Abstract
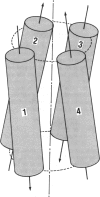
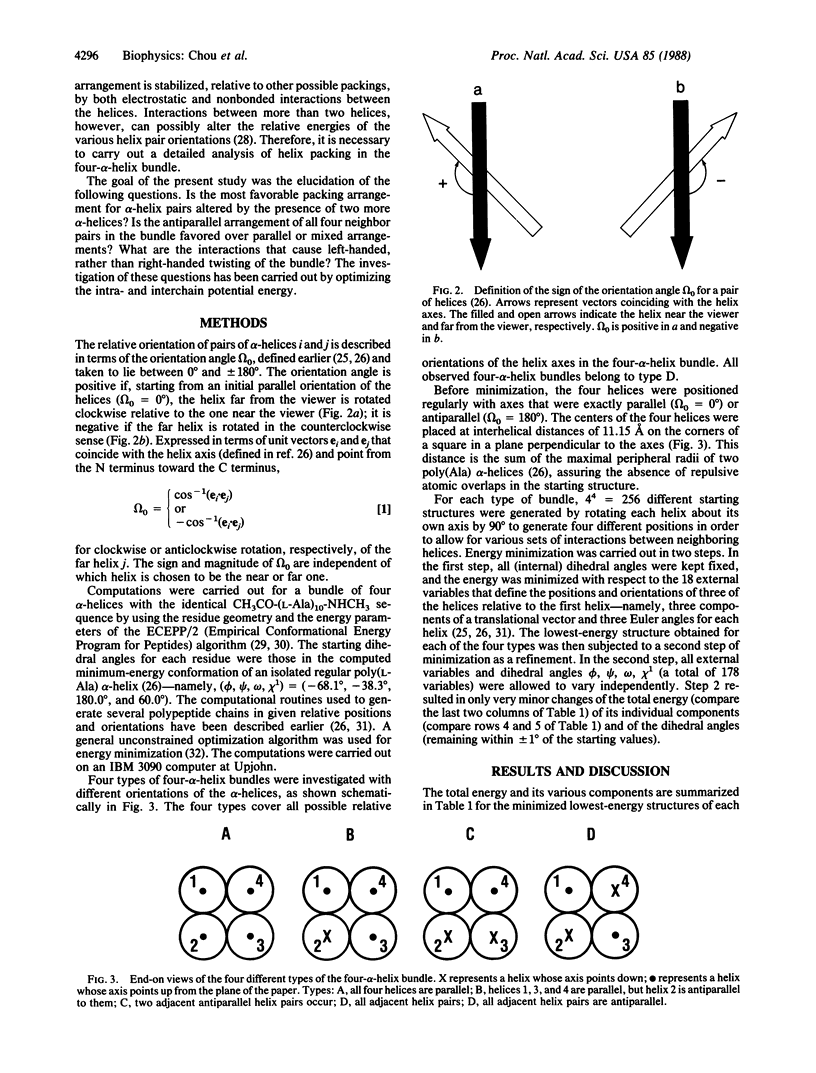
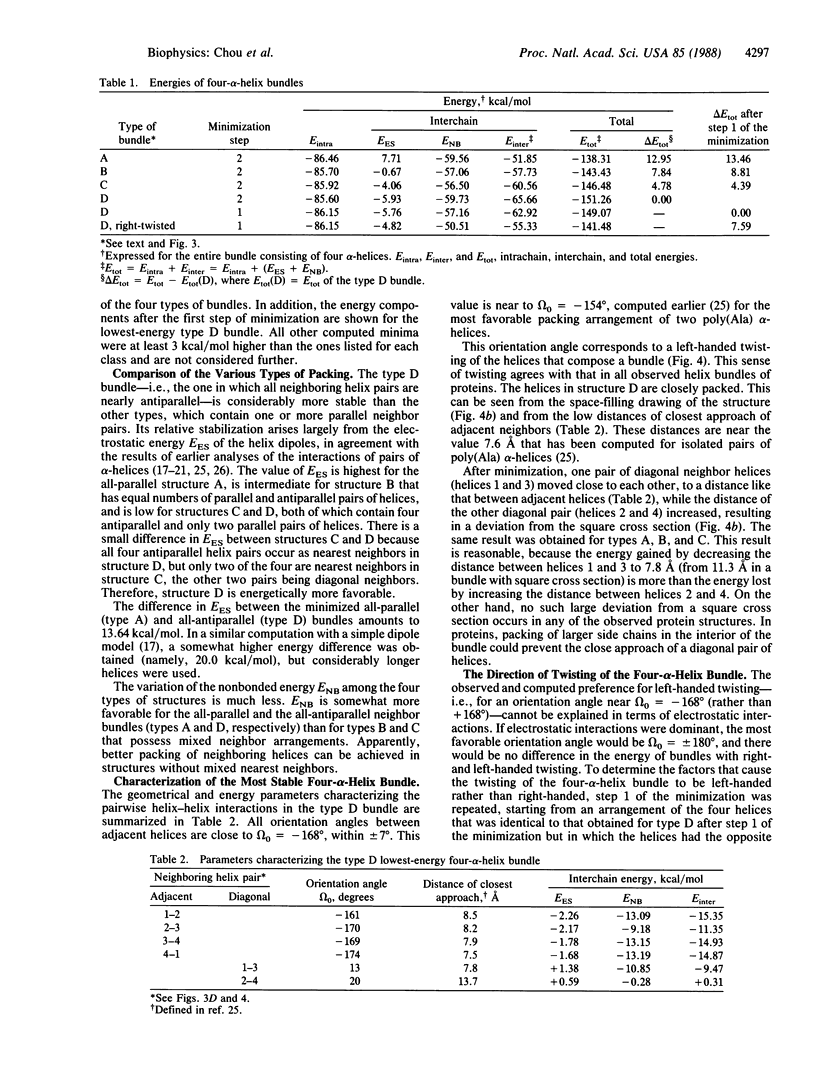
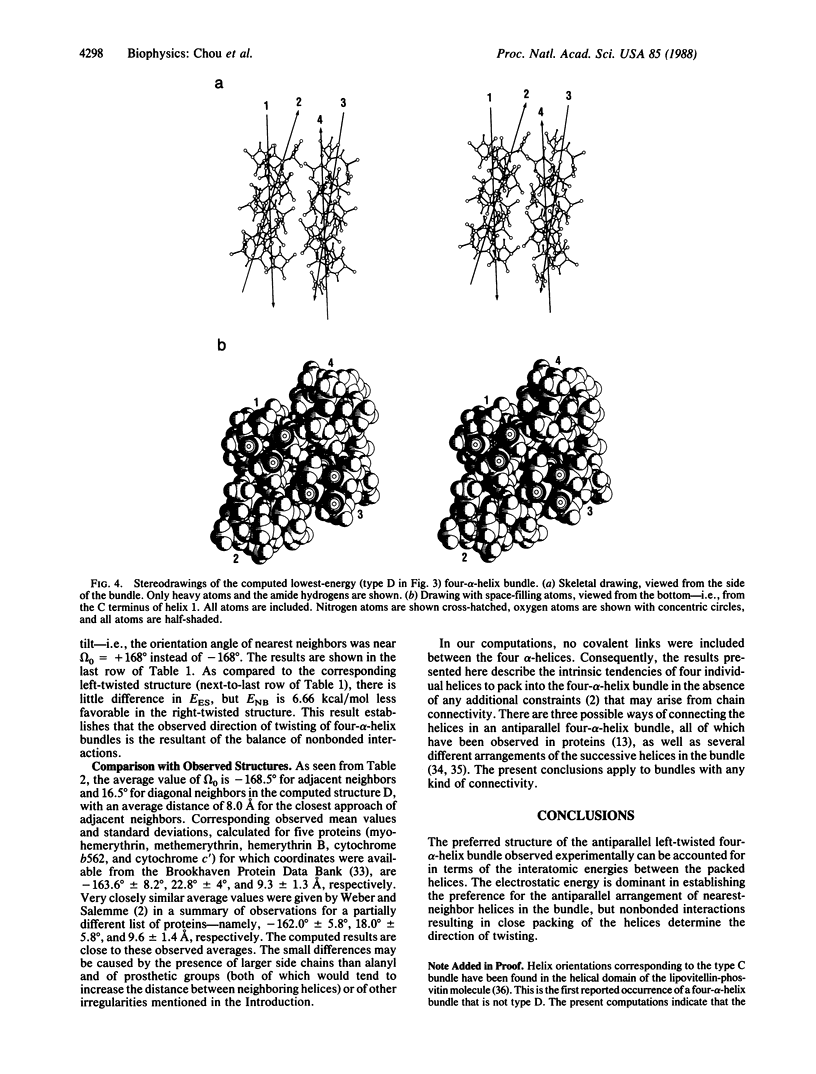
The main features of the four-alpha-helix bundle, one of the characteristic structural elements of many proteins, can be explained in terms of noncovalent interactions between the constituent helices. Conformational energy computations have been carried out on four types of four-alpha-helix bundles, each consisting of four CH3CO-(L-Ala)10-NHCH3 polypeptide chains, with various combinations of parallel and antiparallel orientations of the helices. In the bundle with the most favorable energy, all pairs of neighboring helices are oriented antiparallel--i.e., in the orientation that is favored by electrostatic interactions between the helices. In this structure, the orientation angle between neighboring helix axes is -168 degrees, within +/- 7 degrees, in close agreement with the orientation angles observed in proteins and with the value that we computed earlier for the most favorable packing of pairs of interacting alpha-helices. This orientation corresponds to a left-handed twisting of the helical bundle. The preferred handedness of this twisting arises as a result of favorable nonbonded interactions between the alpha-helices.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Meguid S. S., Shieh H. S., Smith W. W., Dayringer H. E., Violand B. N., Bentle L. A. Three-dimensional structure of a genetically engineered variant of porcine growth hormone. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6434–6437. doi: 10.1073/pnas.84.18.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6162–6166. doi: 10.1073/pnas.84.17.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Rossmann M. G., Johnson J. E. A four-helical super-secondary structure. Biochem Biophys Res Commun. 1977 Mar 7;75(1):83–86. doi: 10.1016/0006-291x(77)91292-x. [DOI] [PubMed] [Google Scholar]
- Banner D. W., Kokkinidis M., Tsernoglou D. Structure of the ColE1 rop protein at 1.7 A resolution. J Mol Biol. 1987 Aug 5;196(3):657–675. doi: 10.1016/0022-2836(87)90039-8. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blow D. M., Irwin M. J., Nyborg J. The peptide chain of tyrosyl tRNA synthetase: no evidence for a super-secondary structure of four alpha-helices. Biochem Biophys Res Commun. 1977 Jun 6;76(3):728–734. doi: 10.1016/0006-291x(77)91560-1. [DOI] [PubMed] [Google Scholar]
- Brandhuber B. J., Boone T., Kenney W. C., McKay D. B. Three-dimensional structure of interleukin-2. Science. 1987 Dec 18;238(4834):1707–1709. doi: 10.1126/science.3500515. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Structure of proteins: packing of alpha-helices and pleated sheets. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C., Némethy G., Rumsey S., Tuttle R. W., Scheraga H. A. Interactions between an alpha-helix and a beta-sheet. Energetics of alpha/beta packing in proteins. J Mol Biol. 1985 Dec 5;186(3):591–609. doi: 10.1016/0022-2836(85)90133-0. [DOI] [PubMed] [Google Scholar]
- Clegg G. A., Stansfield R. F., Bourne P. E., Harrison P. M. Helix packing and subunit conformation in horse spleen apoferritin. Nature. 1980 Nov 20;288(5788):298–300. doi: 10.1038/288298a0. [DOI] [PubMed] [Google Scholar]
- Cohen F. E., Kosen P. A., Kuntz I. D., Epstein L. B., Ciardelli T. L., Smith K. A. Structure-activity studies of interleukin-2. Science. 1986 Oct 17;234(4774):349–352. doi: 10.1126/science.3489989. [DOI] [PubMed] [Google Scholar]
- Gerritsen M., Chou K. C., Némethy G., Scheraga H. A. Energetics of multihelix interactions in protein folding: application to myoglobin. Biopolymers. 1985 Jul;24(7):1271–1291. doi: 10.1002/bip.360240714. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A., Klippenstein G. L., Ward K. B. Tertiary structure of myohemerythrin at low resolution. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2160–2164. doi: 10.1073/pnas.72.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A., Ward K. B. Atomic models for the polypeptide backbones of myohemerythrin and hemerythrin. Biochem Biophys Res Commun. 1975 Oct 27;66(4):1349–1356. doi: 10.1016/0006-291x(75)90508-2. [DOI] [PubMed] [Google Scholar]
- Hol W. G., Halie L. M., Sander C. Dipoles of the alpha-helix and beta-sheet: their role in protein folding. Nature. 1981 Dec 10;294(5841):532–536. doi: 10.1038/294532a0. [DOI] [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Mathews F. S., Bethge P. H., Czerwinski E. W. The structure of cytochrome b562 from Escherichia coli at 2.5 A resolution. J Biol Chem. 1979 Mar 10;254(5):1699–1706. [PubMed] [Google Scholar]
- Mornon J. P., Fridlansky F., Bally R., Milgrom E. X-ray crystallographic analysis of a progesterone-binding protein. The C2221 crystal form of oxidized uteroglobin at 2.2 A resolution. J Mol Biol. 1980 Mar 15;137(4):415–429. doi: 10.1016/0022-2836(80)90166-7. [DOI] [PubMed] [Google Scholar]
- Piela L., Némethy G., Scheraga H. A. Proline-induced constraints in alpha-helices. Biopolymers. 1987 Sep;26(9):1587–1600. doi: 10.1002/bip.360260910. [DOI] [PubMed] [Google Scholar]
- Raag R., Appelt K., Xuong N. H., Banaszak L. Structure of the lamprey yolk lipid-protein complex lipovitellin-phosvitin at 2.8 A resolution. J Mol Biol. 1988 Apr 5;200(3):553–569. doi: 10.1016/0022-2836(88)90542-6. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Richards F. M. Packing of alpha-helices: geometrical constraints and contact areas. J Mol Biol. 1978 Mar 15;119(4):537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- Sheridan R. P., Allen L. C. The electrostatic potential of the alpha helix (electrostatic potential/alpha-helix/secondary structure/helix dipole). Biophys Chem. 1980 Apr;11(2):133–136. doi: 10.1016/0301-4622(80)80015-9. [DOI] [PubMed] [Google Scholar]
- Sheridan R. P., Levy R. M., Salemme F. R. alpha-Helix dipole model and electrostatic stabilization of 4-alpha-helical proteins. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4545–4549. doi: 10.1073/pnas.79.15.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A. The alpha-helix as an electric macro-dipole. Adv Biophys. 1976:1–63. [PubMed] [Google Scholar]
- Weber P. C., Bartsch R. G., Cusanovich M. A., Hamlin R. C., Howard A., Jordan S. R., Kamen M. D., Meyer T. E., Weatherford D. W., Nguyen huu Xuong Structure of cytochrome c': a dimeric, high-spin haem protein. Nature. 1980 Jul 17;286(5770):302–304. doi: 10.1038/286302a0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Salemme F. R. Structural and functional diversity in 4-alpha-helical proteins. Nature. 1980 Sep 4;287(5777):82–84. doi: 10.1038/287082a0. [DOI] [PubMed] [Google Scholar]