 0.5 | 1–4 × 0.5 |
| Oxidase | − | − | − | − | − | − | − | − |
| Catalase | − | − | − | − | − | − | − | − |
|
| ||||||||
| Growth at 37°C, pH 7.2 | ||||||||
| 0% NaCl | − | − | − | − | − | − | − | − |
| 2% NaCl | + | + | − | − | − | − | − | − |
| 5% NaCl | + | + | − | − | − | − | + | − |
| 8% NaCl | + | + | − | + | − | − | + | − |
| 10% NaCl | + | + | + | + | + | + | + | + |
| 15% NaCl | + | + | + | + | + | + | + | + |
| 25% NaCl | + | + | + | + | + | + | + | + |
| 30% NaCl | − | − | − | − | − | − | − | − |
|
| ||||||||
| Growth at 37°C | ||||||||
| pH 4.5 | − | − | − | − | − | − | − | − |
| pH 6 | + | + | + | + | + | + | + | + |
| pH 7 | + | + | + | + | + | + | + | + |
| pH 7.5 | + | + | + | + | + | + | + | + |
| pH 8 | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20E: | ||||||||
| Hydrolysis of: | ||||||||
| ONPG | − | − | − | − | − | − | − | − |
| Arginine dihydrolase | + | + | + | + | + | + | + | + |
| Lysine decarboxylase | + | + | + | + | + | + | + | + |
| Ornithine decarboxylase | + | + | + | + | + | + | + | + |
| Citrate utilization | − | + | − | − | − | − | − | + |
| H2S production | − | − | − | − | − | − | − | − |
| Urease | + | + | + | + | + | + | + | + |
| Tryptophan deaminase | − | − | − | − | − | − | − | − |
| Indol production | − | − | − | − | − | − | − | − |
| Voges-Proskauer | − | − | − | − | − | − | + | − |
| Gelatinase activity | − | − | − | − | − | − | − | − |
| Fermentation/oxidation: | ||||||||
| D-glucose | − | − | − | − | − | − | − | − |
| D-mannitol | − | − | − | − | − | − | − | − |
| Inositol | − | − | − | − | − | − | − | − |
| D-sorbitol | − | − | − | − | − | − | − | − |
| L-rhamnose | − | − | − | − | − | − | − | − |
| D-sucrose | − | − | − | − | − | − | − | − |
| D-melibiose | − | − | − | − | − | − | − | − |
| Amygdalin | − | − | + | + | − | − | − | − |
| L-arabinose | − | − | − | − | − | − | − | − |
|
| ||||||||
| API 20NE: | ||||||||
| NO3 reduction | + | + | + | + | − | − | − | − |
| NO2 reduction | − | − | − | − | − | − | − | − |
| Hydrolysis of: | ||||||||
| Aesculin | − | − | − | − | − | − | − | − |
| PNPG | − | − | − | − | − | − | − | − |
| Assimilation of: | ||||||||
| D-mannose | − | − | − | − | − | − | − | − |
| N-acetyl-glucosamine | − | − | − | − | − | − | − | − |
| D-maltose | − | − | − | − | − | − | − | − |
| Potassium gluconate | − | − | − | − | − | − | − | − |
| Capric acid | − | − | − | − | − | − | − | − |
| Adipic acid | − | − | − | − | − | − | − | − |
| Malic acid | − | − | + | + | − | − | − | − |
| Phenylacetic acid | − | − | − | − | − | − | − | − |
|
| ||||||||
| Sampling sites (S)/depth (cm)/ characteristics | 105 S1/0 | 2018 S2/0 | 411 S4/10 | 4115 S4/10 | ||||
|
| ||||||||
| Taxonomical status | Marinococcus sp. | Pontibacillus sp. | Halobacillus sp. | Halorubrum sp. | ||||
| Colonial morphology | Circular | Circular | Circular | Circular | ||||
| Colony size | 1 mm convex | 1 mm flat | 1 mm flat | 0.3 mm slightly raised | ||||
| Colony density | opaque matt | opaque matt | opaque matt | opaque matt | ||||
| Pigmentation | reddish- orange | cream | cream | brick-red | ||||
| Cell shape | pleomorphic rods | pleomorphic rods | pleomorphic rods | pleomorphic cells | ||||
| Cell arrangement | single, paired and irregularly clustered cells | long chains | long chains | single and paired cells | ||||
| Chains | − | + | + | − | ||||
| Motile | − | + | + | + | ||||
| Cell size; length and width (μm) | 1–5 × 2 | 1–4 × 1 | 1–4 × 1 | 1–4 × 1 | ||||
| Oxidase | − | − | − | − | ||||
| Catalase | + | − | − | − | ||||
|
| ||||||||
| Growth at 37°C, pH 7.2 | ||||||||
| 0% NaCl | − | − | − | − | ||||
| 2% NaCl | − | − | − | − | ||||
| 5% NaCl | + | + | + | − | ||||
| 8% NaCl | + | + | + | + | ||||
| 10% NaCl | + | + | + | + | ||||
| 15% NaCl | + | + | + | + | ||||
| 25% NaCl | + | + | + | + | ||||
| 30% NaCl | − | − | − | − | ||||
|
| ||||||||
| Growth at 37°C | ||||||||
| pH 4.5 | − | − | − | − | ||||
| pH 6 | + | + | + | + | ||||
| pH 7 | + | + | + | + | ||||
| pH 7.5 | + | + | + | + | ||||
| pH 8 | − | − | − | − | ||||
|
| ||||||||
| API 20E: | ||||||||
| Hydrolysis of: | ||||||||
| ONPG | − | − | − | − | ||||
| Arginine dihydrolase | + | + | + | + | ||||
| Lysine decarboxylase | + | + | + | + | ||||
| Ornithine decarboxylase | + | + | + | + | ||||
| Citrate utilization | + | + | + | + | ||||
| H2S production | − | − | − | − | ||||
| Urease | + | + | + | + | ||||
| Tryptophan deaminase | − | − | − | − | ||||
| Indol production | − | − | − | − | ||||
| Voges-Proskauer | − | − | − | − | ||||
| Gelatinase activity | − | − | − | − | ||||
| Fermentation/oxidation: | ||||||||
| D-glucose | − | − | − | − | ||||
| D-mannitol | − | − | − | − | ||||
| Inositol | − | − | − | − | ||||
| D-sorbitol | − | − | − | − | ||||
| L-rhamnose | − | − | − | − | ||||
| D-sucrose | − | − | − | − | ||||
| D-melibiose | − | − | − | − | ||||
| Amygdalin | − | − | − | − | ||||
| L-arabinose | − | − | − | − | ||||
|
| ||||||||
| API 20NE: | ||||||||
| NO3 reduction | − | − | − | − | ||||
| NO2 reduction | − | − | − | − | ||||
| Hydrolysis of: | ||||||||
| Aesculin | − | − | − | − | ||||
| PNPG | − | − | − | − | ||||
| Assimilation of: | ||||||||
| D-mannose | − | − | − | − | ||||
| N-acetyl-glucosamine | − | − | − | − | ||||
| D-maltose | − | − | − | − | ||||
| Potassium gluconate | − | − | − | − | ||||
| Capric acid | − | − | − | − | ||||
| Adipic acid | − | − | − | − | ||||
| Malic acid | − | − | − | − | ||||
| Phenylacetic acid | − | − | − | − | ||||
3.3. Colony and Cell Morphology
The dominant bacterial population comprised motile or nonmotile, gram-positive microorganisms, most of which were spore-forming bacteria. Most colonies on Brown agar medium were 0.5–2 mm in diameter after 3 weeks of incubation. These colonies were smooth, circular, low-convex, transparent or translucent, and entire. Cells of all strains isolated were short, long, and swollen rods that occurred in singles, pairs, or short chains. The cells were approximately 0.5–2 μm wide and 2.5–6.5 μm long. The isolates were facultatively anaerobic or aerobic and required yeast extract for growth. The characteristics of all strains are shown in Table 3. Colony pigmentation from these samples ranged from blood-red to pale-pink. Most colonies were 1 to 2 mm in diameter, circular, convex, glistening, and entire. Optimum growth occurred at 25% (w/v) NaCl, 37°C, and pH 7.3, thus suggesting that these isolates should be considered as extremely halophilic according to the definition of Ventosa et al. [1].
3.4. Biochemical Tests
Selected strains were tested in biochemical test media (Table 3). ONPG and gelatin hydrolysis were found to be negative and tryptophan deaminase was not produced, whereas lysine decarboxylase, ornithine decarboxylase, and arginine dihydrolase were found to be positive for the majority of isolates. Most of the strains reduced nitrate to nitrite (Table 3).
3.5. Phylogenetic Analysis
Based on the enzymatic digestion profiles obtained, 16 representative bacteria of the 36 isolates were chosen for taxonomic and phylogenetic studies. To determine their phylogenetic position, the 16S rRNA gene sequence of each strain was analyzed, and a phylogenetic tree was constructed based on 1280 unambiguous bp (Figure 2). The 16S rRNA gene sequences of the strains have been deposited in the GenBank database
Phylogenetic analysis indicated that the majority of strains isolated are related to genera Halomonas or Salicola, whereas the other strains are most closely related to species of genera Halobacillus, Pontibacillus, and Marinococcus (Figure 2). All strains shared more than 97% identity with their closest phylogenetic relative (Table 2), suggesting that they should be considered at the same species level until the results of DNA/DNA hybridization studies can validate their affiliation (work in progress). Only one strain representative of the domain Archaea was identified as Halorubrum sp., but this microorganism was not further characterized. The 16S rRNA gene sequences recovered from the 16 representative isolated bacteria made up 9 taxonomically distinct microorganisms whose closest phylogenetic relatives are Halobacillus salinus, Pontibacillus chungwhensis, Marinococcus halophilus, Salicola marensis, S. salis, Halomonas elongata, H. sinaiensis, H. salina, and H. koreensis.
Table 2.
Distribution and taxonomic characteristics of bacteria and archaea isolated from the 6 sampling sites in El-Djerid Lake.
| Organisms | Number of strains/site | ||||||
|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S6 | Number of strains | |
| Halomonas sp. | 10 | 8 | 2 | 3 | 0 | 3 | 26 |
| Salicola sp. | 0 | 0 | 5 | 1 | 0 | 0 | 6 |
| Pontibacillus sp. | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Marinococcus sp. | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Halobacillus sp. | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Others (Archaea)(a) | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Total strains | 11 | 9 | 7 | 6 | 0 | 3 | 36 |
(a)data not shown.
4. Discussion
Recent decades have seen a surge in studies on extreme environments including hypersaline ecosystems. Both molecular and microbiological studies have revealed the presence of moderately to extremely halophilic microorganisms in a wide range of these saline environments [9, 30–33]. El-Djerid salt lake is a hypersaline environment in southern Tunisia that is considered athalassohaline because its salt composition derives from the dissolution of minerals of continental origin [34]. Similarly to other hypersaline ecosystems, the lake is subjected to drastic physico-chemical conditions including high salinity, high radiation (UV) and strong changes in temperatures and dryness which make it a relevant study target for microbiologists. To our knowledge, this is the first microbiological study on extremely halophilic aerobic bacteria from El-Djerid salt lake.
Table 1 reports the results of physico-chemical analysis of soil samples from the six sites. The samples differ from those of the other hypersaline environments studied so far. Sodium and potassium concentrations are higher at the six sites than in the Dead Sea in Israël [8]. In contrast to the waters of the Dead Sea and the Great Salt Lake in the USA, which are slightly acidic (pH 6 to 7), the pH of sites 5 and 6 is 8.3 and 8.2, respectively, and should therefore be considered weakly alkaline. The pH of Lakes Wadi Natrun and Magadi (in Kenya) is considered as highly alkaline environments (pH 11) [35, 36].
Throughout the course of this work, we isolated 130 extremely halophilic strains and further characterized 36 of these strains showing different pigmentations with colonies on agar plates (Table 2). Phylogenetic analysis indicated that all isolates were members of five genera of the domain Bacteria, including Salicola, Pontibacillus, Halomonas, Marinococcus, and Halobacillus. Members of the genera Salicola, Pontibacillus, Marinococcus, and Halobacillus are considered aerobic microorganisms, whereas members of genus Halomonas are considered facultative anaerobes able to use nitrate as terminal electron acceptor under anaerobic conditions. All these microorganisms may use various organic compounds including sugars as substrates and should be considered chemoorganotrophs. Almost all these isolates were detected on the surface of sediments as well in the first centimetres down (0.1-0.2 m) of each biotope. Halomonas species were distributed in all 5 sites studied and represented the major strains isolated, especially in site 1. Members of this genus together with those of the genera Salicola, Pontibacillus, Marinococcus, and Halobacillus have also been isolated from other saline environments, including athalassohaline and thalassohaline lakes and marine waters [37–40]. It should be underlined that among the halophilic microbes isolated, only one, which originated from site 4, belonged to the domain Archaea (data not shown). However, the limited number of halophilic archaeons detected in El-Djerid lake may be due to the culture media used, which may have favoured bacterial growth and thus do not reflect their real distribution within the lake. All bacterial strains were found as gram-positive rods producing lysine decarboxylase, ornithine decarboxylase, and arginine dihydrolase. Some of the isolates are able also to reduce nitrate to nitrite, suggesting that they may be involved in the global nitrogen cycle within the lake. Since all the isolates are able to grow optimally in the presence of 25% NaCl, they should be considered extremely halophilic [41] and therefore of ecological significance with regard to the biogeochemistry of the El-Djedid Lake in its most saline parts. Surprisingly, despite the fact that site 5 had suitable physico-chemical conditions to allow microbial life, no isolate was recovered from it.
Extensive research on different hypersaline habitats in Spain and Morocco that focused on the screening of new exopolysaccharide-(EPS)-producing bacteria resulted in several strains isolated from saline soils and described as new species belonging to the genus Halomonas [42–46]. Similarly to the observations reported here, a minority of these isolated microorganisms were identified as members of genera Pontibacillus, Marinococcus, and Halobacillus. Several other aerobic or facultatively anaerobic, moderately halophilic bacteria have been classified within genera related to the order Bacillales [47]. The potential industrial use of these microorganisms has been underlined (production of compatible solutes, biopolymers, and bioremediation processes) and reviewed in detail [1, 48, 49], prompting us to screen our collection of halophiles for molecules of industrial interest (work in progress).
Finally, the metabolic features of the extremely halophilic isolates from the El-Djerid salt lake indicated that most of these isolates were able to oxidize organic polymers in the Sebkha and should therefore participate in the mineralization of resident organic matter, similarly to other hypersaline ecosystems [37]. Studies on these bacteria should be reinvestigated as they constitute a source of halostable enzymes (Table 3) that offer potential applications in various pharmacochemical industries [50, 51].
Acknowledgments
The authors thank Mhamed Ben Abid (pôle technologique Sidi Thabet, Tunis) for technical assistance and help with physico-chemical analyses. They thank the French University Cooperation Programme and the Tunisian Ministry for Science, Technology and Development for providing A. Hedi with financial support.
References
- 1.Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiology and Molecular Biology Reviews. 1998;62(2):504–544. doi: 10.1128/mmbr.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kushner DJ. Life in high salt and solution concentration: halophilic bacteria. In: Kushner DJ, editor. Microbial Life in Extreme Environments. London, UK: Academic Press; 1978. pp. 318–358. [Google Scholar]
- 3.Ollivier B, Caumette P, Garcia J-L, Mah RA. Anaerobic bacteria from hypersaline environments. Microbiological Reviews. 1994;58(1):27–38. doi: 10.1128/mr.58.1.27-38.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oren A. The order Halobacteriales . In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. 3rd edition. New York, NY, USA: Springer; 2001. [Google Scholar]
- 5.Oren A. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. Journal of Industrial Microbiology and Biotechnology. 2002;28(1):56–63. doi: 10.1038/sj/jim/7000176. [DOI] [PubMed] [Google Scholar]
- 6.Mouné S, Caumette P, Matheron R, Willison J-C. Molecular sequence analysis of prokaryotic diversity in the anoxic sediments underlying cyanobacterial mats of two hypersaline ponds in Mediterranean salterns. Federation of European Materials Societies Microbiology Ecology. 2003;44(1):117–130. doi: 10.1016/S0168-6496(03)00017-5. [DOI] [PubMed] [Google Scholar]
- 7.Post FJ. The microbial ecology of the Great Salt Lake. Microbial Ecology. 1977;3(2):143–165. doi: 10.1007/BF02010403. [DOI] [PubMed] [Google Scholar]
- 8.Oren A. Ecology of extremely halophilic microorganisms. In: Vreeland RH, Hochstein LI, editors. The Biology of Halophilic Bacteria. Boca Raton, Fla, USA: CRC Press; 1993. pp. 26–53. [Google Scholar]
- 9.Oren A. Molecular ecology of extremely halophilic Archaea and Bacteria . Federation of European Materials Societies Microbiology Ecology. 2002;39(1):1–7. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 10.Oren A. The ecology of the extremely halophilic archaea. Federation of European Materials Societies Microbiology Reviews. 1994;13(4):415–439. [Google Scholar]
- 11.Kamekura M. Diversity of extremely halophilic bacteria. Extremophiles. 1998;2(3):289–295. doi: 10.1007/s007920050071. [DOI] [PubMed] [Google Scholar]
- 12.Mevarech M, Frolow F, Gloss LM. Halophilic enzymes: proteins with a grain of salt. Biophysical Chemistry. 2000;86(2-3):155–164. doi: 10.1016/s0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 13.Oren A. Biological processes in the Dead Sea ace influenced by short-term and long-term salinity changes. Archives of Hydrobiology Special Issues Advances Limnology. 2000;55:531–542. [Google Scholar]
- 14.Tehei M, Franzetti B, Maurel MC, Vergne J, Hountondji C, Zaccai G. The search for traces of life: the protective effect of salt on biological macromolecules. Extremophiles. 2002;6(5):427–430. doi: 10.1007/s00792-002-0275-6. [DOI] [PubMed] [Google Scholar]
- 15.Grant WD, Gemmell RT, Mc Genity TJ. Halophiles. In: Horikoshi K, Grant WD, editors. Extremophiles: Microbial Life in Extreme Environments. New York, NY, USA: Wiley-Liss; 1998. pp. 93–132. [Google Scholar]
- 16.Boone DR, Garrity GM. Bergey's Manual of Systematic Bacteriology. Vol. 1. New York, NY, USA: Springer; 2001. The Archaea and the deeply branching and phototrophic bacteria. [Google Scholar]
- 17.Elevi R, Assa P, Birbir M, Ogan A, Oren A. Characterization of extremely halophilic Archaea isolated from the Ayvalik Saltern, Turkey. World Journal of Microbiology and Biotechnology. 2004;20(7):719–725. [Google Scholar]
- 18.Trussel R, Lenore S, Clesceri A, Greenberg E. American Public Health Association. 17th edition. Baltimore, Md, USA: Port City Press; 1989. Standard methods for the examination of water and wastewater. [Google Scholar]
- 19.Oren A, Ventosa A, Grant WD. Proposed minimal standards for description of new taxa in the order Halobacteriales . International Journal of Systematic Bacteriology. 1997;47(1):233–238. [Google Scholar]
- 20.Kovacs N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 1956;178(4535):703–704. doi: 10.1038/178703a0. [DOI] [PubMed] [Google Scholar]
- 21.Logan NA, Berkeley RCW. Identification of Bacillus strains using the API system. Journal of General Microbiology. 1984;130:1871–1882. doi: 10.1099/00221287-130-7-1871. [DOI] [PubMed] [Google Scholar]
- 22.Winker S, Woese CR. A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal rRNA characteristics. Systematic Applied Microbiology. 1991;13:161–165. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- 23.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 24.Maidak BL, Cole JR, Lilbrum TG, et al. The RDP-II (Ribosomal Database Project) Nucleic Acids Research. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson DA, Boguski MS, Lipman DJ, Oullette BFF, Rapp BA, Weller DL. GenBank. Nucleic Acids Research. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York, NY, USA: Academic Press; 1969. pp. 211–232. [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Ventosa A, Ramos-Cormenzana A, Kocur M. Moderately halophilic Gram-positive cocci from hypersaline environments. Systematic Applied Microbiology. 1983;4:564–570. doi: 10.1016/S0723-2020(83)80014-9. [DOI] [PubMed] [Google Scholar]
- 31.Cayol J-L, Ollivier B, Patel BKC, Prensier G, Guezennec J, Garcia J-L. Isolation and characterization of Halothermothrix orenii gen. nov., sp. nov., a halophilic, thermophilic, fermentative, strictly anaerobic bacterium. International Journal of Systematic Bacteriology. 1994;44(3):534–540. doi: 10.1099/00207713-44-3-534. [DOI] [PubMed] [Google Scholar]
- 32.Demergasso C, Casamayor EO, Chong G, Galleguillos P, Escudero L, Pedros-Alio C. Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. Federation of European Materials Societies Microbiology Ecology. 2004;48(1):57–69. doi: 10.1016/j.femsec.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Ventosa A. Unusual micro-organisms from unusual habitats: hypersaline environments. In: Logan NA, Lappin-Scott HM, Oyston PCF, editors. Prokaryotic Diversity: Mecanisms and Significance. Cambridge, UK: Cambridge University Press; 2006. pp. 223–253. [Google Scholar]
- 34.Rodriguez-Valera F. Introduction to saline environments. In: Vreeland RH, Hochstein LI, editors. The Biology of Halophilic Bacteria. Boca Raton, Fla, USA: CRS Press; 1993. pp. 1–23. [Google Scholar]
- 35.Fritze D. Bacillus haloalkaliphilus sp. nov. International Journal of Systematic Bacteriology. 1996;46(1):98–101. [Google Scholar]
- 36.Jeon CO, Lim J-M, Lee J-M, Xu L-H, Jiang C-L, Kim C-J. Reclassification of Bacillus haloalkaliphilus (Fritze, 1996) as Alkalibacillus haloalkaliphilus gen. nov., a novel halophilic bacterium isolated from a salt lake in China. International Journal of Systematic and Evolutionary Microbiology. 2005;55(5):1891–1896. doi: 10.1099/ijs.0.63456-0. [DOI] [PubMed] [Google Scholar]
- 37.Ventosa A. In: Taxonomy of Moderately Halophilic Heterotrophic Eubacteria. Halophilic Bacteria. Rodriguez-Valera F, editor. Vol. 1. Boca Raton, Fla, USA: CRC Press; 1988. [Google Scholar]
- 38.Javor BJ. Hypersaline Environments. Microbiology and Biogeochemistry. Berlin, Germany: Springer; 1989. [Google Scholar]
- 39.Grant WD, Kamekura M, McGenity TJ, Ventosa A. Class III. Halobacteria class. nov. In: Bonne DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. New York, NY, USA: Springer; 2001. pp. 294–334. [Google Scholar]
- 40.Arahal DR, Ventosa A. The family Halomonadaceae . In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: An Evolving Electronic Resource for the Microbial Community. New York, NY, USA: Springer; 2005. Release 3.20. [Google Scholar]
- 41.Larsen H. Halophilism in the Bacteria. Vol. 4. New York, NY, USA: Academic Press; 1962. [Google Scholar]
- 42.Quesada E, Valderrama MJ, Béjar V, et al. Volcaniella eurihalina gen. nov., sp. nov., a moderately halophilic nonmotile Gram-negative rod. International Journal of Systematic Bacteriology. 1990;40(3):261–267. [Google Scholar]
- 43.Quesada E, Béjar V, Calvo C. Exopolysaccharide production by Volcaniella eurihalina . Experientia. 1993;49(12):1037–1041. [Google Scholar]
- 44.Bouchotroch S, Quesada E, del Moral A, Llamas I, Béjar V. Halomonas maura sp. nov., a novel moderately halophilic exopolysaccharide-producing bacterium. International Journal of Systematic and Evolutionary Microbiology. 2001;51:1625–1632. doi: 10.1099/00207713-51-5-1625. [DOI] [PubMed] [Google Scholar]
- 45.Martínez-Cánovas MJ, Béjar V, Martínez-Checa F, Quesada E. Halomonas anticariensis sp. nov., from Fuente de Piedra, a saline-wetland wildfowl reserve in Malaga, southern Spain. International Journal of Systematic and Evolutionary Microbiology. 2004;54(4):1329–1332. doi: 10.1099/ijs.0.63108-0. [DOI] [PubMed] [Google Scholar]
- 46.Martínez-Cánovas MJ, Quesada E, Llamas I, Béjar V. Halomonas ventosae sp. nov., a moderately halophilic, denitrifying, exopolysaccharide-producing bacterium. International Journal of Systematic and Evolutionary Microbiology. 2004;54(3):733–737. doi: 10.1099/ijs.0.02942-0. [DOI] [PubMed] [Google Scholar]
- 47.Spring S, Ludwig W, Marquez MC, Ventosa A, Schleifer K-H. Halobacillus gen. nov., with descriptions of Halobacillus litoralis sp. nov. and Halobacillus trueperi sp. nov., and transfer of Sporosarcina halophila to Halobacillus halophilus comb. nov. International Journal of Systematic Bacteriology. 1996;46(2):492–496. [Google Scholar]
- 48.Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5:73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- 49.Mellado E, Ventosa A. Biotechnological potential of moderately and extremely halophilic microorganisms. In: Barredo J-L, editor. Microorganisms for Health Care, Food and Enzyme Production. Kerala, India: Research Signpost; 2003. pp. 233–256. [Google Scholar]
- 50.Jones BE. Industrial enzymes: do halophiles and alkaliphiles have a role to play? In: Ventosa A, editor. Halophilic Microorganisms. New York, NY, USA: Springer; 2004. pp. 275–284. [Google Scholar]
- 51.Quesada E, Bejar V, Ferrer MR. Moderately halophilic, exopolysaccharide-producing bacteria. In: Ventosa A, editor. Halophilic Microorganisms. New York, NY, USA: Springer; 2004. pp. 297–314. [Google Scholar]