Abstract
Mutations in the voltage-gated K+ channel Kv1.1 have been linked with a mixed phenotype of episodic ataxia and/or myokymia. Recently, we presented autosomal dominant hypomagnesemia as a new phenotypic characteristic associated with a mutation in Kv1.1 (N255D) (Glaudemans, B., van der Wijst, J., Scola, R. H., Lorenzoni, P. J., Heister, A., van der Kemp, A. W., Knoers, N. V., Hoenderop, J. G., and Bindels, R. J. (2009) J. Clin. Invest. 119, 936–942). A conserved asparagine at position 255 in the third transmembrane segment was converted into an aspartic acid, resulting in a non-functional channel. In this study, we explored the functional consequence of this conserved residue by substitution with other hydrophobic, polar, or charged amino acids (N255E, N255Q, N255A, N255V, N255T, and N255H). Upon overexpression in human embryonic kidney (HEK293) cells, cell surface biotinylation revealed plasma membrane expression of all mutant channels. Next, we used the whole-cell patch clamp technique to demonstrate that the N255E and N255Q mutants were non-functional. Substitution of Asn-255 with other amino acids (N255A, N255V, N255T, and N255H) did not prevent ion conduction, and these mutant channels activated at more negative potentials when compared with wild-type channels, −41.5 ± 1.6, −45.5 ± 2.0, −50.5 ± 1.9, and −33.8 ± 1.3 mV to −29.4 ± 1.1 mV, respectively. The time constant of activation was significantly faster for the two most hydrophobic mutations, N255A (6.2 ± 0.2 ms) and N255V (5.2 ± 0.3 ms), and the hydrophilic mutant N255T (9.8 ± 0.4 ms) in comparison with wild type (13.0 ± 0.9 ms). Furthermore, the voltage dependence of inactivation was shifted ∼13 mV to more negative potentials in all mutant channels except for N255H. Taken together, our data showed that an asparagine at position 255 in Kv1.1 is required for normal voltage dependence and kinetics of channel gating.
Keywords: Biophysics, Membrane/Channels, Membrane/Function, Tissue/Organ Systems/Kidney, Transport, Transport/Potassium
Introduction
Voltage-gated K+ channels (Kv)2 are a diverse family of membrane proteins, with the Shaker-related group (Kv1) representing a major subfamily (1–3). Its members play an important role in excitable cells by setting the resting membrane potential, shaping the action potentials, and controlling the neuronal excitability (4). Kv channels comprise four subunits that encircle a central ion conduction pathway (5, 6). Each subunit consists of six transmembrane-spanning α-helices (S1–S6) with both the N-terminal tail and the C-terminal tail on the intracellular side. The S1–S4 segments form the voltage-sensing domain, whereas S5 and S6 along with the intervening re-entrant P-loop form the pore domain (7, 8). Kv channels are known to switch between the closed and open conformation upon cell depolarization (8, 9). Several molecular mechanisms on voltage-sensing motion have been described, i.e. the “canonical” or “helical screw” model, the “transporter” model, the “paddle” model, and the “twisted S4” model (8). It is generally accepted that the array of positive charges on the S4 helix form the principal structural elements responsible for voltage sensing (8).
Kv1.1 was the first mammalian subunit of the Kv family to be cloned and is abundantly expressed in excitable and non-excitable cells (10, 11). Studies with Kv1.1 knock-out mice showed that deletion of Kv1.1 results in a seizure disorder similar to epilepsy (12). Mutations in Kv1.1 in humans are the cause of periodic episodic ataxia type 1 and/or myokymia (13–18). Electrophysiological analyses of these mutant Kv1.1 channels showed either a significant reduction in current amplitude or altered kinetic properties when compared with wild-type Kv1.1 channels (14, 19, 20).
Recently, a novel mutation, in the third transmembrane segment of Kv1.1 was identified in a family with isolated autosomal dominant hypomagnesemia (21). Surprisingly, hypomagnesemia had thus far not been reported in patients with mutations in Kv1.1. Furthermore, this study demonstrated Kv1.1 expression in the apical membranes of the renal distal convoluted tubule segment, where active Mg2+ reabsorption takes place. The mutation resulted in the single amino acid substitution of an asparagine at position 255 for an aspartic acid (N255D) (21). The mutant channel was non-functional with a dominant negative effect on wild-type channel activity. The aim of the present study is to characterize the N255D mutation in Kv1.1. To examine the importance of this position in channel function, we systematically substituted six amino acids with different chemical and physical properties. The mutant Kv1.1 channels were electrophysiology and biochemically analyzed.
EXPERIMENTAL PROCEDURES
DNA Constructs
Full-length wild-type KCNA1 and N255D mutant were constructed in the pCIneo-IRES-GFP expression vector as described previously (21). Other KCNA1 mutants (N255A, N255E, N255Q, N255H, N255T, N255V) were created using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. All constructs were verified by sequence analysis.
Electrophysiology
HEK293 cells were grown in Dulbecco's modified Eagle's medium (BioWhittaker Europe, Vervier, Belgium) containing 10% (v/v) fetal calf serum, 2 mm l-glutamine, and 10 μg/ml ciproxin at 37 °C in a humidity-controlled incubator with 5% (v/v) CO2. Cells were transiently transfected with the respective constructs using Lipofectamine 2000 (Invitrogen, Breda, The Netherlands), as described previously (22), and electrophysiological recordings were performed 48 h after transfection. Transfected cells were identified by their green fluorescence when illuminated at 488 nm. Non-transfected (green fluorescent protein (GFP)-negative) cells from the same batch were used as controls. Patch clamp experiments were performed in the tight seal whole-cell configuration at room temperature (20–25 °C). An EPC-10 patch clamp amplifier computer was used and controlled by PatchMaster Classic 1.20 software (HEKA Elektronik). Currents were digitized at 20 kHz and digitally filtered at 2.9 kHz. Patch pipettes were pulled from thin-walled borosilicate capillaries (1.5-mm outer diameter, 1.17-mm inner diameter); pipette resistance was typically between 2 and 4 megaohms. The liquid junction potential was not corrected. The pipette solution contained (in mm): 140 KCl, 1 MgCl2, 0.1 CaCl2, 2 EGTA, 10 HEPES/KOH (pH 7.3), and 1 Na2-ATP. The bath solution contained (in mm): 138 NaCl, 5.4 KCl, 1.2 MgCl2, 1 CaCl2, 10 EGTA, 10 HEPES/NaOH (pH 7.3), and 10 glucose. Recordings of Kv1.1 were obtained by voltage steps, applied every 10 s, consisting of 150-ms steps from −100 mV to +50 mV (10-mV increments). The holding potential was −80 mV. Equimolar pipette and bath solutions of K+ (140 mm) were used to determine the voltage dependence of activation. For steady-state inactivation, cells were held at −80 mV and then subjected to steps from −90 mV to +30 mV (10-mV increments) for 10 s followed by a depolarizing step to +30 mV. Linear leak and capacitance currents were corrected with a P/5 leak subtraction procedure (23). The analyses of patch clamp data were performed using Igor Pro software (WaveMetrics, Lake Oswego, OR). Current densities were obtained by normalizing the current amplitude to the cell membrane capacitance.
Cell Surface Biotinylation
Cell surface labeling with biotin was performed as described previously (24). HEK293 cells were transiently transfected with 1 μg of wild-type or mutant Kv1.1 constructs using Lipofectamine 2000 (Invitrogen) in 6-well plates (1.5 million cells/plate). At 48 h after transfection, the biotinylation assay was performed using the sulfo-NHS-LC-LC-biotin (Pierce, Etten-Leur, The Netherlands). Cells from each 6-well plate were homogenized in 1 ml of lysis buffer as described previously (24). Next, 5% of the total protein amount was collected as an input sample. Subsequently, biotinylated proteins (plasma membrane fraction) were precipitated using NeutrAvidin-agarose beads (Pierce). Kv1.1 expression was analyzed by immunoblot analysis for the input and the plasma membrane fraction using the monoclonal Kv1.1 antibody (Neuromab, Davis, CA).
Sequence Analysis and Structure Modeling
The structural model of Kv1.1 was built based on the three-dimensional structure of a chimeric Kv1.2–Kv2.1 channel (26) (Protein Data Bank (PDB) file 2R9R). To obtain optimal modeling results, we used a re-refined version of this template from the PDB_REDO data bank (27). The sequences of the template and Kv1.1 share 75% sequence identity. The Centre for Molecular and Biomolecular Informatics (CMBI) WHAT IF server was used for model building, and Yasara (28) was used for loop building, energy minimization, and subsequent mutation analysis. The tetrameric model was obtained by superposing four models on the biological subunit of PDB file 2R9R followed by an energy minimization in Yasara.3
Statistical Analysis
Data are shown as mean ± S.E. values. Statistical significance was determined using analysis of variance followed by Tukey's test. Differences in means with p < 0.05 were regarded as statistically significant. Statistical analysis was performed using Prism (GraphPad, San Diego, CA) software.
RESULTS
Structure Analysis of Kv1.1 Asn- 255
A few years ago, the crystal structure of the mammalian Kv channel, Kv1.2, was solved (PDB file 2A79). This crystallized structure has been used to construct a paddle-chimera channel where the Kv2.1 voltage sensor paddle (S3b and S4 helices) has been transferred to Kv1.2 (5, 26). Based on this latter chimeric Kv1.2-Kv2.1 structure, homology modeling of Kv1.1 was performed (Fig. 1B).3 The sequence identity between Kv1.2-Kv2.1 and Kv1.1 was 75%, which is enough to build a good homology model (29). Kv1.1 subunits consist of six transmembrane α-helices with both the N-terminal and the C-terminal tails of the protein at the intracellular side (Fig. 1C). Recently, a missense mutation in Kv1.1 was found in patients with isolated autosomal dominant hypomagnesemia, converting the highly conserved asparagine at position 255 (Fig. 1A) into an aspartic acid (21). This mutation is positioned in the third transmembrane segment (S3) close to the intracellular compartment (Fig. 1, B and C). Electrophysiological analysis of the Kv1.1 N255D channel expressed in HEK293 cells demonstrated a significantly reduced current amplitude when compared with the wild-type Kv1.1 expressing cells (72.0 ± 3.2 pA/pF versus 398 ± 83 pA/pF) (21). Importantly, both channels were expressed at the plasma membrane in equal amounts (21). To examine the importance of Asn-255 in Kv1.1 channel function, we substituted the asparagine by six different amino acids with distinct chemical and physical properties (N255E, N255Q, N255A, N255T, N255V, and N255H). We used the homology model to study the effect of these mutations on the structure of the channel (Fig. 1D).3The asparagine was converted into a glutamic acid or histidine to investigate the involvement of charge in channel function. Further, glutamine was used as a control to test the possible steric hindrance of the extra CH2 group in glutamic acid. Next, we introduced alanine and valine as non-polar amino acids that are not able to participate in hydrogen bonding. Threonine was used as a control for residue size as alanine is a smaller amino acid.
FIGURE 1.
Structural analysis of mutations in Kv1.1 at position 255. A, multiple alignment analysis shows conservation of the Asn-255 amino acid (black bar) among species and human family members Kv1.2 and Kv2.1. Light gray- and dark gray-colored letters represent conserved and non-conserved amino acids, respectively. B, the predicted three-dimensional structure model of the tetrameric Kv1.1 channel. C, schematic representation of the Kv1.1 channel, which consists of six transmembrane segments (S1–S6) with S1–S4 functioning as a voltage-sensing domain and a pore-forming region between S5 and S6. Localization of the Asn-255 position is denoted by the light gray dot. D, enlarged view of the predicted three-dimensional structure model showing the side chains of the polar residues surrounding Asn-255 and the mutated amino acids at this position. N (WT) indicates Asn-255 (WT).
Surface Expression of the Kv1.1 Mutants
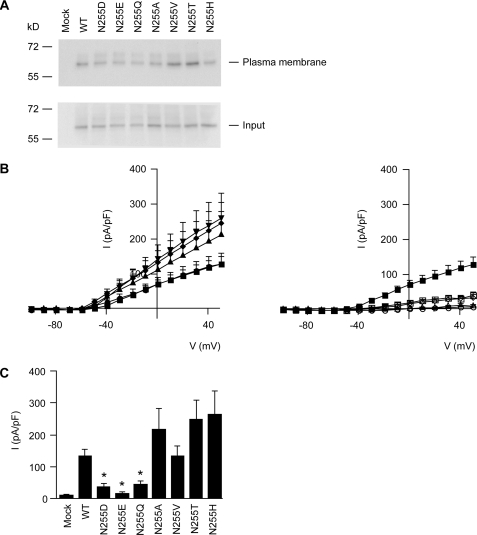
The effect of the substituted amino acids on the amount of Kv1.1 channels at the plasma membrane was examined by cell surface biotinylation experiments. As shown in Fig. 2A, the substitution of Asn-255 by other amino acids did not affect the expression of Kv1.1 channels at the plasma membrane.
FIGURE 2.
Expression of wild-type and mutant Kv1.1 channels. A, cell surface biotinylation of mock, Kv1.1, Kv1.1 N255D, Kv1.1 N255E, Kv1.1, N255Q, Kv1.1 N255A, Kv1.1 N255V, Kv1.1 N255T, and Kv1.1 N255H expressing HEK293 cells. Kv1.1 expression was analyzed by immunoblotting for plasma membrane fraction and input from the total cell lysates. A representative immunoblot of four independent experiments is shown. B, right panel, the I-V relationships of the outward K+ currents in HEK293 cells expressing mock (○), wild-type Kv1.1 (■), Kv1.1 N255D (◇), Kv1.1 N255E (△), Kv1.1 N255Q (□). Left panel, the I-V relationships of the outward K+ currents in HEK293 cells expressing Kv1.1 N255A (▴), Kv1.1 N255V (●), Kv1.1 N255T (♦), and Kv1.1 N255H (▾). Mean ± S.E. (error bars) values are shown. C, histogram presenting averaged current densities at +50 mV of mock (n = 4), wild-type Kv1.1 (WT, n = 11), Kv1.1 N255D (n = 6), Kv1.1 N255E (n = 4), Kv1.1 N255Q (n = 4), Kv1.1 N255A (n = 10), Kv1.1 N255V (n = 7), Kv1.1 N255T (n = 9), and Kv1.1 N255H (n = 9) expressing HEK293 cells. The asterisk indicates significance (p < 0.05) in comparison with wild-type Kv1.1 expressing cells. Mean ± S.E. (error bars) values are shown.
Electrophysiological Characterization of Kv1.1 Mutants
Whole-cell patch clamp recordings from HEK293 cells transiently expressing the wild-type Kv1.1 gave typical delayed rectifying currents in response to a depolarization step from −100 mV to +50 mV (Fig. 2B). We observed in the Kv1.1 mutants a clear difference in current amplitude corresponding to the amino acid substituted. The Kv1.1 N255E and N255Q mutants showed small current amplitudes, similar to mock and the described Kv1.1 N255D mutation (21). Further, Kv1.1 N255A, N255T, and N255H displayed slightly increased current amplitudes when compared with wild-type Kv1.1. The other substituted amino acid (N255V) did not affect the current amplitude of Kv1.1 (Fig. 2, B and C).
To characterize the activation from the functional mutant and wild-type Kv1.1 channels, tail currents were elicited by 150-ms depolarizing pulses from −100 mV to +50 mV in 10-mV increments every 10 s, from a holding potential of −80 mV (Fig. 3A). The voltage dependence of activation was determined by recording tail currents after prepulse voltage steps. The amplitude of the tail currents was normalized to the maximum current and plotted as a function of the conditioning potential. Data points were fitted with a Boltzmann equation to determine the potential of half-maximal activation (V½) and the slope factor or steepness of voltage dependence (k). This revealed that V½ for all mutant channels was shifted to more negative potentials when compared with wild type, with extreme shifts of 15–20 mV for Kv1.1 N255V and N255T (Fig. 3B and Table 1). The slope factor was not significantly changed for the Kv1.1 mutants when compared with wild-type Kv1.1 (Table 1).
FIGURE 3.
Channel activation of wild-type and mutant Kv1.1 channels. A, representative tail currents of wild-type Kv1.1 at −80 mV, recorded in response to a set of 150-ms voltage steps from −100 to +50 mV in 10-mV increments every 10 s. B, the activation curve of the wild-type Kv1.1 (n = 5) and Kv1.1 N255A (A) currents (n = 4), wild-type Kv1.1 (n = 5) and Kv1.1 N255T (T) currents (n = 5), wild-type Kv1.1 (n = 5) and Kv1.1 N255V (V) currents (n = 5), and wild-type Kv1.1 (n = 5) and Kv1.1 N255H (H) currents (n = 4), respectively. Normalized tail currents are plotted as a function of the prepulse potential. The lines reflect the best fits to the averaged current voltage data points, according to the Boltzmann equation: I = Imax/(1 + exp ((V − V½)/k)), where I is the current measured at each test potential, V; Imax is the maximal current; V½ is the voltage of half-maximal activation; and k is the slope factor. Error bars indicate S.E.
TABLE 1.
Electrophysiological characteristics of Kv1.1 wild-type and mutant channels
The voltage-dependent parameters of activation and inactivation (V½ and k) were obtained from the Boltzmann equation as described in the legends for Figs. 3 and 5, respectively. Activation time constants at V½ (τv½) were derived from Fig. 4. Inactivation was measured at 30 mV and is presented as the ratio Ifinal/Ipeak. Data are presented as mean ± S.E., with the number of investigated cells in parentheses.
| WT | N255A | N255T | N255V | N255H | |
|---|---|---|---|---|---|
| Voltage dependence | |||||
| Number of investigated cells | (n = 5) | (n = 4) | (n = 5) | (n = 5) | (n = 4) |
| V½ (mV) | −29.4 ± 1.1 | −41.5 ± 1.6a | −50.5 ± 1.9a | −45.5 ± 2.0a | −33.8 ± 1.3 |
| k (mV) | 12.1 ± 1.0 | 13.7 ± 1.4 | 11.2 ± 1.7 | 13.9 ± 1.8 | 12.3 ± 1.2 |
| Activation | |||||
| Number of investigated cells | (n = 8) | (n = 5) | (n = 4) | (n = 5) | (n = 5) |
| τv½ (ms) | 13.0 ± 0.9 | 6.2 ± 0.2a | 9.8 ± 0.4b | 5.2 ± 0.3a | 14.6 ± 0.7 |
| Inactivation | |||||
| Number of investigated cells | (n = 4) | (n = 5) | (n = 4) | (n = 6) | (n = 3) |
| Ifinal/Ipeak | 0.44 ± 0.07 | 0.49 ± 0.01 | 0.50 ± 0.04 | 0.41 ± 0.04 | 0.40 ± 0.08 |
| Steady-state inactivation | |||||
| Number of investigated cells | (n = 5) | (n = 5) | (n = 5) | (n = 5) | (n = 2) |
| V½ (mV) | −33.5 ± 1.0 | −46.7 ± 1.8a | −46.9 ± 1.8a | −46.8 ± 1.5a | −39.6 ± 1.7 |
a p < 0.01 (when compared with WT).
b p < 0.05 (when compared with WT).
Monoexponential functions were used for fitting K+ current rise to quantify the time dependence of activation (Fig. 4A). Means of the calculated activation time constants were plotted against the test potentials (Fig. 4B), demonstrating that the mutant channels activated ∼2–3 times faster, except for Kv1.1 N225H, which was not different from wild-type Kv1.1 (14.6 ± 0.7 versus 13.0 ± 0.9 ms) (Fig. 4B and Table 1).
FIGURE 4.
Time dependence of activation from wild-type and mutant Kv1.1 channels. A, representative current traces of wild-type Kv1.1 elicited in response to a set of 150-ms voltage steps from −100 to +40 mV in 10-mV increments every 10 s. These activating traces were fitted with a monoexponential function. B, the time constants of activation for wild-type Kv1.1, Kv1.1 N255A, Kv1.1 N255T, Kv1.1 N255V, and Kv1.1 N255H channels were plotted as a function of prepulse potentials and fitted with the equation: τ = τv½exp (V −V½)/k, where τv½ is the time constant at the half-maximal activation voltage (V½) of the channels and k is the slope factor for the voltage dependence of the time constants. Error bars indicate S.E.
The inactivation of the total outward currents was determined using a standard double-pulse protocol (Fig. 5A). With the holding potential of −80 mV, 10-s conditioning potentials were given from −90 to +30 mV in 10-mV increments, every 10 s. Then, the membrane was depolarized to +30 mV for 300 ms (Fig. 5A). The rate of inactivation was quantified by measuring the peak current (Ipeak) and the current at the end of the conditioning pulse (Ifinal). The ratio, Ifinal/Ipeak, showed that inactivation kinetics were not altered in the mutant channels when compared with wild-type channels (Fig. 5B). Voltage dependence of inactivation was investigated by plotting the relative amplitudes of the elicited outward currents at +30 mV as a function of the potentials. The derived steady-state inactivation curve was fitted to the Boltzmann function, demonstrating the voltage dependence of inactivation (Fig. 5C). The N255A, N255T, N255V, and N255H mutant channels showed 50% inactivation at −46.7 ± 1.8, −46.9 ± 1.8, −46.8 ± 1.5, and −39.6 ± 1.7 mV, respectively, when compared with −33.5 ± 1.0 mV for wild-type Kv1.1 (Fig. 5C and Table 1).
FIGURE 5.
The steady-state inactivation of wild-type and mutant Kv1.1 channels. A, the representative wild-type Kv1.1 currents recorded with the double-pulse protocol. Outward currents were evoked on membrane depolarizations to +30 mV after (10-s) conditioning prepulses to potentials between −90 and +30 mV from a holding potential of −80 mV. B, histogram of current amplitudes at the end of the conditioning voltage step (Ifinal) at +30 mV relative to the peak current amplitude at the beginning of the step (Ipeak). T, N255T; V, N255V; A, N255A; H, N255H. Error bars indicate S.E. C, the steady-state inactivation curve of the wild-type Kv1.1 (n = 5) and Kv1.1 N255A currents (n = 5), wild-type Kv1.1 (n = 5) and Kv1.1 N255T currents (n = 5), wild-type Kv1.1 (n = 5) and Kv1.1 N255V currents (n = 5), and wild-type Kv1.1 (n = 5) and Kv1.1 N255H currents (n = 2), respectively. The peak amplitudes of currents at +30 mV evoked from each conditioning potential were measured in individual cells and normalized to the amplitude of the current evoked after the conditioning pulse at −80 mV. Normalized currents are plotted as a function of the conditioning potential. The lines represent the best Boltzman fits to the data points I = Imax/(1 + exp ((V −V½)/k)), where I is the current measured at each test potential, V; Imax is the maximal current; V½ is the voltage of half-maximal inactivation; and k is the slope factor. Error bars indicate S.E.
DISCUSSION
Kv channels are gated in response to changes in transmembrane voltage (30). Kv1.1 is abundantly expressed in excitable and non-excitable cells (3, 10, 11). Recently, a mutation in Kv1.1 (N255D) was found in a large Brazilian family with isolated autosomal dominant hypomagnesemia (21). In the present study, we investigated the nature of the Kv1.1 N255D mutation and demonstrated that the asparagine at position 255 is essential for normal voltage dependence and kinetics of channel gating. First, homology modeling of Kv1.1 was performed, based on 75% sequence identity with the crystallized Kv1.2-Kv2.1 chimera. Subsequently, the Asn-255 was substituted into different amino acids (N255E, N255Q, N255A, N255V, N255T, and N255H), which did not affect the expression of the channels at the plasma membrane. Second, the N255E and N255Q mutant channels displayed current amplitude close to the control situation (mock) and N255D and were considered as non-functional. Third, the other mutants (N255A, N255V, and N255T) showed a negative shift in V½ when compared with wild-type Kv1.1 and had a faster time constant of activation except for N255H. Fourth, the half-maximal inactivation voltage was shifted to more negative potentials for all mutants, with N255H as the exception.
Affected family members from the Brazilian family with inherited hypomagnesemia showed low plasma Mg2+ levels (0.40 mmol/liter; normal range, 0.70–0.95 mmol/liter) and suffered from muscle cramps, tetanic episodes, tremor, and muscle weakness. Remarkably, mutations in Kv1.1 thus far were known to result in a mixed phenotype of episodic ataxia type 1 and myokymia (13–16, 18, 21), a neurological phenotype in which hypomagnesemia has not been reported. Kv1.1 was shown to localize to the apical membrane of distal convoluted tubule cells and postulated to be involved in the generation of a favorable apical membrane voltage as a driving force for Mg2+ entry (21). It is puzzling that distinct mutations in nearby amino acid residues in Kv1.1 can result in phenotypes with dysfunction in two different organs (brain and kidney). A possible reason is that the composition of Kv1.1 channels in brain and kidney is different, due to tissue-specific expression of auxiliary β-subunits or co-assembly with other Kv1 subunits that define the functional characteristics of these channels (31–33). As a result, mutations at close locations within the protein may have tissue-specific effects, giving rise to diverse phenotypic characteristics.
Electrophysiological analyses of the episodic ataxia type 1/myokymia-related mutant Kv1.1 channels showed either a significant reduction in current amplitude or altered kinetic properties when compared with wild-type Kv1.1 channels (14, 19, 20). The mutation identified in the Brazilian family caused substitution of the asparagine at amino acid position 255 into an aspartic acid (N255D). The asparagine at position 255 is highly conserved among species and Kv1 family members, which suggests its importance in channel function. Indeed, we demonstrated that the change of the neutral asparagine into a negatively charged aspartic acid results in a non-functional channel (21). In the present study, we investigated the amino acid substitution at position 255 in relation to channel function more extensively.
The tertiary structure of Kv1.1 was modeled by the WHAT-IF server, based on 75% sequence identity with the crystallized Kv1.2-Kv2.1 chimeric channel (5, 26). The asparagine residue (Asn-255) is located in the third transmembrane segment (S3) close to the S4 voltage sensor element. S4 contains a long array of positive charges that are shown to sense differences in voltage and start the transition from the closed to open conformation by forming stabilizing hydrogen bonds with the external and internal negative clusters in the voltage-sensing domain (34, 35). In general, mutations can affect channel activity via loss-of-function at the plasma membrane, protein instability, or lack of plasma membrane targeting. Importantly, cell surface biotinylation studies showed that all mutants were expressed at the plasma membrane. Thus, the change in amino acid at position 255 had no effect on channel trafficking. However, the amino acid substitution has a clear effect on channel activity as we demonstrated that the N255E and N255Q channels were non-functional. This indicates that next to the addition of a negative charge at position 255, an additional CH2 group in the side chain also influences channel function, likely via affecting conformational rearrangements.
Subsequently, the voltage dependence and kinetics of channel gating of the wild-type functional mutant channels were examined. All mutations stabilized the open state of Kv1.1, measured as negative shifts in the voltage dependence of channel activation. There was no obvious correlation between charge and the magnitude of the shift in V½, and k as the effect was not significantly changed for N255H.
Depending on the polarity of the side chain, amino acids vary in their hydrophilic or hydrophobic character (36). These properties are important determinants of the protein structure, and the physical properties of the side chains influence the interactions of the amino acid residues with other structures, both within a single protein and between proteins. Therefore, the hydrophilic asparagine could be important for structural rearrangements within the channel in response to voltage changes, which can be affected by conversion into hydrophobic amino acids as alanine and valine. Interestingly, the change in activation kinetics was most evident with these two residues as N255A and N255V activated 2–3-fold faster than wild-type channels. Furthermore, these mutants significantly shifted the voltage dependence of activation. However, the magnitude of shift in V½ was highest for N255T, which suggests that channel gating is independent of hydrogen bonding with the residue at position 255.
Inactivation, besides activation, is another important property with respect to channel function. Interestingly, the voltage dependence of inactivation was significantly affected in all mutant Kv1.1 channels when compared with wild-type Kv1.1 except for N255H. Substitution of the asparagine with other amino acids shifted the half-point for inactivation to more negative potentials. The acceleration of the inactivation process is also of interest as it is explained by a constriction mechanism of the outer mouth of the channel vestibule (37, 38). It has been demonstrated that negatively charged clusters in the S2 and S3 segments, together with the positive charges in S4, are involved in the opening and closing of Kv1 channels (26). In line with this, an earlier study showed that mutation of conserved negatively charged residues in the S2 and S3 segments selectively modulate channel gating. Mutation of the aspartic acid at position 258 in Kv1.1 abolished channel activity (25). Therefore, we suggest that an additional negative charge nearby this cluster in S3 could keep the channel in the inactivated state, which may explain the non-functionality of the mutation found in patients with hypomagnesemia (N255D). However, there were no significant changes in inactivation kinetics between wild-type and mutant channels.
Taken together, we have previously described hypomagnesemia as a new phenotypic variability associated with a mutation in Kv1.1 (N255D) (21) In this study, we provided more information about the structural arrangement of Asn-255 and its involvement in channel activity. We have demonstrated that Asn-255 is essential for normal channel function because substitution by other amino acids significantly altered channel activity, voltage dependence, and kinetics of Kv1.1 channels.
Acknowledgments
We thank Dr. KyuPil Lee and Femke van Zeeland for excellent technical assistance and Dr. Gert Vriend for valuable discussion.
This work was supported by grants from the Netherlands Organization for Scientific Research (Grants ZonMw 9120.6110, NWO-CW 700.55.302, and ZonMw 9120.8026), a European Young Investigator award from the European Science Foundation, and the Dutch Kidney foundation (Grants C03.6017 and C08.2252).
H. Venselaar, unpublished data.
- Kv
- voltage-gated potassium channels
- Kv1.1
- voltage-gated potassium channel subtype 1.1
- WT
- wild type
- pF
- picofarads.
REFERENCES
- 1.Christie M. J. (1995) Clin. Exp. Pharmacol. Physiol. 22, 944–951 [DOI] [PubMed] [Google Scholar]
- 2.Dolly J. O., Parcej D. N. (1996) J. Bioenerg. Biomembr. 28, 231–253 [DOI] [PubMed] [Google Scholar]
- 3.Armstrong C. M. (2003) Sci. STKE 2003, re10. [DOI] [PubMed] [Google Scholar]
- 4.Hille B. (2001) Ion Channels of Excitable Membranes, 3rd Ed., pp. 131–158, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 5.Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 6.Grottesi A., Sands Z. A., Sansom M. S. (2005) Curr. Biol. 15, R771–774 [DOI] [PubMed] [Google Scholar]
- 7.Aggarwal S. K., MacKinnon R. (1996) Neuron 16, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 8.Sands Z., Grottesi A., Sansom M. S. (2005) Curr. Biol. 15, R44–47 [DOI] [PubMed] [Google Scholar]
- 9.Swartz K. J. (2004) Nat. Rev. Neurosci. 5, 905–916 [DOI] [PubMed] [Google Scholar]
- 10.Armstrong C. M., Hille B. (1998) Neuron 20, 371–380 [DOI] [PubMed] [Google Scholar]
- 11.O'Grady S. M., Lee S. Y. (2005) Int. J. Biochem. Cell Biol. 37, 1578–1594 [DOI] [PubMed] [Google Scholar]
- 12.Smart S. L., Lopantsev V., Zhang C. L., Robbins C. A., Wang H., Chiu S. Y., Schwartzkroin P. A., Messing A., Tempel B. L. (1998) Neuron 20, 809–819 [DOI] [PubMed] [Google Scholar]
- 13.Browne D. L., Gancher S. T., Nutt J. G., Brunt E. R., Smith E. A., Kramer P., Litt M. (1994) Nat. Genet. 8, 136–140 [DOI] [PubMed] [Google Scholar]
- 14.Eunson L. H., Rea R., Zuberi S. M., Youroukos S., Panayiotopoulos C. P., Liguori R., Avoni P., McWilliam R. C., Stephenson J. B., Hanna M. G., Kullmann D. M., Spauschus A. (2000) Ann. Neurol. 48, 647–656 [PubMed] [Google Scholar]
- 15.Klein A., Boltshauser E., Jen J., Baloh R. W. (2004) Neuropediatrics 35, 147–149 [DOI] [PubMed] [Google Scholar]
- 16.Lee H., Wang H., Jen J. C., Sabatti C., Baloh R. W., Nelson S. F. (2004) Hum. Mutat. 24, 536. [DOI] [PubMed] [Google Scholar]
- 17.Shook S. J., Mamsa H., Jen J. C., Baloh R. W., Zhou L. (2008) Muscle Nerve 37, 399–402 [DOI] [PubMed] [Google Scholar]
- 18.Zuberi S. M., Eunson L. H., Spauschus A., De Silva R., Tolmie J., Wood N. W., McWilliam R. C., Stephenson J. B., Kullmann D. M., Hanna M. G. (1999) Brain 122, 817–825 [DOI] [PubMed] [Google Scholar]
- 19.Chen H., von Hehn C., Kaczmarek L. K., Ment L. R., Pober B. R., Hisama F. M. (2007) Neurogenetics 8, 131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerr P., Adelman J. P., Maylie J. (1998) J. Neurosci. 18, 2842–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaudemans B., van der Wijst J., Scola R. H., Lorenzoni P. J., Heister A., van der Kemp A. W., Knoers N. V., Hoenderop J. G., Bindels R. J. (2009) J. Clin. Invest. 119, 936–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang Q., Hoefs S., van der Kemp A. W., Topala C. N., Bindels R. J., Hoenderop J. G. (2005) Science 310, 490–493 [DOI] [PubMed] [Google Scholar]
- 23.Armstrong C. M., Bezanilla F. (1974) J. Gen. Physiol. 63, 533–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gkika D., Topala C. N., Chang Q., Picard N., Thébault S., Houillier P., Hoenderop J. G., Bindels R. J. (2006) EMBO J. 25, 4707–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planells-Cases R., Ferrer-Montiel A. V., Patten C. D., Montal M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9422–9426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 27.Joosten R. P., Womack T., Vriend G., Bricogne G. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krieger E., Koraimann G., Vriend G. (2002) Proteins 47, 393–402 [DOI] [PubMed] [Google Scholar]
- 29.Sander C., Schneider R. (1991) Proteins 9, 56–68 [DOI] [PubMed] [Google Scholar]
- 30.Sigworth F. J. (1994) Q Rev. Biophys. 27, 1–40 [DOI] [PubMed] [Google Scholar]
- 31.Gulbis J. M. (2002) Novartis Found. Symp. 245, 127–141; discussion 141–145, 165–168 [PubMed] [Google Scholar]
- 32.Sokolov M. V., Shamotienko O., Dhochartaigh S. N., Sack J. T., Dolly J. O. (2007) Neuropharmacology 53, 272–282 [DOI] [PubMed] [Google Scholar]
- 33.Zhu J., Watanabe I., Gomez B., Thornhill W. B. (2003) J. Biol. Chem. 278, 25558–25567 [DOI] [PubMed] [Google Scholar]
- 34.Liman E. R., Hess P., Weaver F., Koren G. (1991) Nature 353, 752–756 [DOI] [PubMed] [Google Scholar]
- 35.Pathak M. M., Yarov-Yarovoy V., Agarwal G., Roux B., Barth P., Kohout S., Tombola F., Isacoff E. Y. (2007) Neuron 56, 124–140 [DOI] [PubMed] [Google Scholar]
- 36.Creighton T. H. (1993) Proteins: Structures and Molecular Properties, pp. 139–165, W. H. Freeman, San Francisco, CA [Google Scholar]
- 37.Panyi G., Sheng Z., Deutsch C. (1995) Biophys. J. 69, 896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yellen G., Sodickson D., Chen T. Y., Jurman M. E. (1994) Biophys. J. 66, 1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]