Abstract
The largest subunit of RNA polymerase II (RNAPII) C-terminal heptarepeat domain (CTD) is subject to phosphorylation during initiation and elongation of transcription by RNA polymerase II. Here we study the molecular mechanisms leading to phosphorylation of Ser-7 in the human enzyme. Ser-7 becomes phosphorylated before initiation of transcription at promoter regions. We identify cyclin-dependent kinase 7 (CDK7) as one responsible kinase. Phosphorylation of both Ser-5 and Ser-7 is fully dependent on the cofactor complex Mediator. A subform of Mediator associated with an active RNAPII is critical for preinitiation complex formation and CTD phosphorylation. The Mediator-RNAPII complex independently recruits TFIIB and CDK7 to core promoter regions. CDK7 phosphorylates Ser-7 selectively in the context of an intact preinitiation complex. CDK7 is not the only kinase that can modify Ser-7 of the CTD. ChIP experiments with chemical inhibitors provide evidence that other yet to be identified kinases further phosphorylate Ser-7 in coding regions.
Keywords: Gene/Regulation, Gene/Transcription, Phosphorylation/Enzymes, Phosphorylation/Kinases/Serine-Threonine, Phosphorylation/Serine/Threonine, Transcription, Transcription/Coactivators, Transcription/RNA Polymerase II
Introduction
In mammalian cells the C-terminal domain of the Rpb1 subunit of RNA polymerase II (RNAPII)2 consists of 52 heptarepeats of the consensus sequence YSPTSPS (for review, see Refs. 1–3). When RNAPII enters the preinitiation complex (PIC) the CTD becomes phosphorylated at least on three serine residues, Ser-2, Ser-5, and Ser-7 (4), at a yet unknown number of repeat elements. In theory, combinations of modifications could generate a complex pattern sometimes referred to as CTD code hypothesis (5), in analogy to the modifications occurring on lysine, arginine, and serine residues within N-terminal tails of histones (6).
Several kinases have been identified that modify the CTD. These are CDK1, CDK2, CDK7, CDK8, CDK9, Erk1/2, and DNA-dependent protein kinase. CDK1 and CDK2 phosphorylate preferentially Ser-5 of the CTD (7, 8). In the case of CDK1 this has been associated with repression of transcription during mitosis (9). The signaling kinases Erk1/2 phosphorylate the RNAPII CTD in response to oxidative and osmotic stress (10). The promoter-associated kinase DNA-dependent protein kinase phosphorylates the CTD at Ser-7 in vitro, although it remains unclear whether this process is relevant to transcription (11, 12). CDK8 and the paralogue CDC2L6 are associated with the Mediator complex (13, 14). CDK8 has mostly been associated with repression of transcription, yet there are also reports that specific genes are activated by it (15, 16). CDK9 is thought to release RNAPII that is paused by negative inhibitory factors 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor and negative elongation factor some 30–40 base pairs downstream of the transcription start sites. RNAPII then resumes elongation of transcription, during which it becomes further phosphorylated at Ser-2. A marked increase of Ser-2 phosphorylation is often seen near the 3′ end of genes (4, 17). It is unclear whether this phosphorylation of the CTD near the polyadenylation site is necessary for efficient elongation by RNAPII per se (17, 18). Instead it may introduce signals for the binding of factors that alter CTD conformation and facilitate the interactions of RNA processing machineries (19–21).
The kinase CDK7 is involved in both transcription and cell cycle control. CDK7 associates with the cyclin H and MAT1 to form an active kinase module, which then functions as an activator of CDK1 during mitosis (CAK function). The kinase module is also part of the general factor TFIIH (22). Upon binding to the PIC CDK7, phosphorylates the CTD of RNAPII at Ser-5 (23, 24). Although Ser-5 phosphorylation by CDK7 is tightly linked to PIC formation, it is not required for RNAPII to clear the promoter and resume elongation (25). Instead Ser-5 phosphorylation seems critical for cap formation at 5′ ends of the newly synthesized mRNA (26, 27). It has further been suggested that RNAPII modified at Ser-5 attracts histone modifying factors such as the trithorax group protein family (28).
CDK7 kinase activity at the CTD is subject to control by the cofactor complex Mediator (PC2). Mediator binds to activators in a complex with RNAPII (29, 30). Mediator then recruits the limiting general factor TFIIB, ultimately leading to efficient preinitiation complex formation (31, 32). In yeast, Mediator enhances the activity of the CDK7 kinase module by a yet unknown mechanism (33). In other investigations Mediator was shown to help to recruit TFIIH and a free CDK7 module (34–36).
In a recent publication the yeast homologue of CDK7 (termed KIN28) was shown to be responsible for phosphorylation of the CTD at Ser-7 at the 5′ end of many yeast genes (37). In mammals Ser-7 phosphorylation has been linked to protein-coding and -noncoding small nuclear RNA genes (4, 38). The underlying mechanism and the responsible kinases are unknown. In agreement with the finding in yeast, we identify human CDK7 as a CTD Ser-7 kinase. Beyond it we characterize the molecular requirements for and investigate the steps during which it is introduced during transcription. We also present evidence for other kinases that target Ser-7.
EXPERIMENTAL PROCEDURES
Preparation and Immunodepletion of Nuclear Extracts
Jurkat nuclear extracts were prepared as described previously (39). Jurkat nuclear extracts were immunodepleted using anti-RNAPII (1C7), MED15 (1H7), and MED25 (VC1) rat monoclonal antibodies, anti-TBP, TFIIH p62, CDK7, and CDK9 rabbit polyclonal antibodies (Santa Cruz sc-273, sc-292, sc- 529, and sc-8338, respectively), and anti-TFIIB (IIB8) mouse monoclonal antibodies (Santa Cruz sc-23875) or anti-CDK8 goat polyclonal antibodies (Santa Cruz sc-1521). Antibodies were loaded onto protein A (p62, CDK7, CDK9, TFIIB)- or protein G-Sepharose (MED15, MED25, CDK8, RNAPII) beads. 1 μg of antibody was loaded for each microliter of Sepharose bead volume. For the depletion 100 μl of Jurkat nuclear extracts were adjusted to 20 mm Tris-HCl, pH 6.8, 150 mm KCl, 0.1% Nonidet P-40, 1 mm DTT, 0.2 mm EDTA, 10% glycerol, 0.2 mm PMSF and incubated twice for 3 h with 25 μl of antibody-loaded protein A- or G-Sepharose beads (1 μg of antibody/μl of Sepharose bead volume) at 4 °C. The protein concentration of nuclear extracts was 4–6 mg/ml.
Immobilized Template Assay
The major-late (ML) promoter template was amplified by PCR from the vector pG5MLT, which contains a G-free cassette downstream of the ML promoter. The promoter comprises five GAL4 binding sites immediately upstream of the ML promoter as described (40). The primers were 5′-CGA TTC ATT AAT GCA GCT GG (biotinylated) and 5′-AAC TCG ACT GCA GCA TAT GTA TCA TAC ACA TAC G. The pGL2-MRG5 promoter templates were amplified from the vector pGL2-MRG5, which in turn stems from pMRG5 plasmid, which contains a Luciferase expression cassette in place of the G-free cassette of pMRG5. The promoter comprises 5 GAL4 binding sites immediately upstream of a synthetic human immunodeficiency virus/ML core promoter as described previously (41). The primers were: 5′-GCA TTC TAG TTG TGG TTT GTC CAA (biotinylated) and 5′-GCC GGG CCT TTC TTT ATG TT. DNA templates were purified on 1% agarose gels and recovered using a gel extraction kit (Qiagen). Biotinylated DNA templates were coupled to paramagnetic streptavidin beads (Promega) as follows. Beads were washed twice in B&W buffer (5 mm Tris-HCl pH 7.5, 1 mm EDTA, 1 m NaCl, 0.003% Nonidet P-40). Subsequently the beads were resuspended in B&W buffer, and 15 ng of biotinylated DNA (in Tris-EDTA with 1 m NaCl) template was added for each microgram of magnetic beads. After shaking for 45 min at room temperature, beads were washed once in B&W buffer containing 0.5 mg/ml BSA (Sigma, fraction V). For blocking, beads were resuspended at a concentration of 1 μg/μl in blocking buffer which is buffer A (150 mm potassium glutamate, 20 mm Hepes, pH 8.2, 5 mm MgCl2, 10 mm DTT, 0.025% Nonidet P-40, 0.5 mg/ml BSA (Sigma, fraction V), 0.2 mm PMSF) plus 5 mg/ml BSA (Sigma, fraction V) and 5 mg/ml polyvinylpyrrolidone (Sigma) and incubated for 15 min at room temperature. Afterward beads were washed three times with buffer A. A typical PIC assembly reaction was conducted in a total volume of 200 μl containing 70 μg of beads coupled to 1050 ng of ML- or pGL2-MRG5 promoter template, 200 ng of GAL-VP16 (comprising the C-terminal 147 amino acids of the GAL4 DNA binding domain linked to the complete VP16 activation domain, amino acids 411–490), 2 μg of poly(dG:dC) competitor DNA, and 100–200 μg of Jurkat nuclear extract. 0.5 m P11 phosphocellulose fractions, recombinant yeast TBP, yeast Toa, human TFIIB (hTFIIB), hTFIIE, and hTFIIF were prepared as described previously (42, 43). Immobilized template assays with the partially purified transcription system contained 100–200 μg of protein provided by a 0.5 m phosphocellulose P11 fraction. In addition, 100 ng of yeast TBP, 10 ng of Toa, and if indicated, 200 ng of hTFIIB were added to the reaction. A typical 200-μl reaction using the purified transcription system contained 100 ng of yeast TBP, 10 ng of Toa, 200 ng of hTFIIB, 20 ng of hTFIIE, and 100 ng of hTFIIF. In addition to the above components, 20 mm Hepes-KOH, pH 8.2, 5 mm MgCl2, 10 mm DTT, 0.025% Nonidet P-40, 0.5 mg/ml BSA (Roche Applied Science), 10% (w/v) glycerol, 0.1 mg/ml polyethylene glycol 8000, 0.2 mm PMSF were present in each PIC assembly reaction. H-8 (N-[2-(methylamino)ethyl]-5-isoquinolinesulfonamide hydrochloride) was purchased from Sigma. The total potassium ion concentration in the reaction was adjusted to 150 mm with potassium glutamate. If PIC formation reactions were carried out under in vitro transcription conditions, the same procedure as described above was followed except that the potassium glutamate was omitted, and all buffers and reactions were adjusted with 3 m potassium chloride to a final salt concentration of 60 mm potassium chloride. After 45 min of incubation at 30 °C the complexes bound to the immobilized templates were concentrated with a magnet and washed with 200 μl of buffer A. Template-bound proteins were incubated in an in vitro transcription reaction, eluted, separated on 8–18% SDS-PAA gels, and analyzed by Western blotting.
In Vitro Transcription Assay
In vitro transcription reactions were carried out in two steps. First a PIC was formed and washed as described above, then ⅛ of a 200-μl PIC formation reaction was resuspended in 25 μl of transcription buffer (20 mm Hepes-KOH, pH 8.2, 60 mm KCl, 5 mm MgCl2, 10 mm DTT, 0.025% Nonidet P-40, 0.5 mg/ml BSA (Roche Applied Science), 10% (w/v) glycerol, 0.1 mg/ml polyethylene glycol 8000, 4 units of RNasin (Promega), 0.2 mm PMSF). Transcription was initiated by the addition of the NTP mix supplemented with 1 μl of [α-32P]UTP (3000 Ci/mmol). Final NTP concentrations were 100 μm for each, ATP and CTP, 20 μm for 3′-O-methyl-GTP, 5 μm for UTP. Transcription reactions were incubated at 30 °C for 30 min and stopped by the addition of 400 μl of transcription stop buffer (7 m urea, 10 mm Tris-HCl, pH 7.8, 10 mm EDTA, pH 8.0, 300 mm sodium acetate, 0.5% (w/v) SDS, 100 mm lithium chloride, 0.4 mg/ml yeast tRNA). Reactions were extracted with phenol/chloroform, and RNAs were precipitated with isopropanol. RNAs were analyzed after gel electrophoresis by autoradiography.
In Vitro Kinase Assay
CDK7, p62, and Mediator were immunopurified as described in the immunodepletion procedure. Protein-loaded IP beads were washed 3 times with 50 column volumes BC150 (20 mm Tris-HCl, pH 7.3, 0.2 mm EDTA, 20% glycerol, 150 mm KCl) supplemented with 0.1% Nonidet P-40 before the kinase assay experiment. Either 10 μl of CDK7, p62, or Mediator-loaded IP beads were included in a 50-μl reaction. As a positive control, 10 μl of Jurkat nuclear extract (adjusted to BC150 supplemented with 0.1% Nonidet P-40) were used in one reaction. In addition a kinase assay reaction contained 20 mm HEPES, pH 8.2, 5 mm MgCl2, 10 mm DTT, 0.025% Nonidet P-40, 0.5 mg/ml BSA (Roche Applied Science), 0.2 mm PMSF, 10 units RNase inhibitor (Promega), 10% (w/v) glycerol, 0.1 mg/ml polyethylene glycol 8000, 25 ng/μl poly(dG·dC). The salt concentration was adjusted to a final concentration of 60 mm KCl. The substrate used in the kinase assay, GST-CTD, was expressed in Escherichia coli and affinity-purified on glutathione-Sepharose as described previously (39). Approximately 0.1 μg of GST-CTD was included in a single 50-μl reaction. Reactions were incubated for 30 min at 30 °C. ATP was added to a final concentration of 100 μm, and the incubation was then continued for another 30 min. Reactions were stopped by adding Laemmli buffer and analyzed by Western blot.
Chromatin Immunoprecipitation (ChIP)
For ChIP experiments, HeLa cells stably transfected with pML53 (39) were grown to 80% confluence. Luciferase gene expression was induced with 1 μg/ml doxycycline (Sigma) for 1 h at 37 °C. When roscovitine (Calbiochem) was used, 30 μg/ml roscovitine was added 30 min before the addition of doxycycline. ChIPs were conducted essentially as described previously (44). Cross-linking was done by incubating 1 × 108 cells with 1% formaldehyde for 10 min at room temperature, and 100 μl of chromatin extract with a DNA concentration of 1 mg/ml was used for a single chromatin IP (antibodies were IgG (Santa Cruz, sc-2027), RNAPII (Santa Cruz, sc-899), CTD Ser-5P (3E8), CTD Ser-7P (4E12)). DNA from input and ChIP samples was purified and eluted in 50 μl of 10 mm Tris-HCl, pH 8.0, and 1 μl of DNA was used for subsequent analysis by real-time PCR. Quantitative real time PCR was carried out with the SYBR Green PCR Master Mix (Applied Biosystems) kit according to the manufacturer's instructions on a Step One Plus Time PCR system (Applied Biosystems). Primers were as follows: pro forward (5′-AGG CGT GTA CGG TGG GAG GCC) and pro reverse (5′-AGG CTG GAT CGG TCC CGG TGT); luc forward (5′-AAT GGA AGA CGC CAA AAA CAT) and luc reverse (5′-TTC ATA GCT TCT GCC AAC CGA); orf forward (5′-CCT CTG GAT CTA CTG GGT TAC CT) and orf reverse (5′-GGA ACA ACA CTT AAA ATC GCA GT), and control forward (5′-GAG TAA TTG GTG ATG AGG ACG AG) and control reverse (5′-TAG ACG ACG CTC AGT GAA TAC AG).
RESULTS
Gradual Phosphorylation of Ser-7 Residues on RNAPII Bound to DNA Templates
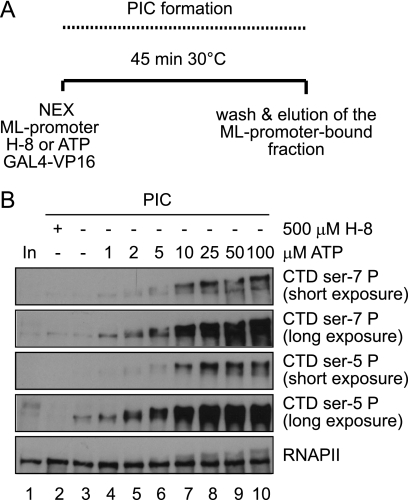
To study phosphorylation of the CTD during preinitiation complex formation, we employed immobilized DNA template assays. Preinitiation complexes were formed on recombinant promoter templates under conditions that facilitate RNAPII transcription in vitro. Here we used an active nuclear extract from human Jurkat T-cells as a source for general factors, RNAPII and CTD kinases. The DNA template comprised five GAL4 binding sites upstream of the adenovirus ML core promoter (39). Templates were then washed briefly, and proteins bound to them were eluted and analyzed in Western blots. In Fig. 1 phosphorylation of the RNAPII CTD at Ser-5 and Ser-7 was analyzed with highly specific monoclonal antibodies (Chapman et al. 4). CTD Ser-7 phosphorylation is first detected at an ATP concentration of about 2 μm, which is slightly higher than the ATP concentration at which we first detect Ser-5 phosphorylation (1 μm). With increasing ATP concentrations, the CTD apparently becomes further phosphorylated, resulting in reduced mobility. We conclude that, like for Ser-5, several residues in the CTD become phosphorylated at Ser-7. Phosphorylation of Ser-7 is dependent on the presence of DNA templates. In the above experiment we also included recombinant GAL-VP16 to promote preinitiation complex formation. The activator is not necessary for CTD phosphorylation, yet GAL4-VP16 usually enhances CDK7 recruitment and CTD phosphorylation (compare Fig. 4C and data not shown). We conclude that the CTD is (hyper)phosphorylated at multiple Ser-7 residues.
FIGURE 1.
Gradual phosphorylation of Ser-7 residues on RNAPII bound to DNA templates. A, shown is a reaction scheme. NEX, nuclear extract. B, preinitiation complexes were formed on ML-promoter templates in the presence of the activator GAL-VP16 and the indicated concentrations of ATP (lanes 3–10). As a control for the presence of residual ATP, the kinase inhibitor H-8 was used in lane 2 in the absence of exogenously added ATP. In, 20% input of the PIC formation reaction. Two exposures are shown to better illuminate the different CTD forms with gradually increasing mobility. Note that minor amounts of phosphorylated RNAPII (Ser-5-P-long exposure) present in nuclear extracts migrate with mobility different from the hypophosphorylated forms generated at low ATP levels.
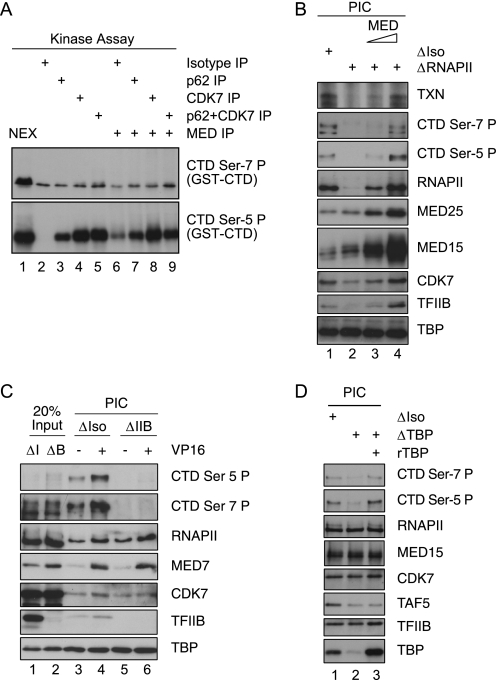
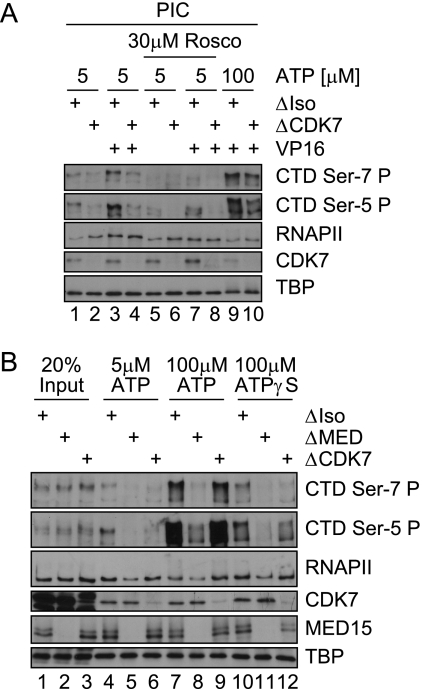
FIGURE 4.
CTD Ser-7 phosphorylation is established in the context of a functional preinitiation complex. A, shown is an in vitro kinase assay using GST-CTD as a substrate. As sources of kinase activity, the indicated immunoprecipitations or Jurkat nuclear extract (NEX) were used. B, shown is an immobilized template assay on ML-DNA templates in the presence of the activator GAL-VP16. Jurkat nuclear extracts were either mock-treated (ΔIso) or depleted for RNAPII (ΔRNAPII). ΔRNAPII extracts in lanes 3 and 4 were supplemented with immunoprecipitated Mediator complexes (high salt-washed MED15 IPs). PICs were washed and either analyzed by immunoblotting or probed in an in vitro transcription assay (TXN). C, shown is an immobilized template assay on pGL2-MRG5 DNA templates in the presence of the activator GAL-VP16. Jurkat nuclear extracts were either mock-treated (ΔIso) or depleted for TFIIB (ΔIIB). D, shown is an immobilized template assay on ML-DNA templates using mock-treated (ΔIso), TBP-depleted (ΔTBP), or TBP-depleted extracts supplemented with recombinant human TBP. PIC formation was carried out under basal conditions in the presence of 1 μm ATP. All assays in this figure were carried out under in vitro transcription conditions.
CTD Ser-7 Phosphorylation Requires Mediator
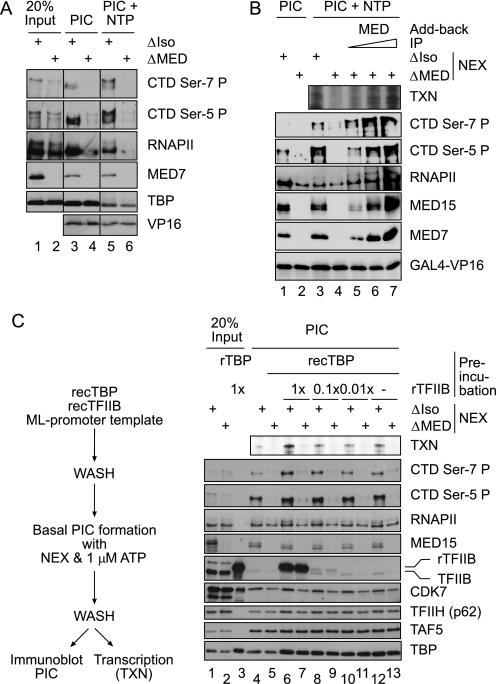
To assess the influence of Mediator, Jurkat nuclear extracts were depleted for the cofactor complex using a highly efficient monoclonal antibody directed against the head subunit MED15 (32). As a control, extracts were mock-depleted (referred to as ΔMED and ΔIso). PIC formation (Fig. 2A) was subsequently conducted in the presence of either a low concentration of ATP (1 μm) or under conditions that facilitate transcription elongation (100 μm NTPs). CTD phosphorylation was fully dependent on the presence of Mediator both during formation of the PIC (Fig. 2A, lanes 3 and 4) and in the presence of NTPs (lanes 5 and 6; note that preinitiation complexes were washed briefly before adding NTPs). In add-back experiments, immunopurified Mediator (stringently washed, see “Experimental Procedures”) fully restored phosphorylation of the CTD on both Ser-5 and Ser-7 in a concentration-dependent manner as well as fully restored transcription (Fig. 2B). In the absence of Mediator, RNA polymerase is recruited but inefficiently phosphorylated (Fig. 2B, lane 4 versus lanes 3 and 5). To further illustrate this difference in activity, we employed a protocol in which recombinant human TBP was prebound to templates together with increasing concentrations of recombinant human TFIIB (see the reaction scheme of Fig. 2C). TFIIB has been shown to be limiting for PIC formation in nuclear extracts (compare transcription analysis on the ML promoter on the top panel). As predicted, the protocol established enhanced transcription, PIC formation, and CTD hypophosphorylation at Ser-7 in the presence of Mediator (Fig. 2C, lanes 12 and 13 versus 6–11). In the absence of Mediator, CTD hypophosphorylation is marginal, and transcription is abrogated. As shown above (Fig. 2A), this experiment further demonstrated that as estimated, less than 50% that of the total RNAPII is associated with Mediator (RNAPII panel, Fig. 2C, lanes 1 versus 2). In the absence of Mediator (all reactions were carried out without GAL4-VP16), substantial amounts of free RNAPII are recruited to templates. Although CDK7 is present, free RNAPII is not modified either on position 5 or on position 7 of the repetition in the CTD. Collectively, we conclude that in the physiological situation of a crude nuclear extract, Mediator-associated RNAPII, but not free RNAPII, is preferentially recruited and phosphorylated at both Ser-5 and Ser-7.
FIGURE 2.
CTD Ser-7 phosphorylation requires Mediator. A, PICs were formed on pGL2-MRG5 promoter templates in the presence of 1 μm ATP and washed and either analyzed directly by immunoblot (lanes 3 and 4) or subjected to a second incubation step in the presence of 100 μm concentrations of each ATP, CTP, GTP, and UTP (lanes 5 and 6). Mock-treated (ΔIso) or Mediator-depleted (ΔMED) Jurkat nuclear extract was used. B, preinitiation complexes were formed in the presence of 1 μm ATP either with mock-treated (ΔIso), Mediator-depleted (ΔMED), or Mediator-depleted extract supplemented with Mediator affinity purified on IP beads as indicated. Mediator IPs were washed stringently at 800 mm KCl before add-back. PICs were either analyzed by immunoblot or probed in an in vitro transcription assay (TXN). NEX, nuclear extract. C, PICs were formed under basal conditions on major-late promoter DNA templates with either mock-treated (ΔIso) or Mediator-depleted (ΔMED) extracts in the presence of 1 μm ATP (lanes 4–13). Before PIC formation, immobilized promoter templates were incubated with recombinant human TBP and recombinant (rec (r)) human TFIIB as indicated. Basal PICs were either analyzed directly by immunoblot or probed in an in vitro transcription assay.
CDK7 Phosphorylates the CTD at Ser-5 and Ser-7
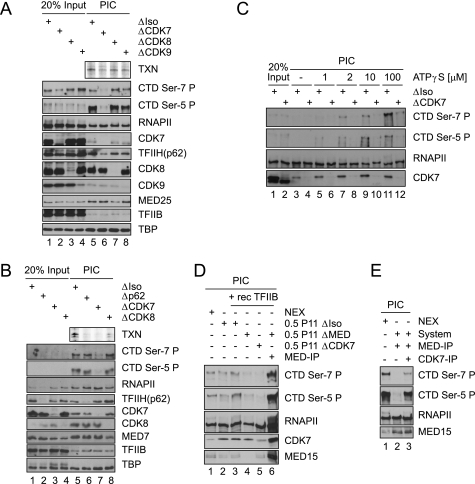
In a semi-systematic approach, the three kinases operating in transcription were analyzed for their impact on Ser-7 phosphorylation. Again, an immunodepletion protocol was applied to crude nuclear extracts. Depletion was efficient for all three kinases as demonstrated in Western blots (Fig. 3A, lanes 1–4). When preinitiation complexes were formed in the presence of 1 μm ATP followed by washing of immobilized templates/complexes, elution of proteins, and analysis in Western blots, Ser-5 and Ser-7 phosphorylation were fully dependent on CDK7. In contrast, depletion of CDK8 had no effect, and CDK9 only moderately reduced CTD phosphorylation (Fig. 3A, lanes 5–8). In this setting a small fraction of CDK7 and CDK9 but significant amounts of CDK8 present in extracts are recruited to templates. Further notable, minor concentrations of phosphorylated RNAPII are found in Jurkat extracts. These are not recruited, as is concluded from their different mobility (Fig. 3A, compare lanes 1–4 and 5–8 in the Ser-5-P panel and Fig. 1A). The experiment above was conducted with templates carrying human immunodeficiency virus TATA instead of the strong ML TATA box (pGL2-MRG5 (45)). However, essentially identical results were obtained when the ML template (used in Fig. 1) was employed (Fig. 3B).
FIGURE 3.
CDK7 is a CTD Ser-7 kinase. A, shown is an immobilized template assay on the pGL2-MRG5 promoter template in the presence of the activator GAL-VP16. Mock-treated or CDK7-, CDK8-, and CDK9-depleted nuclear extracts (lanes 5–8) were used in the PIC formation reactions. Reactions were carried out under standard in vitro transcription conditions in the presence of 1 μm ATP. Lanes 1–4 show 20% of reaction input. B, shown is an immobilized template assay on the ML-promoter template in the presence of GAL-VP16 using mock-treated (ΔIso), core-TFIIH (Δp62), CDK7 (ΔCDK7)- or CDK8-depleted (ΔCDK8) Jurkat nuclear extract. Lanes 1–4 show 20% reaction input. TXN, in vitro transcription analysis. C, shown is an immobilized template assay on ML-promoter DNA templates under basal conditions at physiological salt concentrations. PICs were formed in the presence of the indicated amounts of ATPγS, washed, and then analyzed by immunoblot. D, shown is an immobilized template assay on the ML-promoter DNA template using mock-treated (ΔIso), Mediator-depleted (ΔMED), and CDK7-depleted (ΔCDK7) 0.5 m P11 fractions together with GAL4-VP16, recombinant yeast TBP, Toa, and recombinant human TFIIB as indicated. In lane 6 a high salt-washed MED15-IP was added. PIC formation was carried out under physiological conditions in the presence of 1 μm ATP and 10 μm ATPγS. NEX, nuclear extract. E, immobilized template assay on ML-promoter DNA templates using either Jurkat nuclear extract (lane 1) or a recombinant transcription system consisting of yeast TBP, Toa, human TFIIB, TFIIE, and TFIIF) in combination with a high salt-washed Mediator-IP (which also provides RNAPII; lanes 2 and 3) is shown. In lane 3 a stringently washed CDK7 IP was also added to the PIC formation reactions. PIC formation in D and E was carried out under physiological conditions in the presence of 1 μm ATP and 10 μm ATPγS.
CTD Ser-7 Phosphorylation Is Established in the Absence of Core TFIIH
To assess whether a TFIIH-bound CDK7 kinase was involved, we also depleted core TFIIH with an antibody directed against the p62 subunit. Interestingly, CDK7 depletion but not removal of core TFIIH reduces CTD phosphorylation. The depletion of TFIIH essentially abrogates transcription activity in these extracts (Fig. 3B, top panel, lanes 5 versus 6, the loss of transcription activity in the CDK8-depleted reaction most likely reflects the marked reduction of Mediator). In light of the known activity of TFIIH, these data indicate that Ser-7 phosphorylation starts before DNA is fully opened (see also discussion of Fig. 3D below). The data further show that CTD hypophosphorylation is independent of core TFIIH. Instead, free CAK module is involved in the phosphorylation of the CTD.
CDK7 Is a Ser-7 Kinase
Formation of basal preinitiation complexes (in the absence of GAL4-VP16) was monitored at increasing ATPγS concentrations in the presence or absence of CDK7. We used ATPγS because it proved to select for CDK7 contributions in an extract that also contained other Ser-7 kinases (42). Under these conditions Ser-7 and Ser-5 phosphorylation was established in a CDK7-dependent manner (Fig. 3C). To provide further evidence that CDK7 function on CTD Ser-7 is direct, we employed partially enriched and purified transcription systems. We initially employed a semipurified system consisting of the C-fraction (extracts fractionated on phosphocellulose P11; C-fraction refers to the standard 0.5 m KCl elution step, see Kretzschmar et al. (43)). The C-fraction contains TFIIE, TFIIF, TFIIH, CDK7, Mediator, and RNAPII. It lacks many kinases present in extracts, among them the known CAK targets of CDK7.3 The C-fraction was used in combination with recombinant yeast TFIIA, TBP, and human TFIIB (Fig. 3D). This system efficiently establishes both Ser-5 and Ser-7-P in the presence of ATP (compare lane 1 with lanes 2 and 3). Depletion of either CDK7 or Mediator from the C-fraction diminished CTD phosphorylation (Fig. 3D, left panel, lanes 4 and 5 versus 3). When excess Mediator-RNAPII complex (immunopurified from nuclear extracts) is added to the C-fraction, CDK7 recruitment is strongly enhanced. In parallel, CTD phosphorylation markedly increases (lane 6 versus 3). As in extracts, free RNAPII that is recruited to templates in the absence of Mediator (lane 4) is not efficiently phosphorylated by CDK7. Next we replaced the C-fraction by a reconstituted system consisting of recombinant and purified general transcription factors, TBP, TFIIA, TFIIB, TFIIF, and TFIIE, in combination with immunopurified CDK7 and Mediator-RNAPII. Depending on the CDK7 IP, efficient phosphorylation of Ser-5-P and of Ser-7-P was observed (Fig. 3E). Mediator- and CDK7 IPs are washed extensively (3 times with 50 column volumes). Hence, these data strongly reason for direct phosphorylation of Ser-7 by CDK7. Given that ATP alone establishes CTD phosphorylation in the purified system, we further conclude that Ser-7 phosphorylation by CDK7 begins before transcription is initiated.
CTD Ser-7 Phosphorylation Is Established in the Context of a Functional PIC
To further address the question of whether Ser-7 phosphorylation by CDK7 is direct, we performed in vitro kinase assays using immunoprecipitated CDK7 module (Fig. 4A, lanes 4 and 8) or immunopurified TFIIH complexes (Fig. 4A, lanes 3 and 7). As expected, CDK7 efficiently phosphorylated Ser-5 residues (both the CAK module and the TFIIH complex are functional). Phosphorylation of Ser-7 was moderate but detectable, especially at higher CDK7 concentrations (Fig. 4A, lanes 2 versus 5). It should be noted that the Ser-7 phosphate (Ser-7-P) signal in the Western blots is generally much weaker than the corresponding signal generated by a Ser-5-specific antibody. When GST-CTD is used, higher concentrations are required, leading to cross-reactions of the Ser-7-P monoclonal antibody with GST-CTD and attenuating the relative signal of Ser-7-P.
Of note, the addition of immunopurified Mediator did not increase GST-CTD phosphorylation (Fig. 4A, lanes 2–5 versus 6–9). Thus, although Mediator (or a Mediator-RNAPII complex) is necessary for CTD phosphorylation in the context of PIC formation, it is dispensable for CDK7 function on an isolated CTD.
The Mediator-associated RNAPII contributes substantially to the overall RNAPII found on promoters. More importantly, its presence closely correlated to the formation of active transcription complexes (39, 46). We, therefore, asked whether Mediator-associated RNAPII is phosphorylated at Ser-7. Toward this end, extracts were immunodepleted of RNAPII (Fig. 4B). Subsequently, immunoprecipitated Mediator was added to the depleted extracts, and preinitiation complexes were formed at 1 μm ATP and analyzed in Western blots (Fig. 4B, compare lanes 1–3 and 4). Mediator-associated RNAPII restored transcription activity. Furthermore, Mediator-imported RNAPII was modified at Ser-5 and Ser-7 to a similar extent as the input pool of RNAPII (note that neither the recovery nor the precise ratio of free RNAPII to Mediator-associated RNAPII was determined).
Interestingly, neither CDK7 nor TFIIB was efficiently recruited to the template if RNAPII was removed from the extracts (Fig. 4B, compare lanes 1 and 2). In contrast, Mediator recruitment to the activator GAL4-VP16 was unaffected by RNAPII depletion. When Mediator (with bound RNAPII) was added back to the reaction, both CDK7 and TFIIB were efficiently recruited to promoter templates (Fig. 4B, lane 2 versus lanes 3 and 4). Collectively, we conclude that a complex of Mediator-RNAPII recruits both TFIIB and CDK7 to promoter regions. The recruitment of CDK7 (although not fully excluding an additional activation step of the kinase) explains the increase in CTD phosphorylation. In contrast, binding of the bulk of TBP is fully independent of Mediator-RNAPII complexes (compare Fig. 2C).
Having provided evidence that Mediator-RNAPII complexes directly or indirectly facilitate recruitment of a CDK7 module we next addressed the question of whether CDK7 recruitment is dependent on preinitiation complexes. Toward this end extracts were depleted (or mock-depleted) for TFIIB, an essential component of active preinitiation complexes. Depletion of TFIIB fully abolished CTD Ser-5 and Ser-7 phosphorylation, although CDK7 was found at promoters. We conclude that both Ser-5 and Ser-7 phosphorylation relies on TFIIB and, therefore, likely on the formation of intact preinitiation complexes (Fig. 4C). To further substantiate this hypothesis, we also depleted extracts for TBP, the seeding factor for preinitiation complexes (Fig. 4D). Again, Ser-5 and Ser-7 phosphorylation was reduced (Fig. 4D, lane 1 versus 2). When TBP is depleted, TFIIB and CDK7 are nevertheless found on templates. These data show that the presence of both TFIIB and CDK7 is insufficient to facilitate CTD phosphorylation. Instead intact preinitiation complexes seem necessary.
Together with TBP, we depleted the TBP-associated factors. CTD phosphorylation was fully rescued upon the addition of recombinant TBP. Thus, CDK7 functions independent of TBP-associated factors.
Evidence for Another Ser-7 Kinase
Several of our observations reasoned for further Ser-7 kinases that act independent of CDK7. These became first evident when ATP concentrations were raised during PIC formation (Fig. 5). Although depletion of CDK7 essentially abolishes Ser-5 and Ser-7 phosphorylation at up to 5 μm ATP (Fig. 5, A, lanes 1–4, and B, lanes 4 and 5) the strict dependence is lost at higher concentrations (in lanes 9 and 10, 100 μm ATP was added). Loss of dependence has been confirmed in several independent experiments. As a result we noted that both high ATP and limited preincubation periods (and, therefore, limited opportunity to form a PIC) reduced the relative impact of CDK7 (Fig. 5B). On the other hand, ATPγS as an ATP donor selects for CDK7 (Fig. 5B and Fig. 3C). The reasons for the latter are presently unknown but may hint to further requirements of the missing kinase. Notably, however, the phosphorylation by the missing kinase is also dependent on Mediator. Specific inhibitors to candidate kinases such as DNA-dependent protein kinase (wortmannin) and casein kinases did not suppress Ser-7 phosphorylation in vitro (supplemental Fig. 1). On the other hand, we found CTD Ser-7 phosphorylation to be fully sensitive to the CDK inhibitor roscovitine (Fig. 5A, lanes 1–4 versus 5–8). Roscovitine targets a broad spectrum of kinases (among them many CDKs). Collectively, we presently conclude that, minimally, one additional kinase is missing that functions independent of CDK7 and that is roscovitine-sensitive.
FIGURE 5.
Evidence for another Ser-7 kinase. A, shown is an immobilized template assay using mock-treated (ΔIso) or CDK7-depleted (ΔCDK7) nuclear extracts. PICs were formed in the presence of the indicated amounts of ATP and roscovitine. PICs in lanes 1 and 2 as well as 5 and 6 were formed under basal conditions, and PICs in lanes 3 and 4 as well as in lanes 7–10 were formed in the presence of the activator GAL4-VP16. Physiological salt conditions were used. B, PICs were formed on ML-promoter DNA templates in the presence of the indicated amounts of ATP or ATPγS under physiological salt conditions. PICs were formed with mock-treated (ΔIso), CDK7-depleted (ΔCDK7), or Mediator-depleted (ΔMED) Jurkat nuclear extracts.
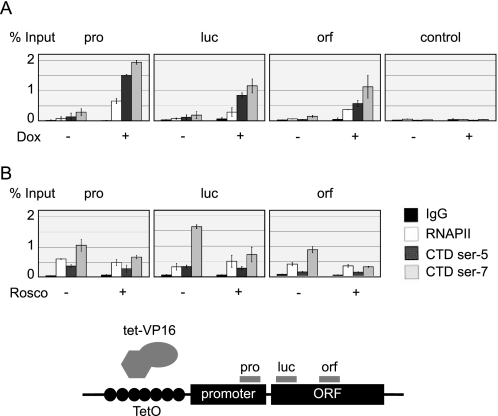
Next we asked whether Ser-7-P is introduced in a promoter-specific manner correlated to the induction of transcription in vivo. Toward this end we employed a stable conditional tet-VP16 reporter system described previously (39) and chromatin IP as a method to detect RNAPII and the phosphorylated serine residues. Indeed, CTD Ser-7 phosphorylation was established together with Ser-5-P upon transcription induction with doxycycline (Fig. 6A). CTD Ser-7 phosphorylation was most pronounced at promoter regions (compare promoter primers (pro) at about −100 with open reading frame primers luc (around +150) with control primers (control). If roscovitine inhibits Ser-7-P in vitro, it should do so in vivo. To assess this question, cells were incubated with 30 μm roscovitine for 30 min before gene induction by doxycycline. ChIP analysis revealed substantially reduced levels of CTD Ser-5 and Ser-7 phosphorylation both at the promoter (relative to the body of RNAPII, Fig. 6B) and downstream of the transcription start site (primers orf are located at +1500). Thus, Ser-7-P occurs both upstream and within the open reading frame. Although the ATP concentrations are much higher inside cells (estimated 1 mm), Ser-7-P is specifically sensitive to roscovitine inside cells.
FIGURE 6.
CTD Ser-7 phosphorylation is roscovitine-sensitive in vivo. A, shown is a ChIP experiment on the doxycycline-induced tet-VP16 system. Induction of cells with 1 μg of doxycycline (Dox) per ml medium was carried out for 60 min. B, ChIP analysis is shown of the tet-VP16-inducible gene system 60 min after induction of Luciferase gene expression with doxycycline (1 μg/ml). If indicated, roscovitine (Rosco) was added to the cell culture medium to a final concentration of 30 μm 30 min before induction.
DISCUSSION
In this study we explore the molecular process of phosphorylation of the Ser-7 residue within the heptarepeat of the CTD of RNAPII at promoter regions in human cells. Despite limited similarity of the environment of the Ser-5 and Ser-7 phosphorylation sites in the heptarepeat YSPTS5PS7, CDK7 is identified as one responsible enzyme. This conclusion is based on direct phosphorylation of isolated GST-CTD by purified CDK7 and the dependence of Ser-7 phosphorylation on CDK7 in crude and highly purified systems. Tests for copurifying kinases in CDK7 IPs were negative. Especially, the known targets of CDK7 (CDK1 and CDK2) were absent in CDK7 IPs. These kinases elute on P11 columns in the 0.3 m elution step and are absent already in the semipurified system (P11 C-fraction based). Clearly, this does not rule out that residual concentrations of an unknown kinase are present. However, we consider it unlikely that minor concentrations suffice in these in vitro systems to mediate efficient CTD phosphorylation. This is best exemplified with CDK7 itself, whose function is concentration-dependent. Mediator functions at least in part via enrichment of CDK7. Furthermore, strong effects indeed require enrichment of the enzyme.
The conclusion that CDK7 can phosphorylate Ser-7 is entirely consistent with the recent identification of yeast CDK7 (Kin28) as the major Ser-7 kinase (37). Beyond it we characterize the process in which the CTD is modified at residue Ser-7. CDK7 can phosphorylate Ser-7 directly in the absence of a functional preinitiation complex. However, in the physiological context of a nuclear extract, phosphorylation by CDK7 is strictly dependent on a series of structural constraints. CDK7 functions dependent on a promoter-bound preinitiation complex that minimally consists of TBP, TFIIB, and RNAPII (we have not tested TFIIF but expect it to be part of it). This is not a trivial finding because CDK7 is found on promoters in the absence of other general factors but in the presence of RNAPII. Indeed, promoter association of CDK7 is fully dependent on Mediator. In line with a recent genetic study in yeast, our data further suggest that Mediator complex recruits a free CDK7 CAK module (36). It is well established that Mediator binds RNAPII at enhancer regions (29, 30). The presence of a Mediator-associated RNAPII is closely linked to transcription activity in vitro (39). Yet, the underlying mechanisms remain unclear. For instance, extracts also contain RNAPII that is not associated with Mediator. In purified systems free RNAPII is fully functional in basal transcription. In a more physiological context RNAPII requires Mediator for efficient incorporation into preinitiation complexes and for both basal and activated transcription (31, 32). Importantly, here we demonstrate for the first time that an isolated Mediator-RNAPII complex suffices to recruit both TFIIB and CDK7 and facilitates the incorporation of RNAPII into preinitiation complexes. Mediator-RNAPII and TBP-TATA complexes form independently of each other. In a reverse stepwise process Mediator-RNAPII recruits TFIIB even in the absence of TBP. Although present, nearby CDK7 does not phosphorylate RNAPII bound to Mediator in the absence of either TFIIB or TBP. This indicates that kinase-substrate interaction relates to or may even require prior relocation of RNAPII to preinitiation complexes. Notably, we never observed Ser-7 phosphorylation in the absence of Ser-5 phosphorylation.
A general conclusion of these findings is that Ser-5 and Ser-7 phosphorylation is tightly controlled during initiation of transcription. Interestingly, core TFIIH is not necessary to initiate Ser-7 phosphorylation by CDK7. These and other data clearly suggested that Ser-7 phosphorylation begins before initiation of transcription. Previous analyses demonstrated that CDK7-mediated phosphorylation is not generally required for transcription (25). Similarly, a requirement of Ser-7 phosphorylation for transcription has not been reported. Nevertheless, our recent and previous ChIP data (4) suggest that Ser-7 occurs coupled to transcription in vivo, therefore, minimally reasoning for a function that is coupled to transcription. ChIP-ing, a conditional model gene, revealed the highest Ser-7-P signals downstream of the start site of transcription. Substantial steady-state levels were also detected further downstream within coding regions.
Last but not least, we have clear evidence for additional Ser-7 kinases. Notably, the latter also require DNA templates and Mediator, but they operate at a higher Km for ATP. Contrary to the molecular constraints observed for CDK7, our unpublished data suggest that at least one kinase functions independently of transcription complex formation.2
Supplementary Material
Acknowledgments
We thank the members of the Meisterernst and Eick laboratories for helpful discussions.
This work was supported by Bundesministerium für Bildung und Forschung Grant QuantPro Grant 0313860D, European Commission Grant EuTRACC 037445, and Deutsche Forschungsgemeinschaft Grants ME 967/2-1 and SPP1356 (to M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
S. Boeing and M. Meisterernst, unpublished observation.
- RNAPII
- RNA polymerase II
- PIC
- preinitiation complex
- CAK
- CDK-activating kinase
- TBP
- TATA-binding protein
- ML
- major-late
- IP
- immunoprecipitation
- GST
- glutathione S-transferase
- CDK
- cyclin-dependent kinase
- hTF
- human TFIIB
- ChIP
- chromatin IP
- CTD
- C-terminal heptarepeat domain
- Erk
- extracellular signal-regulated kinase
- DTT
- dithiothreitol
- PMSF
- phenylmethylsulfonyl fluoride
- BSA
- bovine serum albumin
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P. (2005) Genes Dev. 19, 1401–1415 [DOI] [PubMed] [Google Scholar]
- 2.Phatnani H. P., Greenleaf A. L. (2006) Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 3.Chapman R. D., Heidemann M., Hintermair C., Eick D. (2008) Trends Genet. 24, 289–296 [DOI] [PubMed] [Google Scholar]
- 4.Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- 5.Buratowski S. (2003) Nat. Struct. Biol. 10, 679–680 [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 7.Deng L., Ammosova T., Pumfery A., Kashanchi F., Nekhai S. (2002) J. Biol. Chem. 277, 33922–33929 [DOI] [PubMed] [Google Scholar]
- 8.Gebara M. M., Sayre M. H., Corden J. L. (1997) J. Cell. Biochem. 64, 390–402 [PubMed] [Google Scholar]
- 9.Long J. J., Leresche A., Kriwacki R. W., Gottesfeld J. M. (1998) Mol. Cell. Biol. 18, 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet F., Vigneron M., Bensaude O., Dubois M. F. (1999) Nucleic Acids Res. 27, 4399–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dvir A., Peterson S. R., Knuth M. W., Lu H., Dynan W. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11920–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trigon S., Serizawa H., Conaway J. W., Conaway R. C., Jackson S. P., Morange M. (1998) J. Biol. Chem. 273, 6769–6775 [DOI] [PubMed] [Google Scholar]
- 13.Hengartner C. J., Myer V. E., Liao S. M., Wilson C. J., Koh S. S., Young R. A. (1998) Mol. Cell 2, 43–53 [DOI] [PubMed] [Google Scholar]
- 14.Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., Conaway R. C. (2004) Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 15.Furumoto T., Tanaka A., Ito M., Malik S., Hirose Y., Hanaoka F., Ohkuma Y. (2007) Genes Cells 12, 119–132 [DOI] [PubMed] [Google Scholar]
- 16.Donner A. J., Szostek S., Hoover J. M., Espinosa J. M. (2007) Mol. Cell 27, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes N. P., Bjerke G., Llorente B., Szostek S. A., Emerson B. M., Espinosa J. M. (2006) Genes Dev. 20, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buratowski S. (2005) Curr. Opin. Cell Biol. 17, 257–261 [DOI] [PubMed] [Google Scholar]
- 19.Kim M., Krogan N. J., Vasiljeva L., Rando O. J., Nedea E., Greenblatt J. F., Buratowski S. (2004) Nature 432, 517–522 [DOI] [PubMed] [Google Scholar]
- 20.Meinhart A., Cramer P. (2004) Nature 430, 223–226 [DOI] [PubMed] [Google Scholar]
- 21.Licatalosi D. D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J. B., Bentley D. L. (2002) Mol. Cell 9, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 22.Rossignol M., Kolb-Cheynel I., Egly J. M. (1997) EMBO J. 16, 1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirode F., Busso D., Coin F., Egly J. M. (1999) Mol Cell 3, 87–95 [DOI] [PubMed] [Google Scholar]
- 24.Laybourn P. J., Dahmus M. E. (1990) J. Biol. Chem. 265, 13165–13173 [PubMed] [Google Scholar]
- 25.Kanin E. I., Kipp R. T., Kung C., Slattery M., Viale A., Hahn S., Shokat K. M., Ansari A. Z. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho E. J., Takagi T., Moore C. R., Buratowski S. (1997) Genes Dev. 11, 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Shuman S., Bentley D. L. (1997) Genes Dev. 11, 3306–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng H. H., Robert F., Young R. A., Struhl K. (2003) Mol. Cell 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 29.Blazek E., Mittler G., Meisterernst M. (2005) Chromosoma 113, 399–408 [DOI] [PubMed] [Google Scholar]
- 30.Malik S., Roeder R. G. (2005) Trends Biochem. Sci. 30, 256–263 [DOI] [PubMed] [Google Scholar]
- 31.Baek H. J., Kang Y. K., Roeder R. G. (2006) J. Biol. Chem. 281, 15172–15181 [DOI] [PubMed] [Google Scholar]
- 32.Mittler G., Kremmer E., Timmers H. T., Meisterernst M. (2001) EMBO Rep. 2, 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. (1994) Cell 77, 599–608 [DOI] [PubMed] [Google Scholar]
- 34.Myers L. C., Gustafsson C. M., Bushnell D. A., Lui M., Erdjument-Bromage H., Tempst P., Kornberg R. D. (1998) Genes Dev. 12, 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guidi B. W., Bjornsdottir G., Hopkins D. C., Lacomis L., Erdjument-Bromage H., Tempst P., Myers L. C. (2004) J. Biol. Chem. 279, 29114–29120 [DOI] [PubMed] [Google Scholar]
- 36.Esnault C., Ghavi-Helm Y., Brun S., Soutourina J., Van Berkum N., Boschiero C., Holstege F., Werner M. (2008) Mol. Cell 31, 337–346 [DOI] [PubMed] [Google Scholar]
- 37.Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009) Mol. Cell 34, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egloff S., O'Reilly D., Chapman R. D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007) Science 318, 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlmann T., Boeing S., Lehmbacher M., Meisterernst M. (2007) J. Biol. Chem. 282, 2163–2173 [DOI] [PubMed] [Google Scholar]
- 40.Wu S. Y., Zhou T., Chiang C. M. (2003) Mol. Cell. Biol. 23, 6229–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie J., Collart M., Lemaire M., Stelzer G., Meisterernst M. (2000) EMBO J. 19, 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stelzer G., Goppelt A., Lottspeich F., Meisterernst M. (1994) Mol. Cell. Biol. 14, 4712–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kretzschmar M., Stelzer G., Roeder R. G., Meisterernst M. (1994) Mol. Cell. Biol. 14, 3927–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albert T. K., Grote K., Boeing S., Stelzer G., Schepers A., Meisterernst M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10000–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilfillan S., Stelzer G., Piaia E., Hofmann M. G., Meisterernst M. (2005) J. Biol. Chem. 280, 6222–6230 [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Krutchinsky A., Fukuda A., Chen W., Yamamura S., Chait B. T., Roeder R. G. (2005) Mol. Cell 19, 89–100 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.