Abstract
In the 1950s and 1960s, alphaviruses in the Venezuelan equine encephalitis (VEE) antigenic complex were the most frequently isolated arboviruses in Trinidad. Since then, there has been very little research performed with these viruses. Herein, we report on the isolation, sequencing, and phylogenetic analyses of Mucambo virus (MUCV; VEE complex subtype IIIA), including 6 recently isolated from Culex (Melanoconion) portesi mosquitoes and 11 previously isolated in Trinidad and Brazil. Results show that nucleotide and amino acid identities across the complete structural polyprotein for the MUCV isolates were 96.6 – 100% and 98.7 – 100%, respectively, and the phylogenetic tree inferred for MUCV was highly geographically- and temporally- structured. Bayesian analyses suggest the sampled MUCV lineages have a recent common ancestry of approximately 198 years (with a 95% highest posterior density (HPD) interval of 63 – 448 years) prior to 2007, and an overall rate of evolution of 1.28 × 10−4 substitutions/site/yr.
Keywords: Venezuelan equine encephalitis, alphavirus, Mucambo, phylogeny, mosquito, Trinidad, evolution, Bayesian coalescent theory
Introduction
In the 1950s and 1960s, the Caribbean island of Trinidad, located 10 km off the northeastern coast of Venezuela, was the site of seminal work in arbovirology. This work resulted in the isolation and characterization of hundreds of arboviruses, primarily from the Bush Bush Forest, an approximately 0.85 sq km region within the Nariva swamp in southeastern Trinidad (Downs et al., 1959; Downs et al., 1968; Jonkers et al., 1968a, 1968b; Spence et al., 1969; Spence et al., 1968). Since then, with the exception of work on Dengue virus (DENV) evolution and dispersal (Carrington et al., 2005; Foster et al., 2003; Foster et al., 2004) and several serological studies in the 1970s and early 1980s (Price, 1978a, 1978b; Tikasingh et al., 1974; Tikasingh et al., 1983), there has been very little work performed on Trinidad’s arboviruses. While there are few reports of arboviral disease or infection other than DENV, this may be the result of poor surveillance and reporting, inadequate laboratory diagnosis, and/or sub-clinical infections, rather than an indication of the absence of other arboviral infections. Indeed, increased rates of urbanization, human encroachment on forested areas, tourism, and business travel over the past 30 years in Trinidad, have probably increased the likelihood of emergence/re-60 emergence of both known and unknown arboviruses.
During the Bush Bush studies, alphaviruses in the Venezuelan equine encephalitis (VEE) complex were the most frequently isolated arboviruses (Jonkers et al., 1968a). VEE complex viruses are single-strand positive sense RNA viruses of the family Togaviridae, genus Alphavirus, and are among the most important alphaviral pathogens (Weaver et al., 1999). Their genomes are approximately 11.4 kb in length and encode 4 non-structural proteins (nsP1, nsP2, nsP3 and nsP4) and 3 structural proteins (capsid and envelope glycoproteins E2 and E1). The VEE complex is classified into 6 serotypes (I to VI), but within this serocomplex there are 13 serologically distinct subtypes and varieties (Walton and Grayson, 1988; Young and Johnson, 1969), of which VEE virus (VEEV) subtypes IAB and IC (Oberste et al., 1998; Walton and Grayson, 1988) are associated with equine epizootics and human epidemics of VEE (Walton and Grayson, 1988; Young and Johnson, 1969). Epizootics can involve tens-to-hundreds of thousands of equine cases (Weaver et al., 1999) with up to 80% mortality (Johnson and Martin, 1974). In humans, VEEV epidemic subtypes cause a disease spectrum, including inapparent infection, febrile illness, encephalitis, and in a minority of cases, death (Walton and Grayson, 1988; Weaver et al., 1998). The remaining subtypes are enzootic and equine avirulent (Walton et al., 1973); however, some enzootic VEEVs may be pathogenic to humans and can cause fatalities (Johnson and Martin, 1974; Zárate et al., 1970).
As a consequence of its high morbidity and mortality, VEEV subtype I has been extensively studied. It has been isolated in Texas, Mexico, Guatemala, El Salvador, Nicaragua, Panama, Colombia, Ecuador, Peru, Venezuela, and Trinidad (Weaver and Barrett, 2004). Phylogenetic analyses in combination with in vivo and in vitro experiments have yielded important insights into the evolution (Brault et al., 2002) and pathogenesis (Walton and Grayson, 1988) of VEEV, including the identification of amino acid changes associated with virulence and the emergence of epizootic strains from enzootic progenitors (Anishchenko et al., 2006; Wang et al., 1999). Although there have been several isolations of other VEE complex subtypes in Florida, Brazil, Peru, Argentina, and Trinidad, there are very little sequence data available for these subtypes (Weaver and Barrett, 2004). For example, whereas there are 120 GenBank entries for VEEV (subtype I), there are only 18 for subtypes II–VI.
Herein, we report on the isolation, partial nucleotide sequencing, and phylogenetic analyses of 17 VEE complex subtype IIIA viruses (Mucambo virus; MUCV), 6 of which were isolated recently from pools of Cx.(Melanoconion) portesi collected during a mosquito surveillance between July 2007 and July 2008 in the Aripo Savannah Scientific Reserve (ASSR) in Trinidad. The remaining 11 isolates were from the World Reference Center for Emerging Viruses and Arboviruses collection at the University of Texas Medical Branch (UTMB) and were originally isolated in Trinidad (n=8) and Brazil (n=3) between 1961 and 1972. We used these newly derived sequences, together with the corresponding sequence from the only MUCV isolate represented in GenBank, to infer phylogenetic trees and determine the nature of evolutionary pressures acting on MUCV. Additionally, using a Bayesian phylogenetic approach, we estimated its evolutionary rate and dates of divergence.
Materials and Methods
The Study Site
The study was carried out within the ASSR, situated in the northeast of Trinidad. The region encompasses various ecologically diverse areas including savannahs, and both primary and secondary forests. The topology is depressed and many areas are submerged periodically in the wet season (Ahmad and Jones, 1969). Additionally the soil, which is mainly composed of Aripo Fine Sand (Plinthaquult) (Ahmad and Jones, 1969), tends to be pitted such that small pools of water (adequate for breeding of Culex and Aedes mosquitoes) form after rainfall, particularly in the savannahs. At least 186 vertebrate species (including 26 mammalian and 128 avian) ranging in size from the grass mouse (Akodon urichi) to the brocket deer (Mazana americana) (Schwab, 1988) have been recorded in the reserve, some of which have been implicated as reservoirs for VEEV in studies in Trinidad (Jonkers et al., 1968a).
Mosquito Collection
Mosquitoes were collected on average once a week using CDC mini light traps (John Hock, USA) baited with CO2 and operated for 16 h periods (1600 to 0800 h) along 5 transects, and set approximately 1.5 m above the ground. Mosquitoes were transported to the laboratory at the University of the West Indies on dry ice (or in a minority of cases on wet ice) and then stored at −70°C until taxonomic identification on a refrigerated table maintained at 4°C. After identification, specimens were divided into pools of 1 – 50 mosquitoes according to species and then stored at −70°C until assayed for virus.
Virus Isolations
Mosquito pools were triturated in 750 µL of diluent, (5% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 µg/ml), and gentamycin (0.05 mg/ml)) with a TissueLyser (Qiagen, Valencia, CA) using a 3 mm stainless steel bead at 26,000 motions per minute for 4 minutes and then centrifuged at 10,000 rpm for 5 min at 4°C. Subsequently, each sample was assayed for virus using African green monkey kidney cell (Vero cell) monolayers. Briefly, 100 µL of supernatant was inoculated onto Vero cell monolayers in a 24-well plate format, incubated at 37°C for one hour and then 1 mL of overlay media (2% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml), gentamycin (0.05 mg/ml), and amphotericin B (5.6 µg/ml)) was added to the cells. The cells were observed daily for cytopathic effect (CPE) for up to 14 days. If CPE was detected, the assay was repeated for confirmation and the cell culture supernatant was then removed and stored at −70°C for further characterization by serology and nucleotide sequencing. Repeat assays were performed distal to each other to avoid the possibility of cross-contamination.
Virus Identification, Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Nucleotide Sequencing
The serological group affinities of the isolated viruses were determined by the complement fixation microtechnique adapted from Fulton and Dumbell (1949), using supernatants from infected Vero cells as the source of antigens. The test was performed using two units of complement and hyperimmune mouse ascitic fluids prepared against Group A, Group C, Group Patois, Caraparu and Guama group viruses and primary incubation overnight at 4°C. This resulted in the identification of 6 alphaviruses (Group A) and 10 orthobunyaviruses (Caraparu-like Group C and Guama) (Table 1).
Table 1.
Viruses used in the study and their associated information.
| Isolate ID | Source | Location | Date trapped |
Passage history |
Serological classification |
Virus identity based on sequence similarity |
Accession numbers |
|---|---|---|---|---|---|---|---|
| BeAn159704 | Monkey | Brazil | 02/20/69 | SM2, Vero | VEEV subtype IIIA | MUCV | FJ792625 |
| BeAn166609 | Cx. (Mel.) portesi | Brazil | 1969 | SM1, Vero | VEEV subtype IIIA | MUCV | FJ792624 |
| BeAn8* | Monkey | Brazil | 1954 | Unknown | VEEV subtype IIIA | MUCV | AF075253 |
| Span15600 | Unknown | Brazil | 1969 | ?, Vero | VEEV subtype IIIA | MUCV | FJ792623 |
| TRI1109 | Cx. (Mel.) portesi | Aripo, Trinidad | 02/19/08 | Vero | Caraparu- like Group C | - | - |
| TRI1122 | Cx. (Mel.) portesi | Aripo, Trinidad | 02/19/08 | Vero | Caraparu- like Group C | - | - |
| TRI1753 | Cx. (Mel.) portesi | Aripo, Trinidad | 04/22/08 | Vero | Guama group | - | - |
| TRI482 | Cx. (Mel.) portesi | Aripo, Trinidad | 11/27/07 | Vero | Caraparu- like Group C | - | - |
| TRI488 | Cx. (Mel.) portesi | Aripo, Trinidad | 11/27/07 | Vero | Caraparu- like Group C | - | - |
| TRI495 | Cx. (Mel.) portesi | Aripo, Trinidad | 01/21/08 | Vero | Caraparu- like Group C | - | - |
| TRI550 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/10/07 | Vero | Group A | MUCV | FJ792614 |
| TRI551 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/10/07 | Vero | Group A | MUCV | FJ792613 |
| TRI570 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/10/07 | Vero | Group A | MUCV | FJ792612 |
| TRI587 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/10/07 | Vero | Group A | MUCV | FJ792611 |
| TRI665 | Cx. . (Mel.) pedroi | Aripo, Trinidad | 11/22/07 | Vero | Caraparu- like Group C | - | - |
| TRI673 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/8/07 | Vero | Group A | MUCV | FJ792610 |
| TRI675 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/8/07 | Vero | Group A | MUCV | FJ792609 |
| TRI684 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/08/07 | Vero | Caraparu- like Group C | - | - |
| TRI712 | Cx. (Mel.) portesi | Aripo, Trinidad | 10/30/07 | Vero | Caraparu- like Group C | - | - |
| TRI953 | Cx. (Mel.) portesi | Aripo, Trinidad | 02/12/08 | Vero | Caraparu- like Group C | - | - |
| TRVL114272 | Sentinel mouse | Aripo, Trinidad | 1971 | SM1 | VEEV subtype III | MUCV | FJ792617 |
| TRVL115847 | Cx. (Mel.) portesi | Aripo, Trinidad | 1972 | SM1 | VEEV subtype III | MUCV | FJ792618 |
| TRVL38997 | Rodent | Trinidad | 1961 | ?, Vero | VEEV subtype III | MUCV | FJ792622 |
| TRVL39141 | Rodent | Trinidad | 1961 | ?, Vero | VEEV subtype III | MUCV | FJ792621 |
| TRVL52049 | Rodent | Trinidad | 1963 | ?, Vero2 | VEEV subtype III | MUCV | FJ792620 |
| TRVL62655 | Mosquito | Trinidad | 08/24/65 | ?. Vero | VEEV subtype III | MUCV | FJ792619 |
| TRVL86196 | Sentinel mouse | Turure, Trinidad | 1969 | SM10 | VEEV subtype III | MUCV | FJ792616 |
| TRVL98383 | Sentinel mouse | Turure, Trinidad | 1970 | SM20 | VEEV subtype III | MUCV | FJ792615 |
MUCV = Mucambo virus (VEEV subtype IIIA virus)
indicates reference strain; dates are written as mm/dd/yy.
After serological identification, RNA was extracted from 140 µL of cell culture supernatant using Qiagen viral RNA extraction kits (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RT-PCR was then performed on the alphaviruses using genus-specific primers that anneal to the nsP1 gene (Pfeffer et al., 1997), and the resulting 434 bp PCR amplificons were then subjected to direct sequencing. The derived nucleotide sequences were used for homology searches on the online database GenBank and were subsequently identified as MUCV.
To facilitate a more detailed phylogenetic characterization, a 3777 bp fragment encoding the complete structural polyproteins of the isolated alphaviruses (3764 nt) was amplified from each isolate and sequenced together with 11 isolates from the World Reference Center for Emerging Viruses and Arboviruses Collection at the UTMB (see Table 1). The sequence of interest (3777 bp) was resolved by 2 overlapping fragments of 2.8 kb and 2.4 kb using 2 primer pairs, MUCV26S/MUCV6kE1 and MUCV5’E2/MUCV3’E1, respectively, with an overlap of 1.3 kb.
PCR reactions were performed using the Titan One Tube RT-PCR kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s instructions except for the following adjustments: 3 mM MgCl2, 2 µL of enzyme mix, and 0.5 µL of RNAsin. For MUCV26S/MUCV6kE1, thermocycling was performed as follows: 45°C 45 min, 95°C for 2 min, and 36 cycles of (95°C 10 sec, 55°C for 30 sec and 68°C for 4.5 min) with a final extension at 68°C for 2 min. The same conditions were used for MUCV5’E2 /MUCV3’E1 except that the annealing temperature was 56°C. PCR products were separated by electrophoresis and visualized on a 1% agarose gel stained with ethidium bromide. The amplicons were excised and purified using the QiaQuick gel extraction kit (Qiagen, Valencia, CA) before being subjected to direct sequencing using BigDye Terminator v3.1 cycle sequencing kit (Roche) and an Applied Biosystems 3100 Genetic Analyzer Sequence cycling conditions were as follows: 96°C for 2 min, 35 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 2 min. A total of 24 primers were used for sequencing (Table 3). Ambiguous nucleotides were resolved by comparison of reads from overlapping fragments and where necessary the RT-PCR amplification and sequencing of a given fragment was repeated. Nucleotide sequences derived in this study were submitted to GenBank under Accession numbers FJ792609 – FJ792625.
Table 3.
Primers used for amplification and sequencing of MUCVa.
| Primer nameb | Genomic position (No. of nucleotides) |
Sequence (5’ → 3’) | Estimated melting temperature (°C) |
|---|---|---|---|
| MUCV 26S (+)c | 7429 – 7445 (17) | TAGTYCAGTCCGCAACC | 57.1 |
| MUCV 6kE1(−)c | 9876 – 9890 (15) | GTGTTCGTAGGCGCC | 58.7 |
| MUCV 5’E2 (+)c | 8561 – 8575 (15) | CGACGGGCAYGACGG | 66.7 |
| MUCV 3’E1 (−)c | 11183 – 11209 (23) | TAATTATGTTTCTGGTTGGTCAG | 59.4 |
| MUCV 7637 (−) | 7637 – 7654 (18) | TTAGCAGGTGGCCCCTCG | 68.5 |
| MUCV 7611 (+) | 7611 – 7626 (16) | TCCAATGCAACCAGCG | 62.4 |
| MUCV 7930 (+) | 7930 – 7947 (18) | TTGCCTCCCTGAAGACGA | 64.7 |
| MUCV 8138 (−) | 8138 – 8155 (18) | TCTAGTATGGGGCGTCCA | 61.7 |
| MUCV 8432 (+) | 8432 – 8449 (18) | CCGGCAGACAGAAGAGAT | 60.5 |
| MUCV 8638 (−) | 8638 – 8655 (18) | CTCTGCTCTTAACGTTCC | 54.9 |
| MUCV 8639 (−) | 8639 – 8659 (21) | ACTCTGCTCTTAACATTCCCG | 68.9 |
| MUCV 8811 (+) | 8811 – 8828 (18) | TCCTGTTCAGTGCCTTAT | 55.5 |
| MUCV 8942 (+) | 8942 – 8959(18) | AAGATGCGTATGTGGAGA | 56.5 |
| MUCV 9128 (−) | 9128 – 9145(18) | TACGCGCGGCATTGTGAT | 68.7 |
| MUCV 9132 (−) | 9132 – 9149 (18) | TACGCGCGGCATTGTGAT | 68.7 |
| MUCV 9311 (+) | 9311 – 9328 (18) | ACGGAATCCGACATATTT | 56.7 |
| MUCV 9430 (+) | 9430 – 9447 (18) | GTTTGGGGTAATCACCCG | 62.6 |
| MUCV 9638 (−) | 9638 – 9655 (18) | GGAGTCAAACAAGATACT | 48.0 |
| MUCV 9641 (−) | 9641 – 9658 (18) | CGGAGTCAAACAAGATAC | 52.6 |
| MUCV 9812 (+) | 9812 – 9829 (18) | CCGCTTGCTGAAGTGCAT | 65.3 |
| MUCV 10137 (−) | 10137 – 10154 (18) | GATACACCCCCGTGAAGA | 60.9 |
| MUCV 10311 (+) | 10311 – 10328 (18) | GCGGTAGTGTACGTGAAT | 55.9 |
| MUCV 10624 (−) | 10624 – 10641 (18) | TGTACTTTAATGATGGTG | 48.2 |
| MUCV 10822 (+) | 10822 – 10839 (18) | GGATCGCCACTGTCAAGT | 61.4 |
MUCV = Mucambo virus (VEEV subtype IIIA virus).
(+), positive sense primer; (−), negative sense primer.
Amplification and sequencing primers.
Data Sets
Nucleotide sequences derived in this study were aligned with the single MUCV structural polypeptide sequence available in GenBank (Accession No. AF075253) using Clustal X (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/), and then, manually adjusted using Se-Al (http://tree.bio.ed.ac.uk/software/seal/). A second data set of 59 taxa representing all the available structural polypeptide sequences of each VEE complex serotype and variety was similarly prepared and sequences were trimmed to a common length of 3435 bp. All isolates used in this study were confirmed as non-recombinant using the Recombination Detection Program version 3 (http://darwin.uvigo.es/rdp/rdp.html) (Martin et al., 2005) and the GARD recombination detection tool available in Datamonkey (Kosakovsky Pond and Frost, 2005).
Phylogenetic Analyses
Phylogenetic trees were constructed using distance and maximum likelihood (ML) methods. The General Time Reversible (GTR+Γ4+I) model was used for phylogeny reconstruction having been identified among the best-fit models of nucleotide substitution using MODELTEST version 3.7 (Posada and Crandall, 2001). Bootstrapping was performed to assess the robustness of tree topologies using 1,000 replicate neighbor-joining (NJ) trees under the ML substitution model. All analyses were performed with PAUP* version 4.0b (Sinauer Associates, Inc., Sunderland, MA, USA). Sequence comparison was performed using MegAlign from the DNAStar package (DNAstar, Inc., Madison, WI). To determine the nature of selective pressures acting on the structural polyprotein of MUCV, we employed two ML methods: Single Likelihood Ancestor Counting (SLAC) and Fixed Effects Likelihood (FEL), which are available within the Datamonkey web module of the HYPHY package (Kosakovsky Pond and Frost, 2005).
Inference of Evolutionary Rates and Divergence Dates
The BEAST (v1.4.8) package was used to infer substitution rates and times to common ancestry for the MUCV sequences studied. Analyses were carried out using the Bayesian Skyline Plot (Drummond et al., 2005) under three molecular clock models: the strict clock, the uncorrelated exponential (UCED) relaxed clock, and the uncorrelated lognormal (UCLN) relaxed clock. The latter two models allow rate variation among lineages to be estimated (Drummond et al., 2006). A chain length of 50 million, with sampling every 5000 states, was used to ensure convergence of the effective sample size (ESS) for each parameter. The program Tracer version 1.4 (http://tree.bio.ed.ac.uk/software/tracer/) was used to confirm convergence, and to perform model comparisons for the three molecular clocks tested, via Bayes Factor (BF) calculations. For Ln BF of 2.3 – 3.4 is strong evidence of supporting the preferred model, ln BF 3.4 – 4.6 is very strong evidence and ln BF of >4.6 is decisive. The software program TreeAnnotator version 1.4.8 (http://beast.bio.ed.ac.uk/software/TreeAnnotator) was used to summarize the data output from BEAST. The initial 2.5 million generations were discarded, a posterior probability limit of 0.0 was used and the node heights were set to illustrate mean heights. FigTree version 1.1.0 was used to visualize the results.
Results
Mosquito Collection and Virus Isolations
A total of 2,029 pools containing 30, 739 mosquitoes and representing 29 species was collected and screened for arboviruses. Aedes (Och.) serratus (17.2%), Cx. aikeni (19.1%), Cx. portesi (23.8%) and Cx. pedroi (26.4%) were most frequently collected, with the other 25 species accounting for the remaining 13.5% (Table S1). Sixteen cytopathic viruses were isolated, of which 6 were serologically determined to be alphaviruses and 10 orthobunyaviruses (Caraparu-like Group C and Guama) (Table 1). The alphaviruses produced CPE within 2 to 3 days, and the bunyaviruses between 2 to 10 days. Homology searches based on the nsP1 fragment (data not shown) of the alphaviruses confirmed that all were MUCV. Of the 16 viruses isolated, 15 were from Cx. portesi and one from a Cx. pedroi pool, with all of the MUCV isolates being derived from Cx. portesi mosquitoes trapped within 2 days of each other (Table 1). Figure 1 shows the variation in the number of mosquitoes collected (all species) and the number of Cx. portesi collected per trap night for each month with an indication of when the viruses were isolated. There was a significant correlation between number of viruses isolated (all species) and the mean number of Cx. portesi collected per trap night (Pearson’s correlation; p = 0.017).
Figure 1.
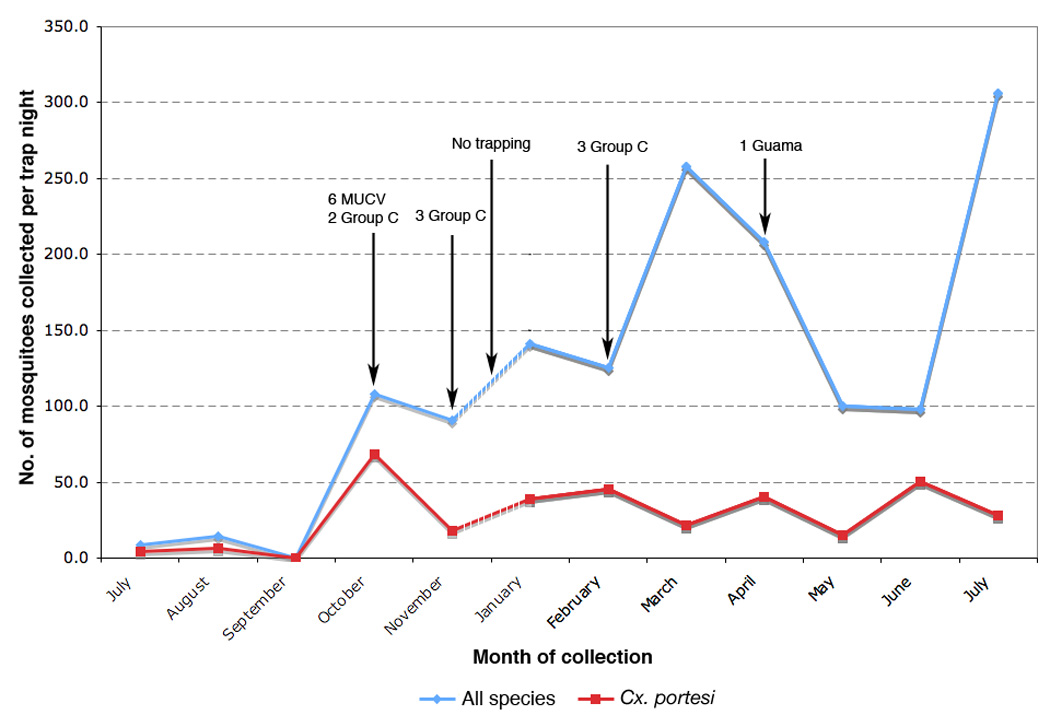
Total number of mosquitoes collected per trap per month and total number of Cx. portesi mosquitoes collected per trap per month between July 2007 and July 2008 in the Aripo Savannah Scientific Reserve (ASSR). Graph labels indicate which viruses were isolated from the mosquito collections.
Sequence Divergence and Phylogenetic Trees
The nucleotide and amino acid identities across the complete structural polyprotein for the MUCV isolates (Table 2) ranged from 96.6 – 100% and 98.7 – 100%, respectively, with the most divergent isolate being BeAn166609, a 1969 Brazilian virus isolated from Cx. portesi mosquitoes. Amino acid changes between isolates were observed at 32 sites within the complete structural polyprotein. The maximum likelihood phylogeny inferred from the 18 MUCV sequences (Fig. 2) demonstrates at least 3 distinct clades (NJ bootstrap values >95%). Clade 1 consisted of the 6 Trinidad isolates derived from the present study with a 1965 Trinidad isolate at its base, all having originated from mosquitoes. Seven other Trinidad viruses isolated between 1961 and 1972, of which 6 originated from rodents and 1 from a mosquito, formed another distinct group (Clade 2) that is more closely related to Clade 1 than Clade 3. The latter clade consisted of Brazilian isolates from both vertebrates and mosquitoes; 1 isolate from 1954 and 3 from 1969. Amino acid changes that define clades with high bootstrap values are also shown in Fig. 2. A second maximum likelihood tree including all other VEEV serotypes is shown in Figure 3. The overall tree topology was similar to those described in previous reports (Kinney et al., 1998; Aguilar et al., 2004). The 17 MUCV isolates that were sequenced in this study grouped together with the MUCV reference isolate BeAn8 and are supported by a 100% bootstrap value.
Table 2.
Nucleotide and amino acid sequence identities for MUCV strainsa.
| Percentage nucleotide sequence identity nucleotide (%) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Percentage amino acid sequence identity (%) |
Isolate ID |
TRI 550 |
TRI 551 |
TRI 570 |
TRI 587 |
TRI 673 |
TRI 675 |
TRVL 115847 |
TRVL 114272 |
TRVL 86196 |
TRVL 98383 |
TRVL 38997 |
TRVL 39141 |
TRVL 52049 |
BeAn 8 |
BeAn 159704 |
BeAn 166609 |
Span 15600 |
TRVL 62655 |
|
TRI550 |
- | 99.8 | 99.8 | 100 | 99.8 | 99.7 | 98.2 | 98.2 | 98.1 | 98.2 | 98.1 | 98.1 | 98.0 | 96.9 | 96.7 | 96.6 | 96.9 | 98.5 | |
|
TRI551 |
99.8 | - | 100 | 99.8 | 99.9 | 99.9 | 98.3 | 98.3 | 98.3 | 98.3 | 98.2 | 98.3 | 98.1 | 97.0 | 96.8 | 96.8 | 97.0 | 98.6 | |
|
TRI570 |
99.8 | 99.9 | - | 99.7 | 99.9 | 99.9 | 98.3 | 98.3 | 98.2 | 98.3 | 98.2 | 98.2 | 98.1 | 97.0 | 96.8 | 96.8 | 97.0 | 98.6 | |
|
TRI587 |
99.9 | 99.8 | 99.7 | - | 99.7 | 99.7 | 98.2 | 98.2 | 98.1 | 98.2 | 98.1 | 98.1 | 98.0 | 96.8 | 96.6 | 96.6 | 96.9 | 98.5 | |
|
TRI673 |
99.8 | 99.9 | 99.8 | 99.7 | - | 99.8 | 98.3 | 98.3 | 98.3 | 98.3 | 98.2 | 98.3 | 98.2 | 97.1 | 96.9 | 96.8 | 97.1 | 98.7 | |
|
TRI675 |
99.8 | 99.9 | 99.8 | 99.7 | 99.8 | - | 98.2 | 98.2 | 98.1 | 98.2 | 98.1 | 98.1 | 98.0 | 97.0 | 96.7 | 96.7 | 97.0 | 98.5 | |
|
TRVL115847 |
99.2 | 99.4 | 99.3 | 99.1 | 99.3 | 99.3 | - | 100 | 99.9 | 100 | 99.8 | 99.9 | 99.8 | 97.7 | 97.4 | 97.4 | 97.8 | 99.1 | |
|
TRVL114272 |
99.2 | 99.4 | 99.3 | 99.1 | 99.2 | 99.3 | 100 | - | 99.9 | 100 | 99.8 | 99.8 | 99.7 | 97.7 | 97.4 | 97.3 | 97.8 | 99.1 | |
|
TRVL86196 |
99.1 | 99.3 | 99.2 | 99.0 | 99.3 | 99.2 | 99.9 | 99.9 | - | 99.9 | 99.8 | 99.8 | 99.7 | 97.7 | 97.4 | 97.3 | 97.8 | 99.0 | |
|
TRVL98383 |
99.2 | 99.4 | 99.3 | 99.1 | 99.3 | 99.3 | 100 | 100 | 99.9 | - | 99.8 | 99.9 | 99.8 | 97.7 | 97.4 | 97.4 | 97.8 | 99.1 | |
|
TRVL38997 |
99.1 | 99.3 | 99.2 | 99.0 | 99.2 | 99.2 | 99.8 | 99.3 | 99.7 | 99.8 | - | 99.9 | 99.8 | 97.6 | 97.3 | 97.3 | 97.6 | 99.0 | |
|
TRVL39141 |
99.2 | 99.4 | 99.3 | 99.1 | 99.3 | 99.3 | 99.8 | 99.8 | 99.8 | 99.8 | 99.9 | - | 99.9 | 97.7 | 97.4 | 97.3 | 97.7 | 99.0 | |
|
TRVL52049 |
99.2 | 99.4 | 99.3 | 99.1 | 99.3 | 99.3 | 99.8 | 99.8 | 99.8 | 99.8 | 99.9 | 100 | - | 97.6 | 97.2 | 97.2 | 97.6 | 98.9 | |
|
BeAn8 |
98.8 | 99.0 | 98.9 | 98.7 | 99.0 | 98.9 | 99.1 | 99.1 | 99.0 | 99.1 | 99.0 | 99.1 | 99.1 | - | 99.2 | 99.1 | 99.9 | 97.5 | |
|
BeAn159704 |
98.8 | 99.0 | 98.9 | 98.7 | 98.9 | 98.9 | 99.1 | 99.1 | 99.0 | 99.1 | 99.0 | 99.1 | 99.1 | 99.4 | - | 99.8 | 99.2 | 97.3 | |
|
BeAn166609 |
98.8 | 99.0 | 98.9 | 98.7 | 98.9 | 98.9 | 99.1 | 99.1 | 99.0 | 99.1 | 99.0 | 99.1 | 99.1 | 99.4 | 99.8 | - | 99.2 | 97.3 | |
|
Span15600 |
98.8 | 99.0 | 98.9 | 98.7 | 99.0 | 98.9 | 99.3 | 99.3 | 99.2 | 99.3 | 99.0 | 99.1 | 99.1 | 99.7 | 99.4 | 99.4 | - | 97.6 | |
|
TRVL62655 |
99.3 | 99.4 | 99.4 | 99.2 | 99.4 | 99.4 | 99.6 | 99.6 | 99.5 | 99.6 | 99.2 | 99.5 | 99.6 | 99.2 | 99.2 | 99.2 | 99.2 | - | |
MUCV = Mucambo virus (VEE complex subtype IIIA).
Figure 2.
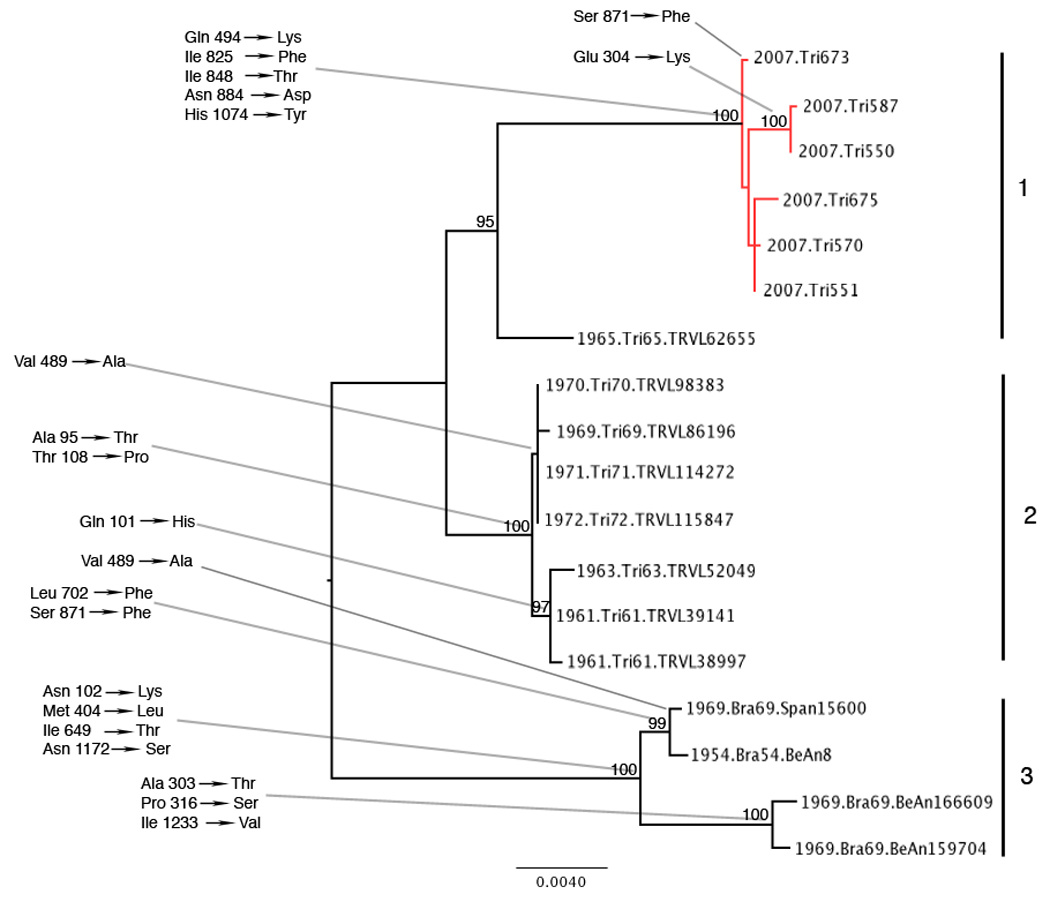
Midpoint rooted maximum likelihood phylogeny based on a 3,777 nt fragment of the 18 MUCV isolates used in the study. Taxon labels include date of isolation, location of isolation, and virus strain (pool number); accession numbers are shown in Table 1. Amino acid changes that defined each cluster are shown at the relevant nodes. Amino acids numbers shown refer to the complete structural polypeptide. Isolates that were derived in this study are shown in red. Bootstrap values above 95% are indicated on the tree, and the major clades are numbered 1–3. The scale bar indicates percent divergence. *positively selected amino acid changes.
Figure 3.
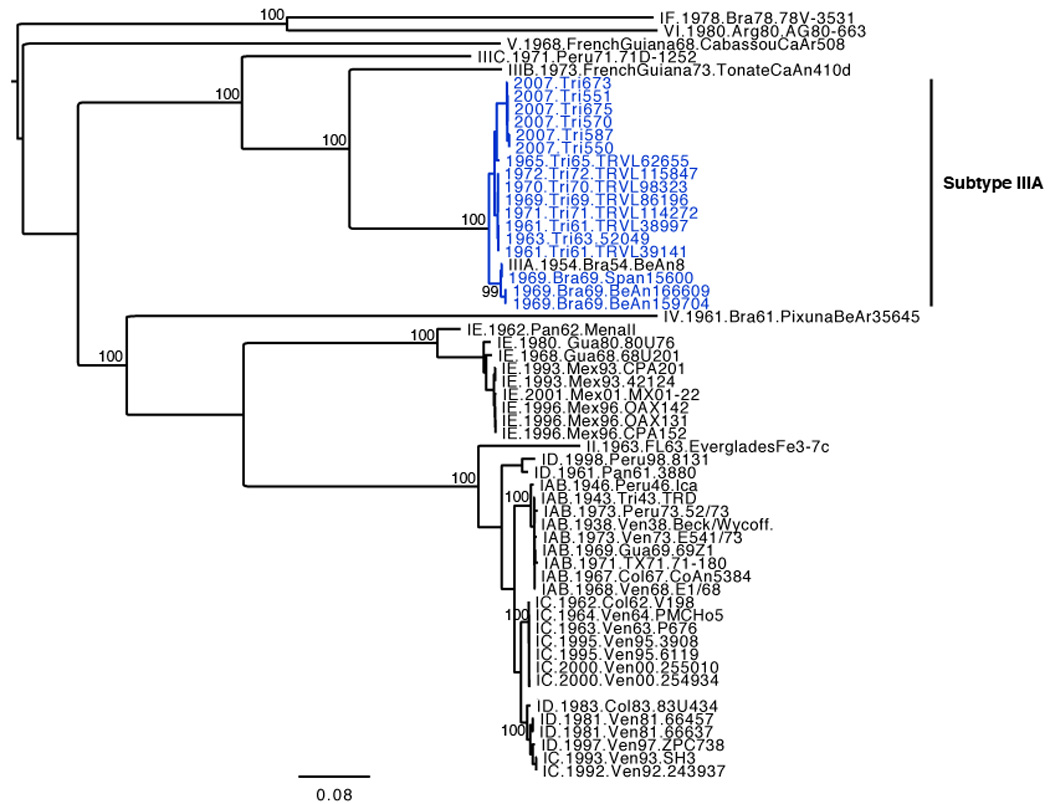
Midpoint rooted maximum likelihood phylogeny based on a 3,435 nt fragment of all VEEV subtypes available from GenBank. Taxon labels include: subtype, date of isolation, location of isolation, and virus strain (pool number). GenBank accession numbers are shown in Table 1. Bootstrap values above 95% are indicated on the tree, and the scale bar indicates percent divergence. Subtype IIIA isolates that were sequenced during this study are shown in blue.
Selection Pressures and Evolutionary Dynamics
Using the FEL algorithm, there was weak evidence of positive selection (p =0.075) at amino acid position 155 V → A of the E2 glycoprotein. This change defined a subclade within Clade 2 containing the Trinidad isolates collected between 1969 and 1972. However, using the same algorithm, negative selection was detected at 43 sites, and the global mean dN/dS ratio (as determined using the SLAC algorithm) was 0.1; indicating purifying selection is the predominant driving force for MUCV evolution.
The Bayesian skyline plot was used to infer evolutionary rates and dates of divergence for the entire data set of 18 taxa. The relaxed clocks had very similar likelihoods and were both much better (BF > 3.4) than the strict clock (BF for UCED vs. strict clock=5.4, UCLN vs. strict clock=4.4). Although the difference between the two relaxed clocks was only marginal (BF = 1), the better-supported UCED clock was used to estimate the above-mentioned parameters. The mean substitution rate estimated for the entire data set was 1.28 × 10−4 (with a 95% highest posterior density (HPD) interval of 0.21– 2.53) substitutions per site per yr (subs/site/yr), similar to previously estimated rates for vector-borne RNA viruses (Jenkins et al., 2002). The inferred age of the entire tree and times to common ancestry for individual lineages of MUCV are shown in Table 4. The results suggest that all sampled MUCV arose from a common ancestor that existed about 200 years ago (95% HPD, 63 – 448 yrs) with individual clades having arisen within the last 100 yrs. The mean estimate for the most recent common ancestor for the isolates collected during the current study (i.e., Trinidad 2007 isolates) suggests that it existed around 1991 (95% HPD, 1973 – 2005).
Table 4.
Dates of divergence for the tree and individual lineages estimated using UCEDa relaxed clock and Bayesian skyline plot.
| Lineages (n) | Years since divergenceb | Dates of MRCAb,c |
|---|---|---|
| All MUCVd (18) | 197.7 (63.0 – 447.8) | 1809 (1559 – 1944) |
| Clade I and II (14) | 103.2 (50.3 – 208.3) | 1904 (1799 – 1957) |
| Clade I (7) | 73.3 (43.6 – 144.0) | 1934 (1863 – 1963) |
| Clade II (7) | 63.0 (47.0 – 95.6) | 1944 (1911 – 1960) |
| Clade III (4) | 86.9 (53.4 – 159.3) | 1920 (1848 – 1954) |
| Trinidad 2007 isolates (6) | 15.5 (2.0 – 34.3) | 1991 (1973 – 2005) |
UCED, uncorrelated exponential.
95% highest posterior density (HPD) intervals are shown in parentheses.
MRCA, most recent common ancestor.
MUCV, Mucambo virus.
Discussion
The ASSR is a protected area, however there has been considerable encroachment by squatter communities. Our study confirms the presence of VEEV complex viruses as well as unidentified Caraparu-like Group C and Guama orthobunyaviruses close to these communities. The overall minimum infection rate (MIR) of mosquitoes during this study was 0.05%, which is comparable to the rate of 0.07% observed by Jonkers et al. (1968a) in Trinidad between 1959 and 1964 in the Bush Bush Forest. The MIR for MUCV was 0.02%, which is slightly lower than the rate of 0.05% reported in the same Bush Bush study. Although isolation of VEE complex alphaviruses have been previously reported from several mosquito species (Jonkers et al., 1968b), the only productive mosquito species in our study was Cx. portesi. This may suggest higher vector competence of Cx. portesi for these viruses, or a closer association with rodent reservoir hosts, when compared to other mosquito species that are collected from the same areas.
The 6 MUCV isolates were all derived from mosquitoes collected within 2 days of each other at 2 trap sites approximately 2 km apart. This was despite the fact that trapping was continued on a weekly basis for an entire year and Cx. portesi were continually trapped in large numbers. While this might suggest a localized outbreak originating from a single viremic host (i.e., the viruses arose from a common ancestor that existed immediately prior to or during the sampling period), the estimated age of the lineage, i.e. approximately 16 years (95% HPD 2.0 – 34.3) prior to 2007, argues against this. For a single vertebrate to have been the source it would have to have been viraemic for at least 2 years (i.e. the lower bound of the estimated age of the lineage) or alternatively several mosquitoes infected by it would have to have lived for at least 2 years. These scenarios are unlikely given that arboviruses cause only short-lived viraemia in vertebrate hosts and mosquitoes have a relatively short life span. Instead, the data suggest different sources of infection for the mosquitoes from which the MUCV was isolated, and detection of the virus in October may be a consequence of the increased Cx. portesi population density at that time (Fig. 1). Further studies over longer periods of time will be required to ascertain the relationship between Cx. portesi population density and MUCV prevalence and to confirm whether MUCV activity is seasonal. Additionally, larger data sets will allow for greater certainty about estimated evolutionary rates and divergence dates (i.e. smaller HPDs).
Our study increases the number of MUCV isolates for which sequence data are available from one to 18, and phylogenetic analysis together with representatives of all VEE complex subtypes confirms MUCV as a distinct species (Fig 3). The MUCV phylogeny inferred is highly geographically structured, with Clades 1 and 2 from Trinidad, and Clade 3 from Brazil. Within Trinidad, there is temporal structure that is suggestive of lineage turnover (Clade 2 consists of 1961 to 1972 isolates and Clade 1 contains 2007 isolates and a single 1965 isolate (Tr65) at its base); however, additional isolates that fill the large gap in our time line would be required to confirm this pattern. In fact Tr65 appears to be genetically closer to clade 2 than to clade 1 viruses, which is also consistent with the dates of isolation and thus Tr65 may represent a fourth clade.
Clades 1 and 2 also appear to be defined largely by species of origin, since all but one of the Clade 2 isolates were from rodents and Clade 1 isolates were all derived from mosquitoes. This is suggestive of in vivo host specific adaptation to mosquitoes and rodents (or in vitro adaptation, in the case of Clade 2 isolates, some of which underwent several passages in mammalian cells), however the observed pattern is more likely a reflection of the isolation method that predominated during each time period, with mouse brain inoculations having been used extensively in earlier studies (Jonkers et al., 1968a).
There are several non-conservative amino acid changes (including changes in polarity and / or charge) on branches leading to the three clades but none of these were identified as being under positive selection. This could indicate that any adaptive mutations occurred outside the structural polypeptide, or that adaptive mutations within the structural protein genes involved unique substitutions, which cannot be detected using the ML methods we employed.
Our estimated mean substitution rate for MUCV (1.28 × 10−4 substitutions/site/yr; 95% HPDs = 0.21– 2.53) is 2 – 3 fold lower than previously estimated for continuously circulating enzootic subtype ID (2.9 – 3.5 × 10−4) and subtype IC lineages during a 1962– 1964 outbreak (4.0 × 10−4), and about fourteen fold lower than estimated for a 1995 IC outbreak (1.84 × 10−3) (Brault et al. 2001). Thus the rate estimated for MUCV is lower than, but of the same order of magnitude as enzootic ID and epizootic IC from the 1962 – 64 outbreak, and one order of magnitude less than 1995 IC. It is possible that these subtypes have fundamentally different evolutionary patterns particularly since they have different reservoir hosts and epidemiological patterns, however it is difficult to draw conclusions about the significance of differences in the absence of data on more VEEV type I subtypes and given the relatively large 95% HPD for our estimate.
The estimated rate of evolution for these viruses translates into a relatively recent origin for the isolates sampled, with the most recent common ancestor estimated to have existed around 1809 (95% HPD 1559 – 1944). The most recent common ancestors for Clades 1, 2 and 3 were estimated to exist around 1934, 1944 and 1917 respectively, with Clades 1 and 2 diverging from each other at the beginning of the 20th century. The MRCA for the 2007 isolates is estimated to have existed about 16 (95% HPD 2 – 34) years prior to 2007. This suggests a relatively recent MUCV population bottleneck or introduction from another region, presumably both within Trinidad given the existence of a Trinidad 1965 isolate at the base of this clade.
Our results confirm that VEE complex viruses and both Group C and Guama group orthobunyaviruses, each of which contain members that cause disease in humans, exist in close proximity to human populations in Trinidad. MUCV is known to cause human disease following laboratory infections as well as natural infection, but is not associated with major epidemics. This probably reflects limited endemic circulation near sylvatic, enzootic foci and a lack of equine amplification potential. However, given the evolutionary relationship between epizootic and enzootic subtype I strains within the VEEV serocomplex, further phylogenetic studies are warranted. In particular, larger data sets comprising VEE complex viruses from other geographic origins that fill temporal gaps in the phylogeny will facilitate better understanding of the evolution, phylodynamics, and migratory history of VEEV.
Supplementary Material
Acknowledgements
This work was supported by grants from the Trinidad and Tobago Government Research Fund, the Caribbean Health Research Council and by NIH grant AI071192. AJA was supported by a scholarship from the University of the West Indies, St. Augustine Campus Research and Publications Fund. SMV was supported by the Training Program in Emerging And Re-Emerging Infectious Diseases, NIH T32 grant AI007536. and NCA was supported by the Biodefense Training Program, NIH T32 grant AI060549. The authors also thank Dunstan Arrindell and Orchid Allicock for assistance with mosquito collections.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar PV, Greene IP, Coffey LL, Medina G, Moncayo AC, Anishchenko M, Ludwig GV, Turell MJ, O’Guinn ML, Lee J, Tesh RB, Watts DM, Russell KL, Hice C, Yanoviak S, Morrison AC, Klein TA, Dohm DJ, Guzman H, Travassos da Rosa AP, Guevara C, Kochel T, Olson J, Cabezas C, Weaver SC. Endemic Venezuelan equine encephalitis in northern Peru. Emerg Infect Dis. 2004;10(5):880–888. doi: 10.3201/eid1005.030634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N, Jones RL. A Plinthaquult of the Aripo Savannas, North Trinidad: I. Properties of the Soil and Chemical Composition of the Natural Vegetation. Soil Sci Soc Am J. 1969;33:762–765. [Google Scholar]
- Anishchenko M, Bowen RA, Paessler S, Austgen L, Greene IP, Weaver SC. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad Sci U S A. 2006;103(13):4994–4999. doi: 10.1073/pnas.0509961103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Powers AM, Medina G, Wang E, Kang W, Salas RA, De Siger J, Weaver SC. Potential sources of the 1995 Venezuelan equine encephalitis subtype IC epidemic. J Virol. 2001;75(13):5823–5832. doi: 10.1128/JVI.75.13.5823-5832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Powers AM, Holmes EC, Woelk CH, Weaver SC. Positively charged amino acid substitutions in the E2 envelope glycoprotein are associated with the emergence of Venezuelan equine encephalitis virus. J. Virol. 2002;76:1718–1730. doi: 10.1128/JVI.76.4.1718-1730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79(23):14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs WG, Aitken TH, Spence L. Eastern equine encephalitis virus isolated from Culex nigripalpus in Trinidad. Science. 1959;130:1471. doi: 10.1126/science.130.3387.1471. [DOI] [PubMed] [Google Scholar]
- Downs WG, Aitken TH, Worth CB, Spence L, Jonkers AH. Arbovirus studies in Bush Bush Forest, Trinidad, W. I., September 1959- December 1964. I. Description of the study area. Am J Trop Med Hyg. 1968;17(2):224–236. doi: 10.4269/ajtmh.1968.17.224. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho S, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22(5):1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Foster JE, Bennett SN, Carrington CV, Vaughan H, McMillan WO. Phylogeography and molecular evolution of dengue 2 in the Caribbean basin, 1981–2000. Virology. 2004;324(1):48–59. doi: 10.1016/j.virol.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Foster JE, Bennett SN, Vaughan H, Vorndam V, McMillan WO, Carrington CV. Molecular evolution and phylogeny of dengue type 4 virus in the Caribbean. Virology. 2003;306(1):126–134. doi: 10.1016/s0042-6822(02)00033-8. [DOI] [PubMed] [Google Scholar]
- Fulton F, Dumbell KR. The serological comparison of strains of influenza virus. J Gen Microbiol. 1949;3(1):97–111. doi: 10.1099/00221287-3-1-97. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 2002;54(2):156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Martin DH. Venezuelan equine encephalitis. Adv. Vet. Sci. Comp. Med. 1974;18:79–116. [PubMed] [Google Scholar]
- Jonkers AH, Spence L, Downs WG, Aitken TH, Tikasingh ES. Arbovirus studies in Bush Bush forest, Trinidad, W. I., September 1959-December 1964. V. Virus isolations. Am J Trop Med Hyg. 1968a;17(2):276–284. doi: 10.4269/ajtmh.1968.17.276. [DOI] [PubMed] [Google Scholar]
- Jonkers AH, Spence L, Downs WG, Aitken TH, Worth CB. Arobovirus studies in Bush Bush Forest, Trinidad, W. I., September 1959- December 1964. VI. Rodent-associated viruses (VEE and agents of groups C and Guama): isolations and further studies. Am J Trop Med Hyg. 1968b;17(2):285–298. [PubMed] [Google Scholar]
- Kinney RM, Pfeffer M, Tsuchiya KR, Chang GJ, Roehrig JT. Nucleotide sequences of the 26S mRNAs of the viruses defining the Venezuelan equine encephalitis antigenic complex. Am J Trop Med Hyg. 1998;59(6):952–964. doi: 10.4269/ajtmh.1998.59.952. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S.L, Frost SDW. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Martin D, Williamson C, Posada D. RDP2: recombination detection and analysis from sequences alignments. Bioinformatics. 2005;21(2):260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Oberste MS, Fraire M, Navarro R, Zepeda C, Zarate ML, Ludwig GV, Kondig JF, Weaver SC, Smith JF, Rico-Hesse R. Association of Venezuelan equine encephalitis virus subtype IE with two equine epizootics in Mexico. Am. J. Trop. Med. Hyg. 1998;59:100–107. doi: 10.4269/ajtmh.1998.59.100. [DOI] [PubMed] [Google Scholar]
- Pfeffer M, Proebster B, Kinney RM, Kaaden OR. Genus-specific detection of alphaviruses by a semi-nested reverse transcription-polymerase chain reaction. Am J Trop Med Hyg. 1997;57(6):709–718. doi: 10.4269/ajtmh.1997.57.709. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 2001;50(4):580–601. [PubMed] [Google Scholar]
- Price JL. Serological evidence of infection of Tacaribe virus and arboviruses in Trinidadian bats. Am J Trop Med Hyg. 1978a;27:162–167. doi: 10.4269/ajtmh.1978.27.162. [DOI] [PubMed] [Google Scholar]
- Price JL. Isolation of Rio Bravo and a hitherto undescribed agent, Tamana bat virus, from insectivorous bats in Trinidad, with serological evidence of infection in bats and man. Am J Trop Med Hyg. 1978b;27:153–161. doi: 10.4269/ajtmh.1978.27.153. [DOI] [PubMed] [Google Scholar]
- Schwab S. Faunal checklist of the Aripo Savannas (Scientific Reserve) Living World (Journal of the Trinidad and Tobago Field Naturalists’ Club) 1988:6–13. [Google Scholar]
- Spence L, Jonkers AH, Casals J. Dengue type 3 virus isolated from an antiguan patient during the 1963–64 Caribbean epidemic. Am J Trop Med Hyg. 1969;18(4):584–587. doi: 10.4269/ajtmh.1969.18.584. [DOI] [PubMed] [Google Scholar]
- Spence L, Jonkers AH, Grant LS. Arboviruses in the Caribbean Islands. Prog Med Virol. 1968;10:415–486. [PubMed] [Google Scholar]
- Tikasingh ES, Ardoin P, Williams MC. First isolation of Catu virus from a human in Trinidad. Trop Geogr Med. 1974;26(4):414–416. [PubMed] [Google Scholar]
- Tikasingh ES, Jonkers AH, Spence L, Davies JB, Aitken TH. Arbovirus studies in an evergreen seasonal marsh forest in Trinidad, West Indies. West Indian Med J. 1983;32(4):223–231. [PubMed] [Google Scholar]
- Walton TE, Alvarez O, Buckwalter RM, Johnson KM. Experimental infection of horses with enzootic and epizootic strains of Venezuelan equine encephalomyelitis virus. J. Infect. Dis. 1973;128:271–282. doi: 10.1093/infdis/128.3.271. [DOI] [PubMed] [Google Scholar]
- Walton TE, Grayson MA. Venezuelan equine encephalomyelitis. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. Vol. IV. Boca Raton, Florida: CRC Press; 1988. [Google Scholar]
- Wang E, Barrera R, Boshell J, Ferro C, Freier JE, Navarro JC, Salas R, Vasquez C, Weaver SC. Genetic and phenotypic changes accompanying the emergence of epizootic subtype IC. Venezuelan equine encephalitis viruses from an enzootic subtype ID progenitor. J. Virol. 1999;73:4266–4271. doi: 10.1128/jvi.73.5.4266-4271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2(10):789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Powers AM, Brault AC, Barrett AD. Molecular epidemiological studies of veterinary arboviral encephalitides. Vet J. 1999;157(2):123–138. doi: 10.1053/tvjl.1998.0289. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Tesh RB, Shope RE. Alphaviruses (VEE) In: Guerrant RL, Krogstad DJ, Maguire JH, Walker DH, Weller PF, editors. Tropical Infectious Diseases: Principles, Pathogens and Practice. New York: Churchill Livingstone; 1998. [Google Scholar]
- Young NA, Johnson KM. Antigenic variants of Venezuelan equine encephalitis virus: their geographic distribution and epidemiologic significance. Am. J. Epidemiol. 1969;89:286–307. doi: 10.1093/oxfordjournals.aje.a120942. [DOI] [PubMed] [Google Scholar]
- Zárate ML, Scherer WF, Dickerman RW. Venezuelan equine encephalitis virus as a human infection determinant. Description of a fatal case occurring in Jaltipan, Ver., in 1965. Rev Invest Salud Publica. 1970;30(4):296–302. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


