Abstract
Proteasome activity is regulated by sequestration of its proteolytic centers in a barrel-shaped structure that limits substrate access. Substrates enter the proteasome by means of activator complexes that bind to the end rings of proteasome α subunits and induce opening of an axial entrance/exit pore. The PA26 activator binds in a pocket on the proteasome surface using main chain contacts of its C-terminal residues and uses an internal activation loop to trigger gate opening by repositioning the proteasome Pro-17 reverse turn. Subunits of the unrelated PAN/19S activators bind with their C termini in the same pockets but can induce proteasome gate opening entirely from interactions of their C-terminal peptides, which are reported to cause gate opening by inducing a rocking motion of proteasome α subunits rather than by directly contacting the Pro-17 turn. Here we report crystal structures and binding studies of proteasome complexes with PA26 constructs that display modified C-terminal residues, including those corresponding to PAN. These findings suggest that PA26 and PAN/19S C-terminal residues bind superimposably and that both classes of activator induce gate opening by using direct contacts to residues of the proteasome Pro-17 reverse turn. In the case of the PAN and 19S activators, a penultimate tyrosine/phenylalanine residue contacts the proteasome Gly-19 carbonyl oxygen to stabilize the open conformation.
Introduction
The proteasome (1–3), also known as 20S proteasome/CP/core particle, is a proteolytic complex that is abundant in the cytosol and nucleus of eukaryotes, where it performs most of the housekeeping and regulatory proteolysis. Substrate proteins are typically targeted for proteasomal degradation by the posttranslational attachment of polyubiquitin chains. Proteasomes degrade numerous and diverse substrate proteins, which is a necessary function for many cellular processes, including DNA repair, transcription, cell cycle regulation, signal transduction, and antigen presentation. Proteasomes are also found in archaea and some eubacteria.
Proteasomes avoid indiscriminate degradation by virtue of their multimeric architecture, which sequesters the active sites inside a barrel-like assembly of four coaxially stacked heptameric rings, with two outer α rings surrounding two inner β rings (4, 5). Archaeal proteasomes, such as that of Thermoplasma acidophilum, comprise one type of α subunit and one type of β subunit and therefore possess an exact seven-fold axis of symmetry (4). In contrast, eukaryotic proteasomes contain seven distinct α subunits (α1–α7) and seven distinct β subunits (β1–β7) that occupy unique positions in their respective rings (6). The various α and β subunits are structurally related and typically share 15–30% sequence identity, with α subunits distinguished by an N-terminal extension of ∼35 residues that functions in activator binding and entrance/exit pore gating. Despite the differences in subunit composition, proteasome structure is highly conserved between archaea, yeast, and mammals (4, 6–8).
Substrate access to the proteasome interior is restricted to proteins that are largely unfolded by a constricted entrance pore through the α subunit ring (4). Access is further restricted by α subunit N-terminal residues, which in eukaryotic proteasomes adopt an ordered asymmetric structure that seals the pore (6). These residues are flexible in the symmetric archaeal proteasomes but nevertheless provide an effective barrier to passage of protein substrates (9, 10). Thus, a critical step in proteasome activation is stabilization of the entrance gate in an open conformation that allows substrates passage into the catalytic chamber. Gate opening is achieved when activator complexes bind to the α subunit rings, and three distinct activator complexes have been identified to date: 19S/PA700/RP/PAN2 (11–15), 11S/PA28/REG/PA26 (16–18), and PA200/Blm10 (19, 20). The eukaryotic 19S activator comprises at least 19 subunits, including a heterohexameric ring of ATPases (Rpt1–Rpt6) that binds the proteasome α subunits and induces gate opening (21, 22). 19S recognizes and unfolds polyubiquitin-conjugated substrates and translocates them into the proteasome for degradation. Archaea such as Methanocaldococcus jannaschii encode a simpler, homohexameric ATPase 19S homolog called PAN (proteasome-activating nucleosidase) (15, 23, 24) that assembles as a trimer of dimers (25, 26). Although the physiological role of 19S in mediating proteolysis is well defined, the biological role of 11S and PA200/Blm1 activators is less clear (27) and, unlike 19S, neither of them recognizes ubiquitin nor utilizes ATP. Nevertheless, they both bind proteasomes on the same surface as 19S and induce opening of the entrance/exit pore (28–30).
A detailed model of proteasome gate opening is provided by crystal structures of PA26 (proteasome activator 26 kDa), a divergent 11S activator of Trypanosoma brucei (18), in complex with the Saccharomyces cerevisiae and T. acidophilum proteasomes (10, 28, 31). Importantly these crystal structures demonstrated that upon proteasome binding, the PA26 activation loop (32) repositions each Pro-17 reverse turn to trigger gate opening. Biochemical and electron microscopic studies suggest that C-terminal residues of PAN/19 activators bind to proteasome in the same manner as PA26 (31, 33, 34). Unlike PA26, however, PAN/19S do not require an internal activation loop to trigger gate opening. Instead these activators can induce gate opening using only interactions from their C-terminal residues, which include a critical penultimate tyrosine (33, 34). In an effort to better understand how C-terminal residues of PAN/19S can provide both binding and gate-opening functions in the absence of an internal activation loop, we have determined proteasome complex crystal structures with a series of PA26 constructs that display modified C-terminal residues and performed associated binding studies. These data indicate a role for the penultimate PAN/19S side chain in both binding and stabilizing the proteasome Pro-17 residues in an open conformation.
EXPERIMENTAL PROCEDURES
Structure Determinations
See the supplemental material for a more complete description of the experimental procedures. Briefly, PA26 variants were constructed using standard methods, and the PA26 proteins and T. acidophilum and S. cerevisiae proteasomes were prepared by modifications of the published procedures (5, 31, 35). PA26 complex crystals were obtained essentially as described (31). Structures were isomorphous to the parent PA26-proteasome complex (31) and were refined to Rfree values of 22.5%-24.1% against 2.9–3.2 Åresolution data (supplemental Table S1).
Biosensor Binding Assays
Human IgG (Sigma-Aldrich) was immobilized on all six flow cells of a GLM sensor chip in the ProteOn XPR36 system (Bio-Rad) using standard amine coupling chemistry. Two g of pulverized yeast extract was dissolved in 10 ml of 20 mm Tris, pH 7.4, 0.1 mm EDTA, 0.5 mm dithiothreitol for 10 min and then centrifuged for 30 min at 14,000 × g. The clarified lysate was captured on the IgG surface at five different densities (100–1000 response units). Resulting surfaces were washed with a 10-min injection of running buffer (10 mm Tris, pH 7.5, 10 mm KCl, 190 mm NaCl, 1.1 mm MgCl2, and 0.1 mm EDTA) containing first 500 mm NaCl and then 1 m NaCl. PA26 proteins were tested in a 3-fold dilution series using the highest concentration for each sample as follows: PA26 (1 μm), C7PAN (333 nm), C7PAN Y230A (1 μm), V230F (111 nm), and V230Y (111 nm). Data were collected at 25 °C. Sensor data were processed using ScrubberPro6 (BioLogic Software Pty. Ltd., Campbell, Australia) and globally fit to a 1:1 interaction model.
RESULTS
Design of Chimeric PA26 Constructs
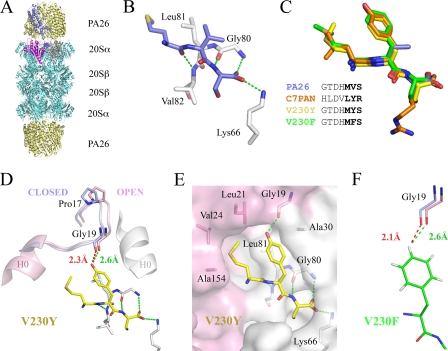
To obtain atomic resolution models of interactions between PAN/19S C-terminal residues and the proteasome, we constructed PA26 variants that displayed altered C termini. This effort was guided by the previously reported crystal structure complexes of PA26 with S. cerevisiae and T. acidophilum proteasomes (10, 28, 31). These structures demonstrated that the 8 C-terminal residues project away from the body of PA26, with the first 5 of these residues being loosely structured and the 3 C-terminal residues forming main chain hydrogen bonds with the proteasome α subunit residues 80–82 and with the PA26 C-terminal carboxylate binding the invariant proteasome α subunit Lys-66 side chain (T. acidophilum proteasome residue numbering is used throughout) (Fig. 1, A and B). Because biochemical and electron microscopic data suggest that PAN/19S C-terminal residues bind in the same manner as PA26 (31, 33, 34), we predicted that substitution of the C-terminal residues of PA26 for those of PAN would reveal how the penultimate tyrosine side chain interacts with α subunits and contributes to gate opening (33).
FIGURE 1.
Structures of PA26-proteasome complexes. A, overall structure with one PA26 subunit colored blue and its three C-terminal residues in space-filling representation. Proteasome subunits that form the corresponding binding pocket are pink and gray (white in subsequent panels). B, close-up showing main chain hydrogen-bonding interactions of the PA26 C-terminal residues (31). C, sequences and structures of PA26 (31) and variants described in this work shown after overlap on the proteasome structures. D, the penultimate Tyr-230 residue contacts Gly-19 to reposition the Pro-17 turn. Distances between Tyr-230-OH and Gly-19-O in closed (red; modeled) and open (green; observed) conformations are shown. Pro-17 undergoes an apparent movement of ∼1 Å. H0, helix 0. E, binding pocket with proteasome shown as a semitransparent molecular surface. Conserved residues whose side chains contact the activator C-terminal residues are indicated. F, V230F penultimate phenylalanine interactions observed with Gly-19 in the open conformation (pink) and modeled in the closed conformation (blue). To best describe contacts, riding hydrogen atoms were included in determined structures and assessed with MolProbity (36).
We crystallized three chimeric PA26 constructs in complex with the T. acidophilum proteasome. One of the constructs displayed the 7 C-terminal residues of M. jannaschii PAN (C7PAN), and two displayed changes only in the penultimate residue (V230Y, V230F) (Fig. 1C). We also attempted to generate PA26 variants with altered activation loop residues that would bind to the proteasome but would fail to induce the ordered open gate conformation. Unfortunately, these constructs either failed to bind the proteasome or bound to an ordered open conformation (data not shown). We utilized one of these activation loop mutations, E102A, in one of the crystallized complexes, V230Y. This mutation does not have structural consequences but in this case gave higher quality crystals.
Crystal Structures
The chimeric PA26-proteasome complex structures were isomorphous to the published PA26-proteasome structure (31), with one PA26 activator and one half-proteasome in the asymmetric unit, and a complete complex generated by applying two-fold crystallographic symmetry. The three new structures and the parent PA26-proteasome structure are all similar to each other, and in all cases, the proteasome α subunit N-terminal residues adopt the open conformation. The 3 C-terminal residues of the PA26 constructs exhibit well defined electron density, and their main chain binding interactions are closely superimposable in all four of the PA26-proteasome complex structures (Fig. 1C). Protein Data Bank codes associated with each complex are 3JTL (C7PAN), 3JSE (V230F), and 3JRM (V230Y).
Penultimate Tyrosine
Our V230Y and C7PAN structures provide a model supporting a critical structural role for the penultimate tyrosine residue of PAN/19S activators. In addition to forming main chain interactions with one of the α subunits that forms the binding pocket, the side chain extends toward Pro-17 of the preceding α subunit to form a hydrogen bond with the Gly-19-O (Fig. 1, D and E). This interaction stabilizes Gly-19, and hence the rest of the Pro-17 reverse turn, in the open conformation. Our previous structural studies have demonstrated that this displacement of the Pro-17 reverse turn triggers gate opening (10, 28, 31). Moreover, simple modeling indicates that the tyrosine side chain would be incompatible with the closed conformation due to steric clash with Gly-19-O (Fig. 1D).
Because the displacement of Pro-17/Gly-19 needed to trigger gate opening is modest (∼1.0 Å), it is necessary that the penultimate tyrosine side chain be constrained to its activating conformation and not allowed to relax away from this position. The structures reveal that this is achieved by the tight fit of the tyrosine and the preceding large hydrophobic residue into the binding pocket. As shown in Fig. 1E, the penultimate tyrosine aromatic ring lies against Leu-81 and is sandwiched between the preceding activator residue (Met in V230Y, Leu in C7PAN) and proteasome helix 0 (Fig. 1, H0), where it contacts Ala-30. Moreover, the preceding hydrophobic residue is buttressed by interactions with the hydrophobic Leu-21, Val-24, and Ala-154 proteasome residues. Notably, these proteasome residues are conserved, Leu-81 is known to be important for gate opening by PAN, and activating peptides require a large hydrophobic side chain at the position preceding the penultimate tyrosine (33).
Penultimate Phenylalanine
Although attention has focused on the penultimate tyrosine residue of PAN, it is striking that this residue is not invariant but is highly conserved as tyrosine or phenylalanine in an alignment of 55 closely related archaeal sequences (23 Tyr, 26 Phe, 6 others; data not shown). Because PAN homologs in archaea are thought to function as homomeric assemblies, this suggests that a penultimate phenylalanine might be functionally equivalent to tyrosine. We therefore visualized the conformation and interactions of a penultimate phenylalanine by determining the structure of the V230F-proteasome complex. As shown in Fig. 1, C and F, the phenylalanine adopts the same conformation as observed for the tyrosine and also contacts the Gly-19-O, with the difference being that the interaction is a van der Waals contact rather than a hydrogen bond. Like the tyrosine, the phenylalanine side chain conformation is tightly restrained by interactions with binding pocket residues, and modeling indicates that a penultimate phenylalanine would clash with the closed conformation and thereby promote the open conformation by repositioning Gly-19-O and hence the rest of the Pro-17 turn.
Contribution of C-terminal Residues to Binding Eukaryotic Proteasome
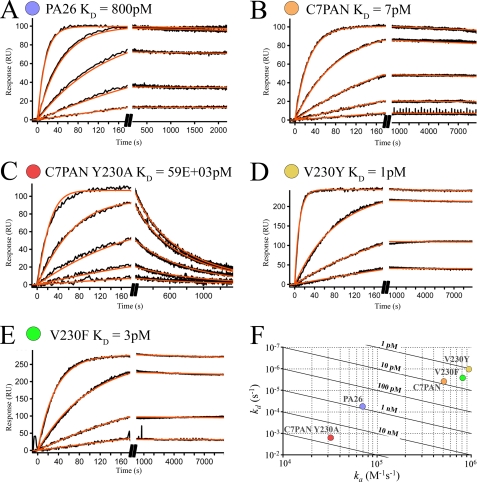
We developed a surface plasmon resonance assay to quantify the importance of C-terminal residues for binding the S. cerevisiae proteasome (Fig. 2). Proteasome displaying a protein-A domain at the C terminus of the β4 subunit (35) was captured from yeast lysate on an IgG-coated biosensor chip. Binding sensorgrams were recorded under a variety of conditions and found to be consistent and to agree well with a 1:1 interaction model. Data were recorded for binding of PA26, C7PAN, and variants of these constructs with altered penultimate residues. The PA26 heptamer bound with a dissociation constant of 800 ± 2 pm and C7PAN bound with a dissociation constant of 7.78 ± 0.02 pm (Fig. 2, A and B). The penultimate tyrosine plays a dominant role in stabilizing this interaction because the C7PAN Y230A variant displayed a dissociation constant of 59 ± 1 nm (Fig. 2C). Similarly, introduction of the penultimate tyrosine/phenylalanine substitutions into the PA26 context increased binding affinity to 1.056 ± 0.004 and 3.303 ± 0.009 pm, respectively (Fig. 2, D and E).
FIGURE 2.
PA26-proteasome binding studies. A–E, biosensorgrams showing concentration-dependent binding of PA26 variants to the S. cerevisiae proteasome and calculated average dissociation constants (KD). Binding to an IgG control surface was negligible (data not shown). RU, response units. F, kinetic plot of ka versus kd where diagonals represent KD, and values for each PA26 variant are shown as spots. The KD standard deviation is less than the size of the spots.
DISCUSSION
Using the well characterized PA26-proteasome complex, we have developed a model system to visualize interactions between C-terminal residues of different proteasome activators and proteasome subunits. Advantages of this approach include the ease of making PA26 constructs and the ability to visualize interactions at atomic resolution by crystallization of complexes with the T. acidophilum proteasome. In principle, other proteasomes could be used in these structural studies, including the S. cerevisiae proteasome, which has been crystallized in complex with PA26 (28), although we have not pursued that possibility at this time because the S. cerevisiae complex crystals are highly irreproducible and diffract only to modest resolution. One limitation of our model system is that only the C-terminal residues of the target binding partner can be explored, although this concern is balanced by structural and biochemical data indicating that these residues of PAN/19S are especially important both for binding and for defining the proteasome gate conformation (33, 34). Our approach allows visualization of interactions corresponding to the binding and gate-opening residues of the ATP-dependent PAN/19S activator.
Models of proteasome binding by the ATP-dependent activators PAN/19S have been somewhat contradictory. On the one hand, biochemical (31, 33) and electron microscopy (34) data indicate that both PA26 and PAN C termini bind in a similar manner. On the other hand, PA26 induces gate opening using its internal activation loop, which directly repositions the Pro-17 reverse turn (10, 28, 31, 32), whereas C-terminal residues of PAN and 19S subunits can activate the proteasome without involvement of an internal loop, and C-terminal PAN peptides are reported to induce gate opening without contacting the Pro-17 reverse turn (34). At least some of these findings are resolved by our proteasome complex structure of C7PAN, which shows that the sequence corresponding to the last 7 residues of PAN can bind superimposably with PA26 to place the critical penultimate tyrosine side chain in a position that stabilizes the Pro-17 turn in the open conformation. This reconciles the earlier implications of equivalent binding among activator C termini with the ability of sequences containing a penultimate tyrosine to autonomously induce gate opening. Our structures also explain the importance of the preceding residue, which is favored to be large and hydrophobic (34), because this residue and the penultimate tyrosine fit snuggly in a pocket that is lined by conserved hydrophobic residues, thereby restricting the tyrosine side chain to an orientation that repositions Gly-19.
Still unresolved is the relevance of proteasome α subunit rocking. On the basis of 6.8 Åresolution electron microscopy reconstructions of peptide complexes (34), it has been suggested that PAN/19S induce gate opening by causing the α subunits to rotate (twist) by ∼5° about a line that is close to and parallel with the seven-fold axis of symmetry of the proteasome. Motions of this sort are not seen in the proteasome crystal structures, although some of the S. cerevisiae α subunits and to a lesser extent the T. acidophilum α subunits undergo an outward displacement upon gate opening by PA26 that can be approximated as rocking of up to 5° about an axis perpendicular to the seven-fold axis of symmetry of the proteasome (31). Regardless of the extent to which twisting of α subunits parallel to the proteasome axis is coupled to gate opening by PAN/19S, we favor the model that direct contacts with the Pro-17 turn induce gate opening by essentially the same mechanism as PA26 (10).
Although it has been established that a penultimate tyrosine is important for activation by PAN/19S (33), it is notable that this position is often substituted by a phenylalanine in PAN homologs from other archaea and is sometimes similarly substituted in ATPase subunits of the eukaryotic 19S activator. Surprisingly, PAN displaying C termini mutated to include a penultimate phenylalanine residue did not stimulate model substrate degradation using the T. acidophilum proteasome (33), although it seems probable that phenylalanine-containing PAN sequences will be functional against proteasomes whose cognate PAN displays a phenylalanine (24). We therefore determined the structure of V230F and found that the penultimate phenylalanine and adjacent C-terminal residues are superimposable with the C7PAN and V230Y structures. Interestingly, like a penultimate tyrosine, the phenylalanine contacts Gly-19-O and hence appears to stabilize the same gate-opening transition of the Pro-17 turn. This substitution of a van der Waals interaction by the phenylalanine for a hydrogen bond by the tyrosine explains how the different side chains might stabilize the same open conformation.
All of the structures reported here are complexes of the archaeal T. acidophilum proteasome, which possesses one type of α subunit, is seven-fold symmetric, and displays a disordered, partly closed conformation in the absence of an activator. In contrast, eukaryotic proteasomes possess seven different α subunits and, in the absence of an activator, adopt an ordered closed conformation in which the entrance pore is sealed primarily by N-terminal residues of α2, α3, α4, and, to a lesser extent, α5. This asymmetric closed conformation explains why some of the S. cerevisiae proteasome Pro-17 reverse turns move as much as 3.5 Åupon binding of PA26, whereas others move less than 1.0 Å. Consistent with their roles in closing the gate, the largest Pro-17 displacements are seen for α2, α3, α4, and α5. Notably, the S. cerevisiae proteasome PA26 complex structure displays density for the PA26 C-terminal residues in the α2/α3, α3/α4, α4/α5, α5/α6 pockets, whereas density in the other pockets is poorly defined or absent (the corresponding Pro-17 is from the first named subunit for each pocket) (31). This pattern of pocket occupancy parallels the conservation of the critical binding residue Lys-66 (from second subunit of each pocket), which among eukaryotes is invariant in α3, α4, α5, and α6, is conserved as Lys/Arg in α2, and α7, and is never Lys in α1 (31). Together, these observations suggest that 19S ATPase subunits likely bind to some or all of these pockets occupied by PA26, and the activating displacement of Pro-17 turns will be promoted by interactions of penultimate tyrosine residues that are equivalent to those seen in our T. acidophilum proteasome complex structures.
As a complement to the structural studies, we developed a quantitative binding assay that uses affinity immobilized S. cerevisiae proteasome. In principle, this assay could be developed for proteasomes from a range of different species, although we have initially focused on yeast because of the potential for using this assay in future studies to identify and characterize novel or candidate proteasome binding factors. We find that the C-terminal residues of PAN promote binding and that a penultimate tyrosine or phenylalanine makes a positive contribution to binding affinity, at least in the context of an activator that already favors the open conformation.
In summary, our structural and binding data support models for proteasome gate opening by 11S and PAN/19S activators that include considerable similarities. In both cases, the activator C-terminal residues are likely to bind through equivalent main chain hydrogen bonds with the activator C-terminal carboxylate forming a salt bridge with Lys-66. The orientation of bound C-terminal residues is constrained by the main chain hydrogen bonding and by packing of the penultimate tyrosine/phenylalanine and the preceding large hydrophobic side chain in a conserved pocket. These interactions dictate that the penultimate tyrosine/phenylalanine side chain contacts Gly-19-O and thereby stabilizes the Pro-17 reverse turn of the proteasome in the open conformation. In the case of PA26 and the other 11S activators, the smaller penultimate residues do not substantially stabilize the open conformation, and the repositioning of Pro-17 is driven by an internal activation loop (10). Thus, we favor the model in which both classes of activator function by the same mechanism of moving Pro-17 to a position that triggers gate opening (28, 31).
Supplementary Material
Acknowledgments
We thank Ira Baci and Heidi L. Schubert for technical assistance. Data collection at the National Synchrotron Light Source (NSLS) was funded by the National Center for Research Resources. Operations of the NSLS are supported by the United States Department of Energy, Office of Basic Energy Sciences, and by the National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM59135 (to C. P. H.).
The atomic coordinates and structure factors (codes 3JTL, 3JSE, and 3JRM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Table S1.
- PAN
- proteasome-activating nucleotidase
- PA26
- proteasome activator 26 kDa.
REFERENCES
- 1.Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 295–317 [DOI] [PubMed] [Google Scholar]
- 2.Zwickl P., Seemüller E., Kapelari B., Baumeister W. (2001) Adv. Protein Chem. 59, 187–222 [DOI] [PubMed] [Google Scholar]
- 3.Goldberg A. L. (2007) Biochem. Soc. Trans. 35, 12–17 [DOI] [PubMed] [Google Scholar]
- 4.Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. (1995) Science 268, 533–539 [DOI] [PubMed] [Google Scholar]
- 5.Seemüller E., Lupas A., Stock D., Löwe J., Huber R., Baumeister W. (1995) Science 268, 579–582 [DOI] [PubMed] [Google Scholar]
- 6.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 7.Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. (2002) Structure 10, 609–618 [DOI] [PubMed] [Google Scholar]
- 8.Groll M., Brandstetter H., Bartunik H., Bourenkow G., Huber R. (2003) J. Mol. Biol. 327, 75–83 [DOI] [PubMed] [Google Scholar]
- 9.Benaroudj N., Zwickl P., Seemüller E., Baumeister W., Goldberg A. L. (2003) Mol. Cell 11, 69–78 [DOI] [PubMed] [Google Scholar]
- 10.Förster A., Whitby F. G., Hill C. P. (2003) EMBO J. 22, 4356–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters J. M., Franke W. W., Kleinschmidt J. A. (1994) J. Biol. Chem. 269, 7709–7718 [PubMed] [Google Scholar]
- 12.Udvardy A. (1993) J. Biol. Chem. 268, 9055–9062 [PubMed] [Google Scholar]
- 13.Hoffman L., Pratt G., Rechsteiner M. (1992) J. Biol. Chem. 267, 22362–22368 [PubMed] [Google Scholar]
- 14.Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. (1994) J. Biol. Chem. 269, 3539–3547 [PubMed] [Google Scholar]
- 15.Zwickl P., Ng D., Woo K. M., Klenk H. P., Goldberg A. L. (1999) J. Biol. Chem. 274, 26008–26014 [DOI] [PubMed] [Google Scholar]
- 16.Ma C. P., Slaughter C. A., DeMartino G. N. (1992) J. Biol. Chem. 267, 10515–10523 [PubMed] [Google Scholar]
- 17.Dubiel W., Pratt G., Ferrell K., Rechsteiner M. (1992) J. Biol. Chem. 267, 22369–22377 [PubMed] [Google Scholar]
- 18.Yao Y., Huang L., Krutchinsky A., Wong M. L., Standing K. G., Burlingame A. L., Wang C. C. (1999) J. Biol. Chem. 274, 33921–33930 [DOI] [PubMed] [Google Scholar]
- 19.Ustrell V., Hoffman L., Pratt G., Rechsteiner M. (2002) EMBO J. 21, 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt M., Haas W., Crosas B., Santamaria P. G., Gygi S. P., Walz T., Finley D. (2005) Nat. Struct. Mol. Biol. 12, 294–303 [DOI] [PubMed] [Google Scholar]
- 21.Glickman M. H., Rubin D. M., Fried V. A., Finley D. (1998) Mol. Cell Biol. 18, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith D. M., Kafri G., Cheng Y., Ng D., Walz T., Goldberg A. L. (2005) Mol. Cell 20, 687–698 [DOI] [PubMed] [Google Scholar]
- 24.Medalia N., Sharon M., Martinez-Arias R., Mihalache O., Robinson C. V., Medalia O., Zwickl P. (2006) J. Struct. Biol. 156, 84–92 [DOI] [PubMed] [Google Scholar]
- 25.Djuranovic S., Hartmann M. D., Habeck M., Ursinus A., Zwickl P., Martin J., Lupas A. N., Zeth K. (2009) Mol. Cell 34, 580–590 [DOI] [PubMed] [Google Scholar]
- 26.Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rechsteiner M., Hill C. P. (2005) Trends Cell Biol. 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 28.Whitby F. G., Masters E. I., Kramer L., Knowlton J. R., Yao Y., Wang C. C., Hill C. P. (2000) Nature 408, 115–120 [DOI] [PubMed] [Google Scholar]
- 29.Ortega J., Heymann J. B., Kajava A. V., Ustrell V., Rechsteiner M., Steven A. C. (2005) J. Mol. Biol. 346, 1221–1227 [DOI] [PubMed] [Google Scholar]
- 30.Iwanczyk J., Sadre-Bazzaz K., Ferrell K., Kondrashkina E., Formosa T., Hill C. P., Ortega J. (2006) J. Mol. Biol. 363, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Förster A., Masters E. I., Whitby F. G., Robinson H., Hill C. P. (2005) Mol. Cell 18, 589–599 [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z., Clawson A., Realini C., Jensen C. C., Knowlton J. R., Hill C. P., Rechsteiner M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2807–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 36.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.