Abstract
Background. With the improvement in genetic testing over time, double-heterozygous mutations are more often found by coincidence in families with hypertrophic cardiomyopathy (HCM). Double heterozygosity can be a cause of the wellknown clinical diversity within HCM families.
Methods and results. We describe a family in which members carry either a single mutation or are double heterozygous for mutations in myosin heavy chain gene (MYH7) and cysteine and glycine-rich protein 3 (CSRP3). The described family emphasises the idea of a more severe clinical phenotype with double-heterozygous mutations. It also highlights the importance of cardiological screening where NT-proBNP may serve as an added diagnostic tool.
Conclusion. With a more severe inexplicable phenotype of HCM within a family, one should consider the possibility of double-heterozygous mutations. This implies that in such families, even when one disease-causing mutation is found, all the family members still have an implication for cardiological screening parallel to extended genetic screening. (Neth Heart J 2009;17:458-63.)
Keywords: cardiomyopathy, hypertrophic, proBNP, genetics, double-heterozygous mutations
Hypertrophic cardiomyopathy (HCM) is the most common inheritable cardiac disorder with a phenotypic prevalence within the general population of 1:500. In 30 to 60% of all cases an autosomal dominant mutation has been found.1-5
The majority of mutations causing HCM are found in genes which encode sarcomeric proteins. The most frequent genes involved in genotyped patients with HCM are cardiac myosin binding protein C (MYBPC3) and myosin heavy chain gene (MYH7), which are mutated in up to 82% of all cases.3,6-8 In addition, in the Netherlands a founder mutation in MYBPC3 accounts for almost one-fourth of all HCM cases.9 Despite the relative uniformity in the underlying genetic cause, HCM is known to exhibit remarkable phenotypic heterogeneity, even within one family, but this well-known clinical heterogeneity is difficult to understand. Several reasons have been proposed in order to explain the differences in clinical phenotype, such as exercise, modifier genes, or the presence of compound or double-heterozygous mutations.10-13
In the Netherlands it is common practice, based on their relative frequency, to start the genetic screening with MYBPC3 and MYH7.14 An important question is whether to screen all known sarcomeric genes in each family, or to stop once a pathogenic mutation has been found. At the moment, no further DNA testing is done after one disease-causing mutation is found.14,15 However, an important caveat is the possibility of double-heterozygous mutations.3,13,16-19 Although this is rarely reported (about 5% of all cases) given the relative common incidence of sarcomeric mutations in general, this may not be such a rare event as is reported, because not all sarcomeric genes are always sequenced.3,12 Missing double heterozygosity will have implications for genetic counselling and cardiac care: family members can be reassured and dismissed from cardiological screening due to failure to recognise clinically relevant double heterozygosity.
Our aim is to find out whether there are clinical differences in HCM manifestations between doubleheterozygous mutation carriers compared with single mutation carriers. Here we describe a family with double heterozygosity for pathogenic mutations in cysteine and glycine-rich protein 3 (CSRP3) and MYH7. This family illustrates that a more severe HCM phenotype is found in double-heterozygous subjects. In addition, it suggests that differences in severity of HCM within a family may be an important factor in deciding whether to search further for double heterozygosity or not.
Methods
Clinical evaluation
Cardiological evaluation of the family members was based on two-dimensional echocardiography (echo), electrocardiogram (ECG), exercise-testing, Holter monitoring, NT-proBNP (Roche Elecsys) and cardiac magnetic resonance imaging (CMR) according to standard procedures for the assessment of HCM.
All CMR images were acquired with a 1.5T Philips Intera scanner (Philips Medical Systems, Best, the Netherlands) with a phased-array cardiac coil placed around the thorax of the patients lying in the decubitus position. An ECG was used to monitor heart rate and for R-wave triggering. Cine and late enhancement images were acquired.
CMR images were analysed using the CASS software (Pie Medical Inc, Maastricht, the Netherlands). End-diastolic and end-systolic volumes were measured by manual delineation of endocardial and epicardial borders on continuous cine short axis images.
Echo was performed using a Sonos 7500 S3 transducer (Philips Medical Systems, Andover, Massachusetts). Echo investigations were performed in the standard parasternal, apical, and subxiphoidal views according to the recommendations of the American Society of Echocardiography.20 Left ventricle enddiastolic diameter (LVEDD), end-systolic diameter (LVESD), end-diastolic thickness of the septum and left ventricle (LV) posterior wall were measured. The LV ejection fraction was calculated from LVEDD and LVESD.
DNA diagnostics
Genomic DNA was extracted from whole blood by standard procedures using the Wizard genomic DNA purification kit (Promega, Leiden, the Netherlands). Mutation analysis was carried out for the full coding exons of known HCM genes. The genes investigated were MYH7, MYBPC3, TPM1, TNNT2, TNNI3, TNNC1, MYL2, MYL3, TCAP, CSRP3, ACTC and PRKAG2. Primers and reaction conditions used to amplify the sarcomeric genes are available upon request. Sequence reaction products were run on the ABI 3100 or the ABI 3730 (Applied Biosystems). Sequence data were analysed in comparison with reference sequences using Vector Nti sequence software.
Results
Identification of the double-heterozygous mutations
The first DNA analyses were carried out in 1995 on the basis of blood from the proband III:2. The following genes were tested, by the (now incomplete) standard of those days: alpha-tropomyosin (TPM1), MYBPC3 and MYH7. After renewed entire gene analyses in 2006, we found a known pathogenic mutation c.1988G>A (p.Arg633His) in exon 18 of MYH7.21 HCM Position Arg633 is highly conserved and assigned as a hotspot for mutations since a founder effect was excluded by haplotype analysis.21-23
In addition, in 2007 we analysed the other known HCM genes and found another novel pathogenic mutation in the CSRP3 gene c.50insGCAGATTTCTT (p.Tyr18GInfsX194). Due to this mutation in the first Zinc-finger, the predicted protein sequence hereafter changes completely and the structure of CSRP3 is therefore disturbed. This mutation was not found in 96 other HCM index patients and not found in 150 control persons.
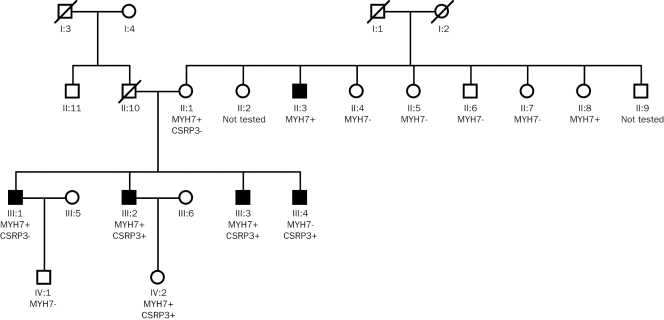
The family was then screened for both mutations in MYH7 and CSRP3 (figure 1). All four male sibs III:1–III:4 and their mother II:1 were carriers of either one of the two mutations or carriers of both mutations. Clinically they all have signs of HCM. III:2 and III:3 are both double-heterozygous mutation carriers and are phenotypically more severely affected. Also the daughter of III:2 carried both mutations, but did not have any signs of HCM.
Figure 1.
Pedigree family with double-heterozygous pathogenic HCM mutations.
Additionally, six of the eight brothers and sisters from the mother were screened genetically and cardiologically. From the tested sibs only one male carried the MYH7 mutation and showed signs of HCM, and only one female carried the MYH7 mutation and showed no signs of HCM. We assume that the CSRP3 mutation is inherited from the father II:10, because II:1 only carries the MYH7 mutation, while more of her children inherited the CSRP3 mutation. II:10 died at the age of 57 due to a non-cardiac illness, it was not possible to test his DNA. He underwent coronary artery bypass grafting at an age of 45. No information is available from his brother (II:11). In both families only one case of resuscitation (II:2) is known, but no detailed clinical information could be obtained.
Clinical characterisation
Characteristics of the double-heterozygous carriers: The index (III:2) was identified after the presentation of a heart murmur at the age of 30, when HCM was diagnosed (table 1). At this time he also had complications of exertional dyspnoea with an NYHA classification II and pre-collapse associated with a sicksinus syndrome. After the implantation of a VVIR pacemaker (PM) he first suffered from paroxysmal atrial fibrillation (AF) and in 2005 he developed permanent AF for which he underwent His ablation. At first presentation the ECG and echo tests both showed signs of left ventricular hypertrophy (LVH) and an enlarged left atrium (LA). III:2 showed a maximal wall thickness of 25 mm on echo.
Table 1.
Clinical features of family members identified as gene carriers.
| Symptoms and clinical profile |
Echo/CMR data |
ECG data |
Lab |
Exercise test |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family member | Age/Sex | Mutations | Age at onset* (years) | NYHA class | Syncope | AF | HBP | Pacemaker | NSVT | Max wall thickness LV# (mm) Echo/CMR | Diameter LA (mm) | EF (%) | SAM | e/a ratio | LVOT gradient | Fibrosis | RE Score | NT-proBNP (pmol/l)‡ | Systolic blood pressure response (mmHg) |
| II:10 | †.M | CSRP? MYH7- | ? | ? | ? | ? | ? | ? | ? | ?/? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| II:1 | 69/F | CSRP- MYH7+ | 54 | - | - | - | - | - | - | 14/13 | 46 | 70 | - | 0.5 | + | - | 5 | 29.1 | 30 |
| III:1 | 51/M | CSRP- MYH7+ | 50 | - | - | - | - | - | - | 11/17 | 49 | 70 | - | 1.1 | + | + | 3 | 11.0 | 130 |
| III:2 | 47/M | CSRP+ MYH7+ | 30 | II-III | - | + | - | + | - | 25/? | 54 | 55 | +/− | 2 | - | ? | 12 | 220 | 15 |
| III:3 | 44/M | CSRP+ MYH7+ | 29 | III | + | + | - | + | - | 21/? | 62 | 62 | + | 3.1 | + | ? | 9 | 139 | ? |
| III:4 | 42/M | CSRP+ MYH7- | 41 | - | - | - | - | - | - | 9/12 | 48 | 63 | - | 2.3 | - | + | 1 | 0.8 | 50 |
| IV:2 | 19/F | CSRP+ MYH7+ | - | - | - | - | - | - | - | 8/8 | 32 | - | - | 1.6 | - | - | 0 | 8.2 | 55 |
AF=atrial fibrillation, CMR=cardiac magnetic resonance imaging, Echo=echocardiography, F=female, HBP=high blood pressure (hypertension), LVOT=left ventricular outflow tract, LV=left ventricle, M=male, NSVT=nonsustained ventricular tachycardia, NYHA=New York Heart Association, RE score=left ventricular hypertrophy score according to Romhilt and Estes, SAM=systolic anterior motion of the mitral valve, VT=ventricular tachycardia, ?=not known i.e. test was not carried out. *Age at onset was defined as age where HCM was measured for the first time. # Maximal wall thickness LV - measured on echo or CMR. ‡ Normal value=0–35 pmol/l. +Presence of a symptom or therapy. -Absence of a symptom or therapy.
The proband's brother (III:3) consulted the cardiologist two years later because of tiredness and atypical chest pain. The echo and ECG indicated signs of LVH, restrictive relaxation pattern, left ventricle outflow tract (LVOT) gradient and an enlarged LA. An elective heart catheterisation showed no signs of coronary atherosclerosis or bridging. Two years after the diagnosis of HCM had been made, he received a DDD pacemaker in order to reduce both his symptoms and the LVOT gradient.24 In 2006 the LVOT gradient worsened and mitral insufficiency occurred. In 2006 he was electrocardioverted twice, once for AF and once for atrial tachycardia. In the last couple of years he collapsed a few times. On echo III:3 showed a maximal wall thickness of 21 mm. Both sibs have high NT-proBNP levels, 220 and 139 pmol/l respectively. The daughter IV:2 of III:2 did not have any symptoms or abnormalities to indicate clinical HCM on ECG, echo, CMR, at the age of 19 years.
Characteristics of the single MYH7 mutation carriers: The mother (II:1) was diagnosed with borderline HCM with a LV wall thickness of 10 mm and diastolic dysfunction without clinical symptoms. Subsequently, echo indicated an asymmetric septum hypertrophy of 14 mm. On CMR, both crypts and a systolic jet in the LVOT were observed. II:1 had symptoms of palpitations and a run of supraventricular extrasystoles was visible on Holter monitoring. III:1 shows no clinical symptoms of HCM, but had asymmetric LVH with a maximal wall thickness septal of 17 mm, a jet in the LVOT and fibrosis on the CMR.
Characteristics of the single CSRP-mutation carrier: III:4 shows no clinical symptoms, but had fibrosis and a maximal wall thickness of 12 mm on CMR.
Discussion
This family illustrates how double-heterozygous mutations aggravate the clinical severity of HCM. Secondly it illustrates that in screening families there are indeed clinical differences between single and double mutation carriers.
In this family, double heterozygosity was associated with increased atrial arrhythmias, more severe LV thickening, an earlier age of onset and also higher values of NT-proBNP (more than four times the normal value). The two male carriers of double mutations expressed the disease at the age of 30. Only IV:2, now 19 years old, did not show any signs of HCM, which could be explained by the known sex-related mitigation of the disease and especially because of her young age.25
The clinical presentation of the MYH7 mutation carriers is comparable with what has been reported earlier for this type of mutation. Gruver et al. described a population of 24 individuals with the same missense mutation in the β-MHC gene, Arg663His. They found a population characterised by a high prevalence of AF and modest LVH. Survival of affected individuals was near normal, although there were family histories of sudden cardiac death (SCD) before the age of 50 years.21
Mutations in the CSRP3 gene have only been described in rare cases of patients with both dilated cardiomyopathy and HCM.7,26,27 Carriers have mild symptoms and a great variation regarding the extent of hypertrophy, but in some cases SCD is described.7,19 The mutation c.50insGCAGATTTCTT (p.Tyr18GInfsX194) in the CSRP3 gene has not been described before. This mutation, when single, appears to induce mild disease with a maximal basal inferoseptal wall thickness of 12 mm and fibrosis on CMR, with no clinical symptoms, low NT-proBNP and no arrhythmias. In the described family the CSRP3 gene mutation was only found in one patient as a single mutation.
In this family the two mutations separately give a mild phenotype, but together they are related to a more severe phenotype. This phenomenon has been described in earlier studies.12 It has been reported that double-heterozygous patients had a higher extent of hypertrophy, earlier onset of disease and premature mortality.12
Failure to recognise double heterozygosity can lead to problems: when only one disease mutation is recognised, the absence of the other mutation could lead to possible false reassurance of family members that carry the other mutation. In this family the identification of the second mutation gave an explanation for the more severe phenotype of the two doubleheterozygous sibs. The Dutch policy is that once a disease-causing mutation is identified, cardiological phenotyping is done only when genotyping reveals a family member who carries the mutation.15 This strategy holds the ability of missing out on cases concerning families where two mutations segregate. The alternative is to perform cardiological phenotyping in all family members.
As described before, it is more cost-effective to start with the screening of MYH7 and MYBPC3 in the DNA of the index before screening the other HCM genes.3 Based on the literature and this family, after one mutation is found, the most cost-effective option is to screen the relatives for the specific mutation in combination with cardiological screening. The costs per gene are € 680 and the costs per ECG and echo are € 270. When an inexplicable discrepancy between phenotype and genotype in a family is found, further DNA analyses of the severely affected ones should be considered. Cardiological screening with echo and ECG is six times more expensive than screening with NT-proBNP and ECG. But more clinical evidence is needed before NT-proBNP can be used as screening tool instead of an echo to identify double-heterozygous mutation carriers.
The described family, although admittedly small, illustrates the usefulness of using a biomarker like NTproBNP in the screening procedure. In this particular family, screening by only NT-proBNP directly seems to identify cases that are more severely affected and are double mutations carriers. This finding is supported by mouse models, where double-heterozygous mice also had an increased expression of BNP.28 Magga et al. have already shown that NT-proBNP is associated with incipient LV remodelling by carriers of a sarcomeric gene mutation.29 NT-proBNP may be a feasible and rapid tool in order to screen families with HCM parallel to the genetic screening. Further clinical studies are needed to confirm our data and to define cut-off values in this specific group of patients.
CMR has already proven to be an important tool for evaluation of patients suspected of having HCM.30,31 CMR is a more sensitive method to measure the maximal wall thickness compared with echo.31 High signal intensity on late gadolinium enhancement imaging, i.e. fibrosis, occurs in up to 80% of the patients with HCM.32 Also typical crypt formation has been found in 81% of presymptomatic HCM mutation carriers without LVH.33 Only the patients with a single mutation underwent a CMR. In 50% of the cases fibrosis was visible on CMR images, one patient showed crypts and the wall thickness was between 8 to 17 mm on CMR.
In this article we describe one family with doubleheterozygous mutations, but the prevalence, clinical diversity and pathogenicity of double mutations in general involved in HCM needs further investigation.
Conclusion
With the improvement of molecular-genetic testing, double-heterozygous mutations are more often found in families with HCM. Double-heterozygous mutations can be a cause of the well-known clinical diversity in HCM families. The family we describe here emphasises the more severe clinical phenotype associated with double-heterozygous mutations. The result also gives an implication of assessing the cardiac phenotype in addition to genetic screening, and suggests that measurement of NT-proBNP could be useful as a rapid screening method.
References
- 1.Maron BJ, Gardin JM, Flack JM, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Nichols PF, 3rd, Pickle LW, et al. Patterns of inheritance in hypertrophic cardiomyopathy: assessment by M-mode and twodimensional echocardiography. Am J Cardiol 1984;53:1087–94. [DOI] [PubMed] [Google Scholar]
- 3.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–32. [DOI] [PubMed] [Google Scholar]
- 4.Van Driest SL, Ommen SR, Tajik AJ, et al. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clinic Proc. 2005;80:463–9. [DOI] [PubMed] [Google Scholar]
- 5.Van Driest SL, Jaeger MA, Ommen SR, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:602–10. [DOI] [PubMed] [Google Scholar]
- 6.Towbin JA. Molecular genetics of hypertrophic cardiomyopathy. Curr Cardiol Reports. 2000;2:134–40. [DOI] [PubMed] [Google Scholar]
- 7.Geier C, Perrot A, Ozcelik C, et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–5. [DOI] [PubMed] [Google Scholar]
- 8.Ho CY, Seidman CE. A contemporary approach to hypertrophic cardiomyopathy. Circulation. 2006;113:e858–62. [DOI] [PubMed] [Google Scholar]
- 9.Alders M, Jongbloed R, Deelen W, et al. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. Eur Heart J. 2003;24:1848–53. [DOI] [PubMed] [Google Scholar]
- 10.Maron BJ, Chaitman BR, Ackerman MJ, et al. Recommendations for physical activity and recreational sports participation for young patients with genetic cardiovascular diseases. Circulation. 2004;109:2807–16. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen L, Chung J, Lam L, et al. Abnormal cardiac response to exercise in a murine model of familial hypertrophic cardiomyopathy. Int J Cardiol. 2007;119:245–8. [DOI] [PubMed] [Google Scholar]
- 12.Tsoutsman T, Bagnall RD, Semsarian C. Impact of multiple gene mutations in determining the severity of cardiomyopathy and heart failure. Clin Exp Pharmacol Physiol. 2008;35:1349–57. [DOI] [PubMed] [Google Scholar]
- 13.Ingles J, Doolan A, Chiu C, et al. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42:e59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Langen IM, Arens Y, Baars H, et al. Concept Multidisciplinaire richtlijn. Genetische diagnostiek en erfelijkheidsadvisering bij Hypertrofische Cardiomyopathie (HCM). Submitted 2009. [Google Scholar]
- 15.Michels M, Hoedemaekers YM, Kofflard MJ, et al. Familial screening and genetic counselling in hypertrophic cardiomyopathy: the Rotterdam experience. Neth Heart J. 2007;15:184–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marian AJ. Modifier genes for hypertrophic cardiomyopathy. Curr Opin Cardiol. 2002;17:242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–10. [DOI] [PubMed] [Google Scholar]
- 18.Alpert NR, Mohiddin SA, Tripodi D, et al. Molecular and phenotypic effects of heterozygous, homozygous, and compound heterozygote myosin heavy-chain mutations. Am J Physiol. 2005;288:H1097–102. [DOI] [PubMed] [Google Scholar]
- 19.Bos JM, Poley RN, Ny M, et al. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol Gen Metabol 2006;88:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiller NB, Shah PM, Crawford M, et al. Recommendations for quantitation of the left ventricle by two-dimensional echocardiography. American Society of Echocardiography Committee on Standards, Subcommittee on Quantitation of Two-Dimensional Echocardiograms. J Am Soc Echocardiogr. 1989;2:358–67. [DOI] [PubMed] [Google Scholar]
- 21.Gruver EJ, Fatkin D, Dodds GA, et al. Familial hypertrophic cardiomyopathy and atrial fibrillation caused by Arg663His betacardiac myosin heavy chain mutation. Am J Cardiol. 1999;83:13H–8H. [DOI] [PubMed] [Google Scholar]
- 22.Greber-Platzer S, Marx M, Fleischmann C, et al. Beta-myosin heavy chain gene mutations and hypertrophic cardiomyopathy in Austrian children. J Mol Cell Cardiol. 2001;33:141–8. [DOI] [PubMed] [Google Scholar]
- 23.Song L, Zou Y, Wang J, et al. Mutations profile in Chinese patients with hypertrophic cardiomyopathy. Clin Chim Acta; Int J Clin Chem. 2005;351:209–16. [DOI] [PubMed] [Google Scholar]
- 24.Sadoul N, Simon JP, de Chillou C, et al. Dual chamber pacemaker therapy in obstructive hypertrophic cardiomyopathy. Circulation. 1995;92:1062–4. [PubMed] [Google Scholar]
- 25.Bos JM, Theis JL, Tajik AJ, et al. Relationship between sex, shape, and substrate in hypertrophic cardiomyopathy. Am Heart J. 2008;155:1128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoll R, Hoshijima M, Hoffman HM, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–55. [DOI] [PubMed] [Google Scholar]
- 27.Mohapatra B, Jimenez S, Lin JH, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Gen Metabol 2003;80:207–15. [DOI] [PubMed] [Google Scholar]
- 28.Tsoutsman T, Kelly M, Ng DC, et al. Severe heart failure and early mortality in a double-mutation mouse model of familial hypertrophic cardiomyopathy. Circulation. 2008;117:1820–31. [DOI] [PubMed] [Google Scholar]
- 29.Magga J, Sipola P, Vuolteenaho O, et al. Significance of plasma levels of N-terminal Pro-B-type natriuretic peptide on left ventricular remodeling in non-obstructive hypertrophic cardiomyopathy attributable to the Asp175Asn mutation in the alpha-tropomyosin gene. Am J Cardiol. 2008;101:1185–90. [DOI] [PubMed] [Google Scholar]
- 30.Hansen MW, Merchant N. MRI of hypertrophic cardiomyopathy: part I, MRI appearances. AJR Am J Roentgenol. 2007;189:1335–43. [DOI] [PubMed] [Google Scholar]
- 31.Germans T, van Rossum AC. The use of cardiac magnetic resonance imaging to determine the aetiology of left ventricular disease and cardiomyopathy. Heart. 2008;94:510–8. [DOI] [PubMed] [Google Scholar]
- 32.Moon JC, Mogensen J, Elliott PM, et al. Myocardial late gadolinium enhancement cardiovascular magnetic resonance in hypertrophic cardiomyopathy caused by mutations in troponin I. Heart. 2005;91:1036–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germans T, Wilde AA, Dijkmans PA, et al. Structural abnormalities of the inferoseptal left ventricular wall detected by cardiac magnetic resonance imaging in carriers of hypertrophic cardiomyopathy mutations. J Am Coll Cardiol. 2006;48:2518–23. [DOI] [PubMed] [Google Scholar]