Abstract
The DNA radical resulting from formal hydrogen atom abstraction from the thymidine methyl group, 5-(2′-deoxyuridinyl)methyl radical, forms interstrand cross-links with the opposing 2′-deoxyadenosine. This is the first chemically characterized, radical mediated cross-link between two opposing nucleotides. In addition, cross-linking between opposing bases in the duplex is less common than between those separated by 1 or 2 nucleotides. The first step in cross-link repair was investigated using the UvrABC bacterial nucleotide excision repair system. UvrABC incised both strands of the cross-linked DNA, although the strand containing the cross-linked purine was preferred by the enzyme in two different duplexes. The incision sites in one strand were spaced 11–14 nucleotides apart, as is typical for UvrABC incision. The majority of incisions occur at the 3rd phosphate from the 3′-side side of the cross-link and 8th or 9th phosphate on the 5′-side. In addition, cleavage was found to occur on both strands, producing double strand breaks in ~25–29% of the incision events. This is the first example of double-strand cleavage during nucleotide excision repair of cross-linked DNA that does not already contain a strand break in the vicinity of the cross-link.
Keywords: DNA damage, interstrand cross-links, DNA repair
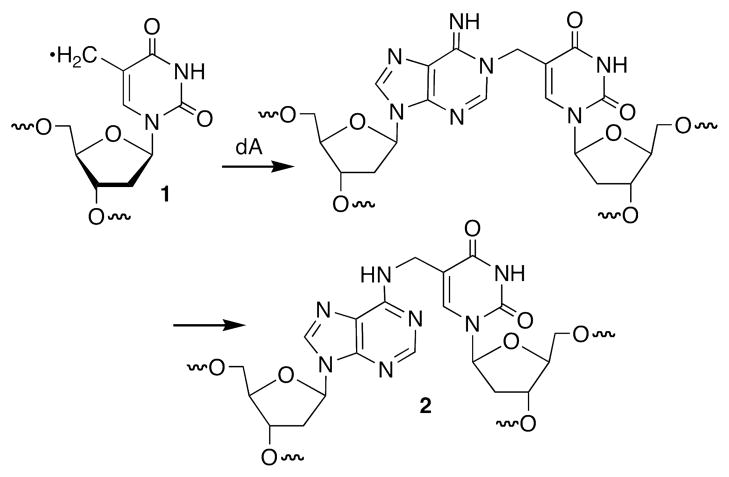
A variety of types of DNA lesions are produced when the biopolymer is exposed to oxidative stress or alkylating agents. These include strand breaks, modified nucleotides, abasic sites, and interstrand cross-links (1–8). Interstrand cross-linked DNA lesions are deleterious because they are absolute blocks to replication and transcription (9, 10). Hence, their repair is vital for survival of the cell (11–14). A number of damaging agents produce DNA interstrand cross-links (15–20). Some of the most well studied cross-links include those produced by mitomycin C, nitrogen mustards, and the photoactive psoralens (21–23). In each instance the damaging agent is incorporated within the cross-link and serves as the covalent bridge between the complementary strands of DNA. Ionizing radiation, which forms a multitude of lesions via various DNA radicals and radical ions, produces low yields of ICLs (24). The chemical constitution of an ICL produced by γ-radiolysis had never been determined until recent studies in which the 5-(2′-deoxyuridinyl)methyl radical (1, Scheme 1) was independently generated in duplex DNA (25–28). Radical 1 forms ICLs by reacting with the opposing 2′-deoxyadenosine, ultimately forming the product in which the thymine’s methyl carbon and the N6-amino group of dA are covalently linked (2). This ICL is structurally distinct from those resulting from the agents mentioned above because the cross-linked nucleotides are Watson-Crick base paired to one another in the precursor DNA. In this article, we describe the repair of this cross-link by a bacterial nucleotide excision repair system.
Scheme 1.
Interstrand cross-link formation from 5-(2′-deoxyuridinyl)methyl radical (1).
The nucleotide excision repair pathway participates in the repair of interstrand cross-links. In bacteria this process is carried out by the three component UvrABC (29). The sequence of events involves recruitment by the UvrA dimer of UvrB to the lesion, which in turn induces a conformational change in the duplex and binds the active endonuclease, UvrC. Hydrolysis of cross-linked substrates typically occurs at the 8th or 9th phosphodiester from the 5′-side of the lesion and ~3 nucleotides on the 3′-side (29–31). Both incisions occur in a single binding event resulting in release of an oligonucleotide segment containing the lesion. After the first round of incisions, the severed oligonucleotide remains tethered to the complementary DNA strand. The resulting gap is believed to be filled in by recombination, and the tethered-oligonucleotide is ultimately removed by a second round of NER (9, 29). The mechanism for recognition of damaged DNA by this multi-protein NER complex is an active area of investigation (32–34). While the UvrAB proteins are believed to recognize macromolecular distortion induced in the DNA helix by the DNA lesion, the precise mechanism of damage recognition is of significant current interest (33–36).
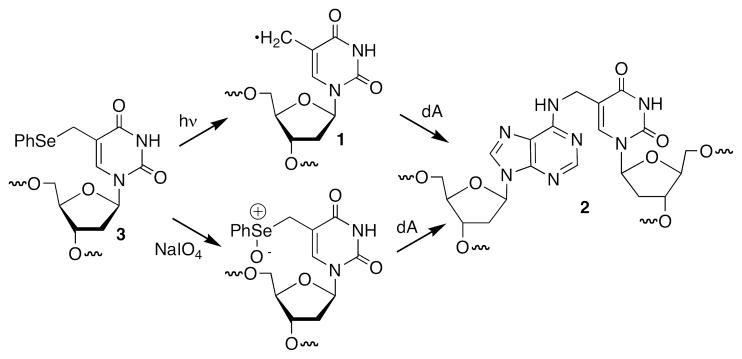
UvrABC incision of the psoralen cross-link is well characterized and structural studies confirm that the cross-linked DNA is locally distorted (9). The sequence surrounding the lesion significantly affects the preference for strand scission (37, 38). In some sequences UvrABC shows a preference for incising the strand cross-linked to the furan ring of psoralen, but in others the two strands are incised more equally. The orientation of a cross-link with respect to the 5′- and 3′-directions of the helix also affects NER incision efficiency and correlates with the distortion and flexibility of the DNA (39). Although the structure of DNA containing 2 has not been characterized by NMR or X-ray crystallography, its formation requires the cross-linked thymidine to adopt the syn-conformation. Additional distortion of the opposing dA from its normal B-form conformation is required in order to form a bond between its N6-amine and the thymidine’s methyl group. The unusual structure of 2 raised the question of whether this ICL would be a substrate for nucleotide excision repair. Furthermore, if the cross-linked product is a substrate for nucleotide excision repair we had no sense as to whether incision of one strand would be preferred over another. We addressed these issues by taking advantage of our ability to generate interstrand cross-linked DNA containing 2 from the phenyl selenide (3) via photochemical generation of 5-(2′-deoxyuridinyl)methyl radical (1) or by mild oxidation (Scheme 2) (25–27).
Scheme 2.
Interstrand cross-link generation via synthetic precursor.
Materials and Methods
Materials and General Methods
Oligonucleotides were prepared on an Applied Biosystems Inc. 394 DNA synthesizer. Commercially available DNA synthesis reagents were obtained from Glen Research Inc. Oligonucleotides containing the cross-link precursor (3) were synthesized as previously described (26, 27, 40). Following denaturing PAGE, the 50mers were purified further via C18-reverse phase HPLC. See below for details. All others were synthesized and deprotected using standard protocols. Synthetic oligonucleotides containing the cross-link precursor (3) were characterized by ESI-MS, which are included in the Supporting Information. UvrABC was obtained as previously described (41, 42). T4 polynucleotide kinase was obtained from New England Biolabs. γ-32P-ATP and α-32P-cordycepin 5′-triphosphate were purchased from Perkin Elmer. C18-Sep-Pak cartridges were obtained from Waters. Analysis of radiolabeled nucleotides was carried out using a Storm 840 Phosphorimager and ImageQuant 5.1 software.
The synthesis and purification of oligonucleotides containing 3
Pac-dA and iPr-Pac-dG phosphoramidites were employed for the synthesis of oligonucleotides containing 3. Pivaloyl anhydride/2,6-lutidine/THF (1:1:8) was used as capping reagent and 1 M t-butyl-hydroperoxide in toluene was used as oxidizing reagent (43). The oxidation time was 40 s and the capping time was 25 s. Deprotection of the nucleobases and phosphate moieties as well as cleavage of the linker were carried out under mild deprotection conditions (28% aq. NH3, room temperature, 3 h). Oligonucleotides were purified by 20% denaturing polyacrylamide gel electrophoresis. Oligonucleotides containing 3 were subjected to additional purification by reversed-phase HPLC on a RP-C18 column (VARIAN, Microsorb-MV 100-5 C18 250 × 4.6 mm). Monitoring was carried out at 260 nm. The peak of interest was collected using the following gradient conditions: 0–5 min 100% A, 5–15 min 0–8% B in A, 15–45 min 8–14% B in A, 45–60 min 14–80% B in A, 60–65 min 80-0% B in A, 65–75 min 100% A, at a flow rate 1.0 mL/min) [A: 0.05 M (Et3NH)OAc (pH 7.0)/MeCN 95:5; B: 0.05 M (Et3NH)OAc (pH 7.0)/MeCN 50:50].
Preparation and purification of cross-linked DNA
The appropriate 32P-labelled oligonucleotide (0.5 μM) and its complementary sequence (0.75 μM) were dissolved in 100 mM NaCl and 10 mM potassium phosphate (pH 7.2). The solution was heated to 65 °C, allowed to cool to room temperature over the course of 2 h, and then kept at 4 °C for 2 h. NaIO4 (5 mM) reactions of DNA duplexes (0.5 μM) were carried out in 10 mM potassium phosphate (pH 7.2) and 100 mM NaCl at 37 °C for 3 h. The cross-linked DNAs formed were purified by 20% denaturing PAGE. The band containing cross-linked product was excised from the gel, crushed, and eluted with 200 mM NaCl, 20 mM EDTA (2.0 mL) for 5 h at room temperature. The crude product was desalted using a C18-Sep Pak and eluted with H2O (3 × 1.0 mL) followed by 1.0 mL MeOH:H2O (3:2, 1.0 mL). The desalted DNA was freeze-dried and stored at −20 °C.
Hydroxyl radical reaction for characterizing cross-links
Fe (II) · EDTA cleavage reactions were carried out in a buffer containing 10 mM sodium phosphate (pH 7.2), 10 mM NaCl, 100 μM EDTA, 10 mM sodium ascorbate, 2 mM H2O2, and 50 μM (NH4)2Fe(SO4)2 (total volume 20 μL) at 25 °C for 3 min, and quenched with 100 mM thiourea (10 μL). Samples were lyophilized, treated with 1.0 M piperidine (20 μL) at 90 °C for 20 min, lyophilized, dissolved in 10 μL H2O: 95% formamide loading buffer (1:1), and subjected to electrophoresis on a 20% denaturing PAGE.
UvrABC reaction
The purified cross-linked DNA was resuspended in 100 mM NaCl and 10 mM potassium phosphate buffer (pH 7.2, 25 μL), and rehybridized by heating to 65 °C (2 min), cooled to room temperature over the course of 2 h, and allowed to equilibrate at 4 °C overnight. The UvrA, UvrB, and UvrC were freshly prepared from stock solutions and heated individually at 65 °C for 10 min before use. They were added sequentially to the reactions. The reaction buffer contains 50 mM Tris-HCl (pH 7.5), MgCl2 (10 mM), KCl (50 mM), and ATP (1.0 mM). The cross-linked DNA (2.0 nM) was incubated with UvrA (20 nM), UvrB (100 nM), and UvrC (50 nM) in a total volume of 20 μL at 55 °C. The concentration of cross-linked DNA was based upon the specific activity of the initially labeled oligonucleotide, which assumed 100% recovery of the oligonucleotide from the Sephadex column used to remove excess γ-32P-ATP. After 60 min the reaction was quenched by precipitation with 5 M NH4OAc (5 μL), 0.25 μg/μL calf thymus DNA (5 μL), and cold ethanol (75 μL). The incision products were separated by 20% denaturing PAGE and visualized using the Phosphorimager. For the time course reaction (100 μL total volume) aliquots (10 μL) were removed at the prescribed times and immediately quenched by precipitation with 5 M NH4OAc (5 μL), 0.25 μg/μL CT DNA (5 μL), and cold ethanol (60 μL).
Results
Oligonucleotide synthesis and interstrand cross-link formation
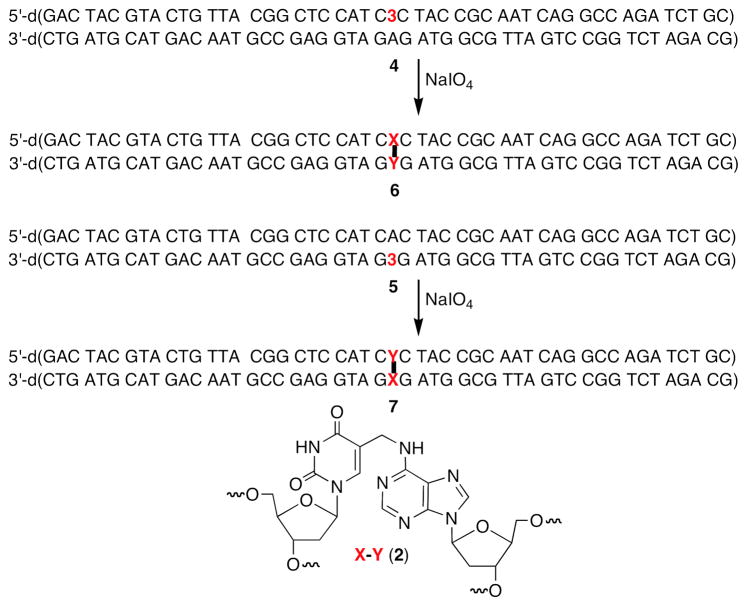
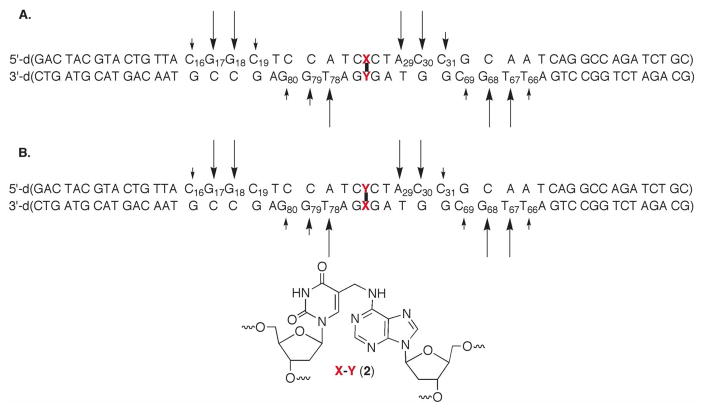
Oligonucleotides containing the cross-link precursor (3) have been synthesized previously (26, 27). The phenyl selenide (3) is susceptible to the oxidation conditions employed during the solid phase synthesis, which gives rise to a small amount of an impurity (5-hydroxymethyl-2′-deoxyuridine) with each synthesis cycle subsequent to its incorporation. The UvrABC system requires a relatively long DNA substrate (50 nucleotides). Hence, the synthetic oligonucleotide containing 3 that is purified by denaturing PAGE contained a significant amount of the oxidative degradation product, 5-hydroxymethyl-2′-deoxyuridine. The oligonucleotide containing this more polar material was separated from the 50mer containing 3 via reverse phase HPLC. The ICL was generated in duplexes containing two flanking sequences (Scheme 3). In one (4), the modified thymidine that becomes cross-linked was flanked by dC. The other substrate (5) flips the orientation of the ICL by incorporating the phenyl selenide opposite the dG’s on the opposing strand, but is identical in all other respects to 4.
Scheme 3.
Formation of Interstrand cross-links used in this study.
The ICL was originally detected following photolysis of duplexes containing 3, which produce 5-(2′-deoxyuridinyl)methyl radical (1, Scheme 3) (27). However, we later found that the identical cross-link product was formed in higher yield when 3 was exposed to mild oxidative conditions (NaIO4) (26, 44). Consequently, the cross-linked substrates used in this study were formed using NaIO4. The yields of cross-linked products were typically 45–65%. The ICLs were purified using denaturing PAGE, desalted, and rehybridized prior to UvrABC incision experiments. Exclusive cross-linking at the dA opposite the modified thymidine was verified in both duplexes using the hydroxyl radical digestion method (45).
Nucleotide selectivity of UvrABC incision of cross-linked DNA
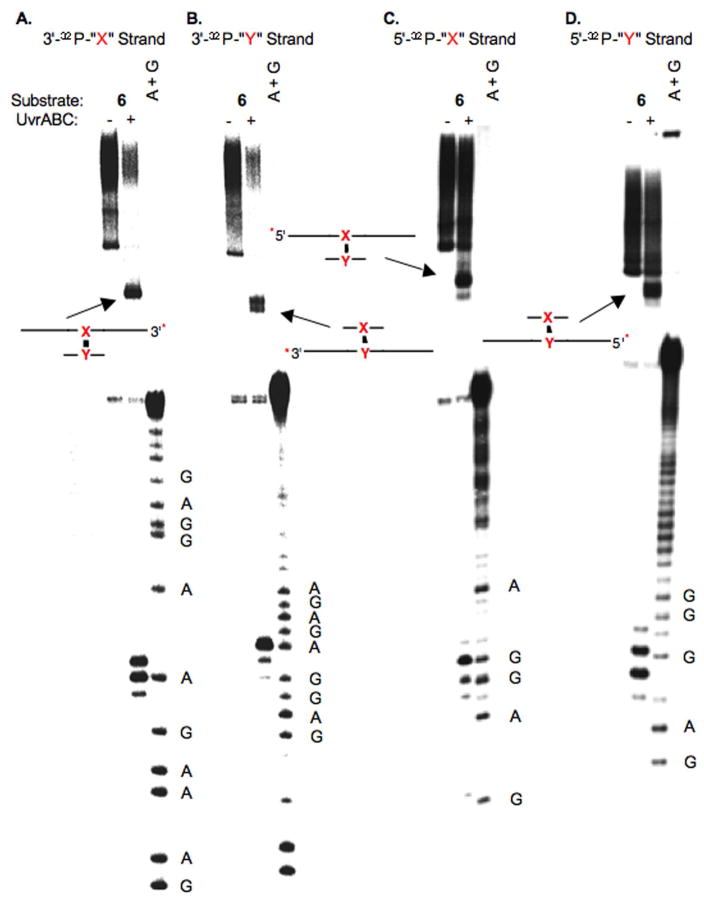
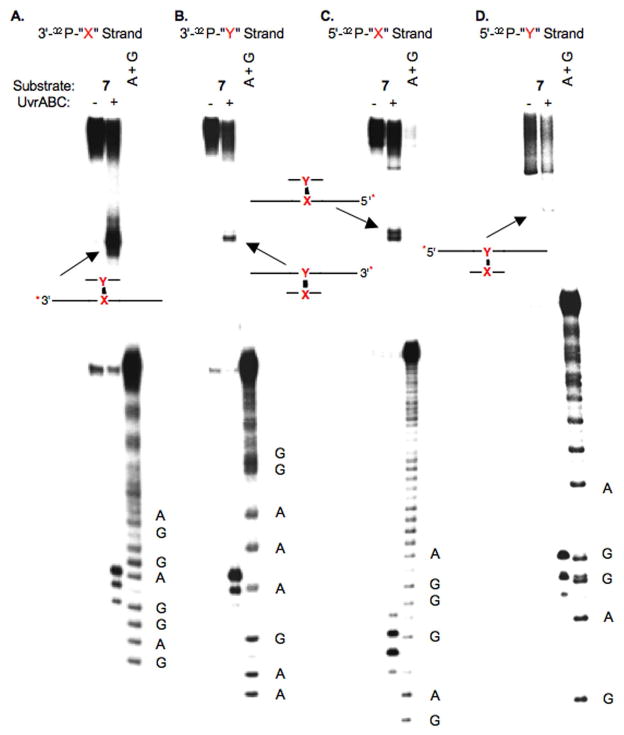
As mentioned above UvrABC incision typically occurs at the 8th or 9th position on the 5′-side of a lesion and at the 3rd to 5th phosphodiester from the 3′-terminus of the lesion (29, 32). Incision sites in 6 (Figure 1) and 7 (Figure 2) were determined by labeling the 5′- and 3′-termini of each strand in separate experiments. For simplicity, the cross-linked nucleotides that make up 2 are referred to as “X-Y” in the duplexes in Figures 1–7 (and Supporting Information), where “X” represents the modified thymidine derived from 3 and “Y” is the cross-linked dA. Histograms that summarize the cleavage sites and relative intensities within a strand (but not relative to the complementary strand) are presented in Figure 3. When “X” was flanked by dCs (4), the product (6) was preferentially incised on the 5′-side of the cross-link at dG17 and dG18, with a modest preference for the latter (Figure 1C, 3A). Much weaker incision was observed at dC16 and dC19. The major incision sites correspond to the 9th and 10th phosphates from the ICL. The same strand was incised on the 3′-side of the ICL at dA29, dC30, and to a lesser extent dC31 (Figure 1A, 3A). The positions of the major incision sites are 3 and 4 phosphates removed from the 3′-side of the cross-link. Overall, the 5′- and 3′-incision sites result in cleavage of an 11–14 nucleotide oligonucleotide segment, which is consistent with UvrABC cleavage of other lesions.
Figure 1.
UvrABC incision of 6. A. Strand containing modified thymidine labeled at its 3′-terminus (3′-32P-“X”-strand) B. Strand containing cross-linked dA labeled at its 3′-terminus (3′-32P-“Y”-strand) C. Strand containing modified thymidine labeled at its 5′-terminus (5′-32P-“X”-strand) D. Strand containing cross-linked dA labeled at its 5′-terminus (5′-32P-“Y”-strand); A + G, sequencing reaction. The cartoons identify the products produced from incision of the unlabeled strand. * indicates position of 32P-label.
Figure 2.
UvrABC incision of 7. A. Strand containing modified thymidine labeled at its 3′-terminus (3′-32P-“X”-strand) B. Strand containing cross-linked dA labeled at its 3′-terminus (3′-32P-“Y”-strand) C. Strand containing modified thymidine labeled at its 5′-terminus (5′-32P-“X”-strand) D. Strand containing cross-linked dA labeled at its 5′-terminus (5′-32P-“Y”-strand); A + G, sequencing reaction. The cartoons identify the products produced from incision of the unlabeled strand. * indicates position of 32P-label.
Figure 7.
Native PAGE analysis of UvrABC incision of 7. A. Strand containing modified thymidine labeled at its 3′-terminus (3′-32P-“X” strand); B. Strand containing cross-linked dA labeled at its 3′-terminus (3′-32P-“Y” strand). Compound 5 is used as a single stranded 50 nt marker. 3′-32P-11 (A) and 3′-32P-12 (B) are used as double strand cleavage markers. C. Sequences of 11 and 12.
Figure 3.
Histograms of UvrABC incision data for: A. 6, B. 7. Arrow length is proportional to relative amounts of incision within one strand.
UvrABC exhibits a similar incision pattern on the cross-linking strand in 6 that contributes the dA (“Y”). The major incision sites on the 3′-side of the cross-link occur 3 and 4 phosphates removed from the cross-linked nucleotide at dT78 and dG79 respectively (Figure 1B, 3A). The former is favored by ~3-fold over incision at dG79. There is also a small amount of incision at dG80. 5′-32P-Labeling of the strand containing the modified dA reveals that dT67 and dG68 are preferably incised in approximately equal amounts (Figure 1D, 3A). These nucleotides (8th and 9th phosphate diesters from the cross-linked nucleotide) are the typical distance from the cross-link. As was observed with the 5′-incision of the modified thymidine containing strand, the major incision sites flanked by nucleotides (dC69, dT66) where minor amounts of cleavage occur.
Exchanging the strands in which the cross-linked nucleotides (7) are in has very little effect on the UvrABC incision pattern (Figures 2 and 3B). The major cleavage sites are dG79, dT78, dG68, and dT67 in the strand of 7 containing the modified thymidine (“X”). These are the same nucleotides at which most of the incision occurs in the respective strand of 6 in which the cross-linked dA resides at this position. Similarly, the major incision sites in the cross-linked dA (“Y”) containing strand of 7 are dG17, dG18, dA29, and dC30. The minor incision sites are also very similar in the corresponding strands of 6 and 7.
Finally, products resulting from incision on the unlabeled strand of the duplexes are observed by denaturing gel electrophoresis (Figures 1 and 2), and are identified as slower moving, less well resolved bands.
Strand selectivity in UvrABC incision of the interstrand cross-link
When NER proteins encounter an interstrand cross-link they must choose between strands. Incision in either strand can be detected by denaturing PAGE in the same experiment. Lesion incision by UvrABC on the two sides of a lesion is typically coupled. Hence, only the cleavage products generated between the lesion and labeled terminus of the radiolabeled strand are typically observed, and these are readily resolved at nucleotide resolution (Figures 1 and 2). In contrast, coupled incision in the unlabeled strand creates products that are longer than either of the parental strand oligonucleotides (50 nucleotides) used to construct the cross-linked substrate (Figures 1 and 2). Products of this length that differ from one another by one to three nucleotides are not resolvable by 20% denaturing PAGE (see Figures 1 and 2). However, those that differ from one another depending on whether they result from incision on one side of the cross-linked nucleotide or from coupled incision are separable from one another. Comparing the migration of the incision product from 7 with those of the independently synthesized hypothetical products (8–10) confirmed that UvrABC incised the DNA on both sides of the lesion in the unlabeled strand. Denaturing PAGE of 5′-32P-8–10 (where the uncleaved strand is radiolabeled) showed that the product resulting from incision 5′ and 3′ on the same unlabeled strand of the cross-linked nucleotide (10) comigrates with the incision product that is observed in the region between single stranded and cross-linked DNA (Figure 4). This experiment also verified that UvrC is required for incision.
Figure 4.
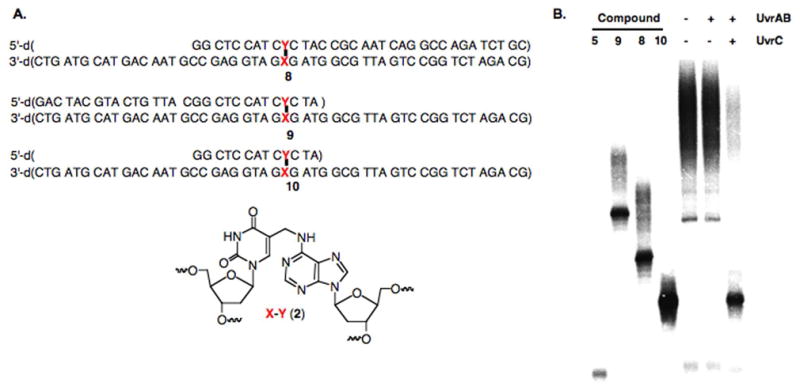
Identification of UvrABC incision product from 6 on unlabeled strand. A. Independently synthesized postulated products. B. Denaturing PAGE analysis of incision product and independently synthesized standards.
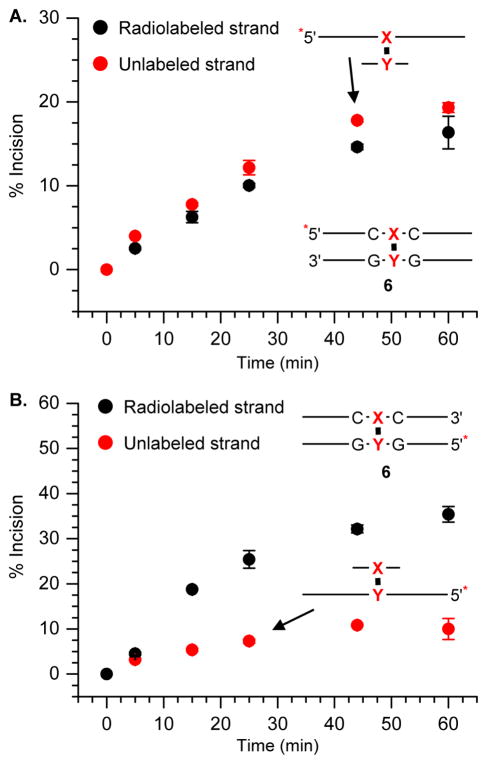
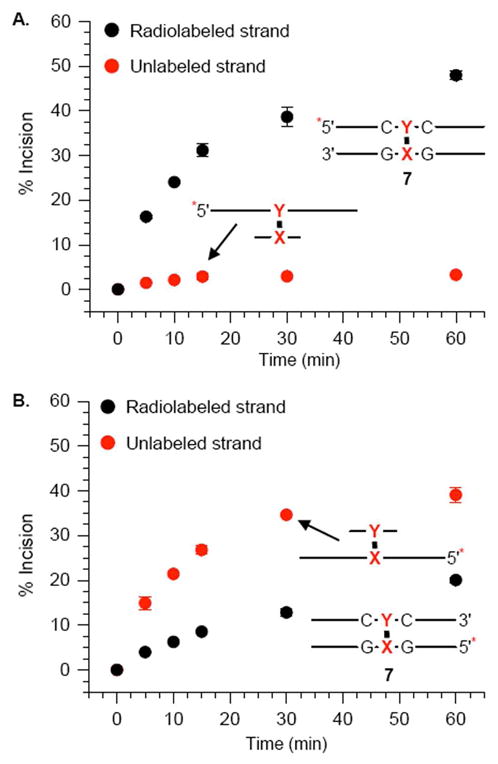
The ability to observe the products resulting from coupled incision on the unlabeled strand of a cross-link, as well as the labeled strand in the same gel enabled us to determine the strand preference for UvrABC in 6 and 7. When the strand containing the cross-linked dA (“Y”) in 6 was labeled, incision on this side of the duplex rose to almost 40% in 60 minutes, and was independent of whether the 5′- (Figure 5B) or 3′-terminus (See supporting information) was labeled. Cleavage of the unlabeled strand containing the modified thymidine (“X”) was significantly less (Figure 5B), rising to less than 10% in 1 h regardless of which terminus was labeled (45). The relative amounts of scission detected on the respective strands in 6 were different when the strand derived from the phenyl selenide (“X”) was labeled (Figure 5A). Cleavage of the radiolabeled strand containing the modified thymidine rose to ~15% over 1 h, whereas incision of the unlabeled strand containing the cross-linked dA (“Y”) was just a few percent higher. These observations were only slightly affected by which terminus of the duplex was radiolabeled (45). Although the observed cleavage ratio on the respective strands depended upon which strand was radiolabeled, the overall amount of incision was very similar. In each instance, the total cleavage of the ICL (6) by UvrABC was approximately 35–45%.
Figure 5.
Time course of UvrABC incision of 32P-6 in which A. 5′-terminus of “X” strand is labeled and B. 5′-terminus of “Y” strand is labeled. 32P-Labeled terminus is indicated by *. Note: Cartoons of products resulting from incision on the unlabeled strand are shown.
The preference for UvrABC incision of the ICL for the strand containing the cross-linked dA (“Y”) was even greater in 7 where the modified thymidine is flanked by dG (Figure 6 and Supporting Informaiton). Approximately 40–45% of the strand containing the cross-linked dA was incised in 1 h when the strand containing it was radiolabeled (Figure 6A). In contrast, only ~5% of the unlabeled strand containing the cross-linked thymidine (“X”) was incised (Figure 6A). The effect of labeling the strand containing the cross-linked thymidine in 7 (Figure 6B) was qualitatively similar to the observations made using 6 (Figure 5). Cleavage of the (“Y”) containing (unlabeled) cross-linked strand was still favored but less so than when this strand was radiolabeled. In addition, the total amount of cleavage was similar irrespective of which strand of the ICL was radiolabeled.
Figure 6.
Time course of UvrABC incision of 32P-7 in which “ A. 5′-terminus of “Y” strand is labeled and B. 5′-terminus of “X” strand is labeled. 32P-Labeled terminus is indicated by *. Note: Cartoons of products resulting from incision on the unlabeled strand are shown.
Double-Strand Break Formation
The dependence of the strand preference on which strand was labeled led us to examine whether UvrABC incised both strands of a single molecule containing the interstrand cross-link (6 or 7). This would result in double strand breaks and would be the first example of such a process in the NER of an interstrand cross-link in which neither strand is cleaved. Strand scission of 7 was examined by 15% native PAGE (Figure 7). Each strand in 7 was 3′-labeled in separate experiments. The overall amount of incision on each strand was quantified by denaturing the sample prior to loading on the gel (Figure 7A, B; lane 3). These products were replaced, albeit not at the same levels, by double strand breaks when the samples were not denatured. The remainder of the material incised on a single strand comigrated with the ICL. Products incised on a single strand are not denatured under these conditions and comigrate with uncleaved substrate. The double strand cleavage product(s) comigrated with independently synthesized products (11, 12, Figures 7A, B, lane 5) whose compositions were anticipated based upon the above analyses of sites of single strand incision. In addition, the amount of double strand breaks based upon the total amount of cleavage was similar (25–29%) regardless of which strand was labeled.
Discussion
The recognition and UvrABC incision of the first interstrand cross-link discovered (2) that results from a DNA radical produced as a result of oxidative stress (e.g. γ-radiolysis, hydroxyl radical) was examined (28). The cross-linked product is incised less rapidly than a 2′-deoxyuridine derivatized at its C5-position by a fluoresceinylated alkyl chain, which is in a way a gold standard for UvrABC reactivity (36). However, the ICL is cleaved with comparable efficiency to a tandem lesion containing thymine glycol and was a slightly poorer substrate than the isolated glycol (46, 47). In addition, the distribution of cleavage sites with respect to the position of the cross-linked nucleotides is very similar to the incision of other lesions by the UvrABC complex. For instance, the oligonucleotide fragment excised from damaged DNA is typically 11–13 nucleotides in length (9, 37, 38). Indeed, the major incision sites in the top strands of 6 and 7 are between 11 and 14 nucleotides apart (Figure 3). Incision on the 5′-side of the cross-link occurs most often at the 8th or 9th phosphodiester from the damaged nucleotide. The majority of the ICLs are cleaved closer (3rd or 4th phosphodiester) to the cross-link position on the 3′-side. These sites are comparable to those incised when UvrABC encounters a psoralen cross-link (37, 38).
The above data do not address the proclivity of the enzyme to cleave either strand. Previous studies demonstrated that the sequence surrounding the psoralen cross-link plays a large role in determining whether only the furan cross-linked strand is cleaved or both strands are by UvrABC (37, 38). Incision on both strands is discernible in any given experiment. We found that the strand containing the cross-linked dA (“Y”) is preferentially incised in the ICLs (6, 7), indicating that the orientation of this cross-link is more important in determining which strand is cleaved, than the surrounding DNA sequence. The observed strand selectivity is independent of whether the 5′- or 3′-terminus of a particular strand (e.g. the oligonucleotide containing the cross-linked dA) is radiolabeled, consistent with the coupled mechanism for strand scission by UvrABC. In contrast, the observed strand selectivity depends strongly on which oligonucleotide in the duplex is radiolabeled.
The preference is more readily apparent when the strand containing the cross-linked dA (“Y”) is radiolabeled (Figures 5 and 6). In these duplexes the dA containing strand is cleaved ~5–10-times more frequently than the one containing the cross-linked thymidine (“X”). More specifically, in 7 the labeled dA (“Y”) containing strand is incised almost 10-fold more frequently than that containing the modified thymidine, “X” (Figure 6A). However, the “Y” strand in 7 is incised only ~2-fold more frequently than the “X” strand (Figure 6B) when the complement is radiolabeled. In 6, the preference for incising the labeled “Y” containing strand is less than in 7. When the “Y” strand is labeled it is incised ~5-times more frequently than the “X” containing complement (Figure 5B), and only ~20% more frequently when the “X” strand is radiolabeled (Figure 5A).
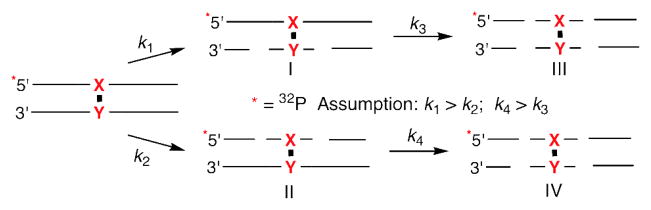
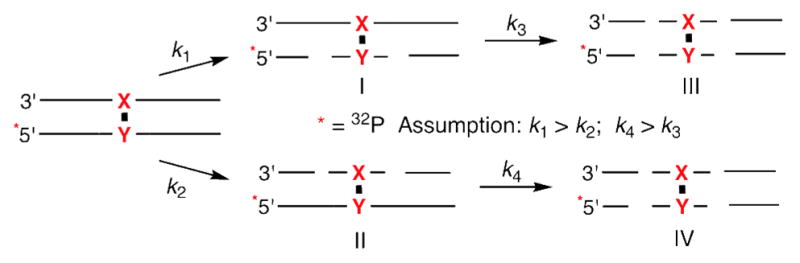
One possible explanation for the above observations is that UvrC incised an ICL molecule twice, resulting in double strand break formation. Double strand cleavage was unambiguously identified via native gel analysis of the UvrABC reactions (Figure 7). DSBs are introduced into ~1 in 4 incised DNA molecules (25–29%). We did not expect double strand cleavage to result from dissociation after the first round of incision and reassociation of the protein, because UvrABC was not expected to bind the ternary complex created following the initial incision (48). Indeed, UvrABC does not cleave independently synthesized preincised substrate containing ICL 2 (45). Instead, we propose that double strand cleavage results from two rounds of incision prior to dissociation (Schemes 4, 5). If so, when the “X” strand is labeled (Scheme 4) products II, III, and IV are indistinguishable by denaturing PAGE, and appear as though the oligonucleotide containing the cross-linked thymine, the less frequently incised strand, was cleaved. Only product I indicates incision of the “Y” containing strand. In contrast, when the “Y” strand is radiolabeled (Scheme 5) products I, III, and IV reflect cleavage of this strand when the reaction is analyzed by gel electrophoresis, and a higher ratio of cleavage on the “Y” strand versus the “X” strand will be observed. This hypothesis explains the plots of incision as a function of time (Figures 5, 6 and See Supporting Information) provided that UvrABC incision on both sides of the lesion in a single strand is coupled and that the following assumptions are valid. The first is that the incision of the original ICL is faster than the second round of cleavage (k1 > k3, k2 > k4). This is consistent with the observation that double strand breaks are the minor products. The other assumption is that incision of the “Y” containing strand is always faster than the “X” strand (k1 > k2, k4 > k3).
Scheme 4.
Repetitive incision of ICL containing radiolabeled “X” strand.
Scheme 5.
Repetitive incision of ICL containing radiolabeled “Y” strand.
Overall, when the “X” containing strand is radiolabeled (Scheme 4), incision of this less favored strand following cleavage of the complementary one (product I) to produce III reduces UvrABC’s observed preference for the dA (“Y”) containing strand, because the second incision prevents one from detecting cleavage on the “Y” containing strand by denaturing PAGE. In contrast, radiolabeling the preferentially incised “Y” strand (Scheme 5) yields a larger observed preference for this strand because assuming that incision of this strand is still favored (k4 > k3) in the initially formed products (I and II), a second cleavage round increases the observed amount of “Y” incised (due to formation of IV) at the expense of “X” cleavage (attributable to II).
Quantification of the UvrABC incision in 7 (Figure 7) by nondenaturing PAGE corroborates this scenario. Analysis of cleaved 7 under denaturing conditions in which the “X” containing strand is radiolabeled revealed 19% cleavage of this strand, and 33% cleavage of the “Y” strand, which produces 10 (Figure 7A). When the sample was not denatured prior to loading, the native gel revealed 15% DSBs. Comparable analysis of 7 at the same concentration (2 nM) in which the “Y” containing strand was radiolabeled (Figure 7B) indicated 47% incision of this strand, 5% cleavage of the unlabeled “X” containing strand (to yield the product comparable to 10), and 13% DSBs. It is gratifying that the differences (14%) in the amounts of “X” strand incision observed when it is labeled (19%) and unlabeled (5%) is approximately equal to the amount of DSBs observed. In addition, comparison of the differences (14%) in “Y” strand cleavage when this strand is labeled (47%) to unlabeled (33%) to the DSB level corroborates this observation.
Summary
Interstrand cross-link repair is vitally important because these lesions are absolute blocks to replication and transcription. Double strand breaks are even more significant because they are the most cytotoxic of lesions. Double strand cleavage could result from nucleotide excision repair of interstrand cross-links, but is extremely rare. For instance, double strand breaks are not formed when DNA cross-linked by N-methylmitomycin or psoralen are treated with UvrABC (14, 37). Recently, we described the first chemical characterization of double strand cleavage resulting from UvrABC repair of a cross-link (49). However, in that instance the cross-link was adjacent to a nick. Double strand cleavage only required UvrABC to incise the strand opposite the nick. To our knowledge, UvrABC mediated double strand cleavage of 6 and 7 is the first example in which both strands of the interstrand cross-linked DNA are incised through dual incisions of phosphate diesters that flank the lesion on its 5′- and 3′-sides.
Supplementary Material
Abbreviations
- ICL
interstrand cross-link
- NER
nucleotide excision repair
- PAGE
polyacrylamide gel electrophoresis
- DSB
double strand break
Footnotes
We are grateful for support of this research by the National Institute of General Medical Sciences (GM-054996).
Supporting Information Available. ESI-MS of oligonucleotides containing 3. Histograms of hydroxyl radical cleavage reactions on 6 and 7. Time course of UvrABC incision of 3′-32P-6 and 3′-32P-7. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Delaney JC, Essigmann JM. Biological Properties of Single Chemical-DNA Adducts: A Twenty Year Perspective. Chem Res Toxicol. 2008;21:232–252. doi: 10.1021/tx700292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dedon PC. The Chemical Toxicology of 2-Deoxyribose Oxidation in DNA. Chem Res Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J, Douki T, Ravanat JL. Oxidatively Generated Damage to the Guanine Moiety of DNA: Mechanistic Aspects and Formation in Cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg MM. Elucidating DNA Damage and Repair Processes by Independently Generating Reactive and Metastable Intermediates. Org Biomol Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 5.Dizdaroglu M, Jaruga P, Rodriguez H. Oxidative damage to DNA: mechanisms of product formation and measurement by mass spectrometric techniques. Crit Rev Oxidative Stress and Aging. 2003;1:165–189. [Google Scholar]
- 6.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA Damage: Mechanisms, Mutation, and Disease. FASEB. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 7.Burrows CJ, Muller JG. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 8.Pogozelski WK, Tullius TD. Oxidative Strand Scission of Nucleic Acids: Routes Initiated by Hydrogen Abstraction from the Sugar Moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 9.Noll DM, Mason TM, Miller PS. Formation and Repair of Interstrand Cross-Links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schärer OD. DNA Interstrand Crosslinks: Natural and Drug-Induced DNA Adducts that Induce Unique Cellular Responses. Chem Bio Chem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 11.Dronkert ML, Kanaar R. Repair of DNA Interstrand Cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 12.Anderson BS, Sadeghi T, Siciliano MJ, Legerski R, Murray D. Nucleotide Excision Repair Genes as Determinants of Cellular Sensitivity to Cyclophosphamide Analogs. Cancer Chemother Pharmacol. 1996;38:406–416. doi: 10.1007/s002800050504. [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide Excision Repair- and Polymerase Eta-mediated Error-prone Removal of Mitomycin C Interstrand Cross-Links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pu WT, Kahn R, Munn MM, Rupp WD. UvrABC incision of N-methylmitomycin A-DNA monoadducts and cross-links. J Biol Chem. 1989;264:20697–20704. [PubMed] [Google Scholar]
- 15.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA Cross-Links Induced by a,β-Unsaturated Aldehydes Derived from Lipid Peroxidation and Environmental Sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beerman TA, Gawron LS, Shin S, Shen B, McHught MM. C-1027, A Radiomimetic Enediyne Anticancer Drug, Preferentially Targets Hypoxic Cells. Cancer Research. 2009;69:593–598. doi: 10.1158/0008-5472.CAN-08-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romano KP, Newman AG, Zahran RW, Millard JT. DNA Interstrand Cross-Linking by Epichlorohydrin. Chem Res Toxicol. 2007;20:832–838. doi: 10.1021/tx700066h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy DR, Lu J, Shen B, Beerman TA. Designer Enediynes Generate DNA Breaks, Interstrand Cross-links, or Both, With Concomitant Changes in the Regulation of DNA Damage Responses. Proc Nat Acad Sci USA. 2007;104:17632–17637. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez AM, Kozekov ID, Harris TM, Lloyd RS. Formation of Inter- and Intrastrand Imine Type DNA-DNA Cross-Links through Secondary Reactions of Aldehydic Adducts. Chem Res Toxicol. 2005;18:1683–1690. doi: 10.1021/tx0500528. [DOI] [PubMed] [Google Scholar]
- 20.Paz MM, Hopkins PB. DNA-DNA Interstrand Cross-Linking by FR66979: Intermediates in the Activation Cascade. J Am Chem Soc. 1997;119:5999–6005. [Google Scholar]
- 21.Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: crosslinking and alkylation of DNA as the molecular basis of their activity. Pharmacology & Therapeutics. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 22.Lage C, de Padula M, de Alencar TAM, Goncalves SRdF, Vidal LdS, Cabral-Neto J, Leitao AC. Mutation Research/Reviews in Mutation Research. Vol. 544. 2003. New Insights on How Nucleotide Excision Repair Could Remove DNA Adducts Induced by Chemotherapeutic Agents and Psoralens Plus UV-A (PUVA) in Escherichia coli Cells; pp. 143–157. [DOI] [PubMed] [Google Scholar]
- 23.Volpato M, Seargent J, Loadman PM, Phillips RM. Formation of DNA Interstrand Cross-links as a Marker of Mitomycin C Bioreductive Activation and Chemosensitivity. Eur J Cancer. 2005;41:1331–1338. doi: 10.1016/j.ejca.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 24.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; London: 1987. [Google Scholar]
- 25.Ding H, Majumdar A, Tolman JR, Greenberg MM. Multinuclear NMR and Kinetic Analysis of DNA Interstrand Cross-Link Formation. J Am Chem Soc. 2008;130:17981–17987. doi: 10.1021/ja807845n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong IS, Ding H, Greenberg MM. Oxygen Independent DNA Interstrand Cross-Link Formation by a Nucleotide Radical. J Am Chem Soc. 2006;128:485–491. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong IS, Greenberg MM. Efficient DNA Interstrand Cross-Link Formation From a Nucleotide Radical. J Am Chem Soc. 2005;127:3692–3693. doi: 10.1021/ja042434q. [DOI] [PubMed] [Google Scholar]
- 28.Ding H, Greenberg MM. gamma-Radiolysis and Hydroxyl Radical Produce Interstrand Cross-Links in DNA Involving Thymidine. Chem Res Toxicol. 2007;20:1623–1628. doi: 10.1021/tx7002307. [DOI] [PubMed] [Google Scholar]
- 29.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic Nucleotide Excision Repair: The UvrABC System. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 30.Cheng S, Van Houten B, Gamper HB, Sancar A, Hearst JE. Use of psoralen-modified oligonucleotides to trap three-stranded RecA-DNA complexes and repair of these cross-linked complexes by ABC excinuclease. J Biol Chem. 1988;263:15110–15117. [PubMed] [Google Scholar]
- 31.Van Houten B, Gamper HB, Hearst JE, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J Biol Chem. 1988;263:16553–16560. [PubMed] [Google Scholar]
- 32.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. ‘Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C. Structural Insights into the First Incision Reaction During Nucleotide Excision Repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A Structural Model for the Damage-sensing Complex in Bacterial Nucleotide Exision Repair. J Biol Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C. Interactions Between UvrA and UvrB: the Role of UvrB’s Domain 2 in Nucleotide Excision Repair. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. The β-Hairpin Motif of UvrB is Essential for DNA Binding, Damage Processing, and UvrC-mediated Incisions. J Biol Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 37.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action Mechanism of ABC Excision Nuclease on a DNA Substrate Containing a Psoralen Crosslink at a Defined Position. Proc Natl Acad Sci. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones BK, Yeung AT. DNA base composition determines the specificity of UvrABC endonuclease incision of a psoralen cross-link. J Biol Chem. 1990;265:3489–3496. [PubMed] [Google Scholar]
- 39.Noll DM, Webba da Silva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Structure, Flexibility, and Repair of Two Different Orientations of the Same Alkyl Interstrand DNA Cross-Link. Biochemisty. 2005;44:6764–6775. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- 40.Hong IS, Greenberg MM. Mild Generation of 5-(2′-Deoxyuridinyl)methyl Radical From a Phenyl Selenide Precursor. Org Lett. 2004;6:5011–5013. doi: 10.1021/ol047754t. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J Biol Chem. 2006;281:15227–15237. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- 42.Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J Biol Chem. 2006;281:26370–26381. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Q, Delaney MO, Greenberg MM. Observation and Elimination of N-Acetylation of Oligonucleotides Prepared Using Fast Deprotecting Phosphoramidites and Ultra-Mild Deprotection. Bioorg & Med Chem Lett. 2001;11:1105–1108. doi: 10.1016/s0960-894x(01)00161-5. [DOI] [PubMed] [Google Scholar]
- 44.Hong IS, Ding H, Greenberg MM. Radiosensitization by a Modified Nucleotide that Produces DNA Interstrand Cross-Links under Hypoxic Conditions. J Am Chem Soc. 2006;128:2230–2231. doi: 10.1021/ja058290c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.See Supporting Information.
- 46.Kow YW, Wallace SS, Van Houten B. UvrABC Nuclease Complex Repairs Thymine Glycol, an Oxidative DNA Base Damage. Mutat Res. 1990;235:147–156. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- 47.Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA Tandem Lesion Repair by Strand Displacement Synthesis and Nucleotide Excision Repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moolenaar GF, Monaco V, van der Marel GA, van Boom JH, Visse R, Goosen N. The Effect of the DNA Flanking the Lesion on Formation of the UvrB-DNA Preincision Complex. Mechanism for the UvrA-Mediated Loading of UvrB onto a DNA Damaged Site. J Biol Chem. 2000;275:8038–8043. doi: 10.1074/jbc.275.11.8038. [DOI] [PubMed] [Google Scholar]
- 49.Sczepanski J, Jacobs AC, Van Houten B, Greenberg MM. Double Strand Break Formation During Nucleotide Excision Repair of a DNA Interstrand Cross-link. Biochemistry. 2009:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.