Abstract
A disintegrin-like and metalloprotease domain with thrombospondin type 1 motifs 9 (ADAMTS9) is a highly conserved metalloprotease that has been identified as a tumor suppressor gene and is required for normal mouse development. The secreted ADAMTS9 zymogen undergoes proteolytic excision of its N-terminal propeptide by the proprotein convertase furin. However, in contrast to other metalloproteases, propeptide excision occurs at the cell surface and leads to decreased activity of the zymogen. Here, we investigated the potential cellular mechanisms regulating ADAMTS9 biosynthesis and cell-surface processing by analysis of molecular complexes formed by a construct containing the propeptide and catalytic domain of pro-ADAMTS9 (Pro-Cat) in HEK293F cells. Cross-linking of cellular proteins bound to Pro-Cat followed by mass spectrometric analysis identified UDP-glucose:glycoprotein glucosyltransferase I, heat shock protein gp96 (GRP94), BiP (GRP78), and ERdj3 (Hsp40 homolog) as associated proteins. gp96 and BiP were present at the cell surface in an immunoprecipitable complex with pro-ADAMTS9 and furin. Treatment with geldanamycin, an inhibitor of the HSP90α family (including gp96), led to decreased furin processing of pro-ADAMTS9 and accumulation of the unprocessed pro-ADAMTS9 at the cell surface. gp96 siRNA down-regulated the levels of cell-surface pro-ADAMTS9 and furin, whereas the levels of cell-surface pro-ADAMTS9, but not of cell-surface furin, were decreased upon treatment with BiP siRNA. These data identify for the first time the cellular chaperones associated with secretion of an ADAMTS protease and suggest a role for gp96 in modulating pro-ADAMTS9 processing.
Introduction
A disintegrin-like and metalloprotease domain with thrombospondin type 1 motifs (ADAMTS)2 proteases are a large family of secreted metalloproteases implicated in diverse biological functions including angiogenesis, hemostasis, connective tissue disorders, inflammation, arthritis, and cancer (1, 2). ADAMTS9 is the most highly conserved member of the family (3). It was recently implicated in regulation of melanoblast development (4) and limb development in mice (5) and was previously identified as a tumor suppressor gene in esophageal and nasopharyngeal cancer (6, 7). Early lethality of the Adamts9-null allele in mice (5, 8) strongly suggests additional critical biological roles. The extracellular matrix proteoglycans aggrecan and versican were previously identified as its substrates (3), and they are relevant to its potential roles in arthritis and embryonic development, respectively (4, 9). Characterization of ADAMTS9 protein showed it to be located at the cell surface in transfected cells, suggesting that despite the absence of a membrane anchor, it could be an operational cell-surface protease (3). Its potential contribution to cell-surface proteolysis is supported by the requirement of its Caenorhabditis elegans ortholog Gon-1 for cell migration during gonadal morphogenesis (10).
Although these observations strongly suggest that ADAMTS9 has considerable biological significance, little is currently known about its cellular trafficking, post-translational modification, and regulatory mechanisms. ADAMTS proteases are synthesized as zymogens with N-terminal propeptides, which like many other metalloprotease propeptides are excised in the secretory pathway by proprotein convertases such as furin. Propeptide excision is usually referred to as “activation,” because the bulky propeptide sterically hinders substrate access to the catalytic domain, and thus the zymogens are usually inactive. Previously, we had investigated the mechanisms by which the ADAMTS9 propeptide was excised and demonstrated two unusual aspects of this process. First, we found that ADAMTS9 was processed by furin extracellularly but not in the secretory pathway (11). Second, we demonstrated that the ADAMTS9 zymogen was catalytically active against versican but that furin processing of the propeptide led to decreased activity (12). Following furin processing, a complex of mature ADAMTS9 and its propeptide was immediately released from the cell surface, possibly as a result of a conformational change (12). Thus, only unprocessed pro-ADAMTS9 is detectable at the surface of cells such as HEK293F, which express furin at robust levels (12). These unusual features led us to investigate further the molecular interactions of pro-ADAMTS9 during biosynthesis and transport through the secretory pathway with emphasis on those that could regulate its activation at the cell surface by furin. The results suggest the participation of the cellular chaperone gp96/GRP94 in the regulation of pro-ADAMTS9 processing by furin. Because furin is implicated in the extracellular processing of other secreted proproteins, these observations may be of broad significance.
EXPERIMENTAL PROCEDURES
Expression Plasmids, Cell Culture, Transfection, and Cell Treatments
Plasmids for the production of full-length ADAMTS9 with a C-terminal FLAG tag, a C-terminally truncated form extending to the 8th thrombospondin type 1 repeat (ADAMTS9(N-L2)), or containing only the signal peptide, propeptide, and catalytic domain (Pro-Cat) with tandem C-terminal Myc and His tags were previously described (3, 11) (Fig. 1A). Plasmid encoding furin was kindly provided by Dr. Nabil Seidah of McGill University. HEK293F cell lines stably transfected with Pro-Cat were maintained as described previously (3, 11). Transient transfections were done using FuGENE6 (Roche Diagnostics, Indianapolis, IN) per the manufacturer's recommendations. Monoclonal rat anti-gp96 (cat. no. MA1-37772) and monoclonal anti-α-actinin-4 (cat. no. MA1-800) were purchased from Affinity Bioreagents (Golden, CO), monoclonal anti-BiP (cat. no. 610979) from BD Biosciences, monoclonal anti-Myc (clone 9E10) from Invitrogen, rabbit polyclonal anti-Myc (cat. no. C3956) and anti-FLAG® M2 Affinity Gel (cat. no. A2220) from Sigma-Aldrich, polyclonal goat antibody to ERdj3 from Santa Cruz Biotechnology (cat. no. sc46945), and monoclonal anti-furin (cat. no. MON-148) from Alexis Biochemicals (San Diego, CA). Polyclonal antibody anti-RP4 recognizing the ADAMTS9 propeptide, and polyclonal antibody 3597 recognizing the linker 2 region were previously described (9, 11).
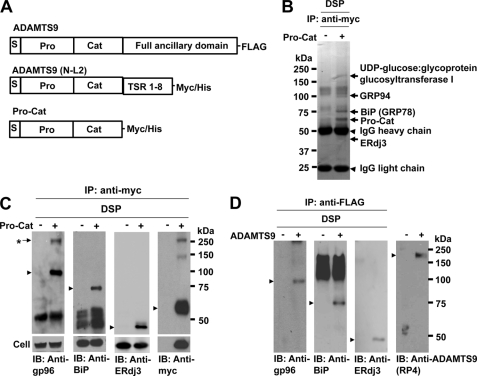
FIGURE 1.
Identification of molecules that interact with ADAMTS9 Pro-Cat. A, domain structure of the previously published ADAMTS9 constructs used in this analysis. TSR, thrombospondin type 1 repeats. B, HEK293 F cells stably expressing ADAMTS9 Pro-Cat were cross-linked with membrane-permeable, thiol-cleavable DSP, and immunoprecipitated with polyclonal anti-Myc antibody as described. The eluted samples were separated by NuPAGE (4–12% gradient gel), and the indicated bands (arrows) were cut out and identified by LC/MS/MS. C, following cross-linking and immunoprecipitation with polyclonal anti-Myc, immunoprecipitates were analyzed by Western blotting with monoclonal anti-gp96 antibody, monoclonal anti-BiP antibody, polyclonal goat anti-ERdj3 antibody, or monoclonal anti-Myc antibody as indicated. The lower panel shows total cellular expression of gp96, BiP, ERdj3, and Pro-Cat in nontransfected (−) or transfected (+) cells. A protein complex (*) is recognized by both anti-gp96 and anti-Myc, but not by anti-BiP and anti-ERdj3. D, full-length ADAMTS9 (panel A) was affinity-isolated using anti-FLAG® M2 Affinity Gel, and the eluted proteins were analyzed by Western blotting with the same antibodies as used above. Anti-RP4 antibody was used to detect ADAMTS9. Arrowheads indicate the individual protein bands corresponding to (from left to right), gp96, BiP, ERdj3, and ADAMTS9.
Cross-linking and Immunoprecipitation
HEK293F cells or HEK293F cells stably expressing Pro-Cat were washed with phosphate-buffered saline three times and cross-linked with the membrane-permeable, thiol-cleavable cross-linker DSP (dithiobis[succinimidylpropionate]) (Pierce) (final concentration; 2 mm), membrane-nonpermeable, thiol-cleavable DTSSP (3,3′-dithiobis[sulfosuccinimidylpropionate]) (Pierce) (final concentration; 2 mm), or membrane-nonpermeable, thiol-noncleavable BS3 (bis(sulfosuccinimidyl)suberate) (Pierce) (final concentration; each 2 mm) for 30 min on ice. The cells were lysed in 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 150 mm NaCl, 1% Triton X-100 (lysis buffer) containing a protease inhibitor mixture (Roche Diagnostics) for 1 h at 4 °C and centrifuged. The soluble portion of the lysate was incubated overnight with appropriate antibodies and protein A or protein G for capturing rat IgG at 4 °C with rotation. The beads were washed six times in lysis buffer and boiled in SDS-sample buffer. The eluted samples were separated on NuPAGE gel (4–12%) (Invitrogen), and the identity of the protein bands was determined by LC/MS/MS (Tufts University Core Facility) or analyzed by Western blotting with appropriate antibodies. As a control to eliminate cell-surface proteins, cells were treated with 0.05% trypsin, 0.53 mm EDTA on ice for 20 min, and trypsin was inactivated by washing the cells three times with 1 mm phenylmethanesulfonyl fluoride in phosphate-buffered saline, prior to cross-linking as above.
Biotinylation of Cell-surface Proteins and Pulse-chase Analysis
Biotinylation of cells was done on ice as previously described (3, 11). As a control, cell-surface proteins were eliminated with trypsin/EDTA prior to biotinylation as described (3, 11). Biotinylated proteins were captured using streptavidin-agarose (Sigma-Aldrich) and eluted by boiling in Laemmli sample buffer, followed by SDS-PAGE and Western blotting with appropriate antibodies. To detect possible biotinylation of intracellular proteins, as a control for validity of cell-surface biotinylation, Western blotting was done for the cell lysate or the biotinylated proteins with monoclonal anti-α-actinin-4 antibody. To examine the role of HSP90 family members in pro-ADAMTS9 processing at the cell surface, biotinylated cells were transferred to serum-free medium in the presence of monensin (50 μg/ml; Sigma-Aldrich) alone, or in the presence of monensin (50 μg/ml) plus 10 μm geldanamycin (InvivoGen, San Diego, CA). Cells and media were collected at sequential time points and analyzed by Western blotting with anti-Myc. For analysis of the trafficking of furin and Pro-Cat to the cell surface, cells were incubated in the presence or absence of 10 μm geldanamycin for up to 5 h. Conditioned media were collected, and cells were biotinylated at sequential time points. Western blotting was done for the streptavidin-captured proteins, conditioned media, and cell lysates with anti-Myc or anti-furin.
RNA Interference
SilencerTM siRNA against gp96 (siRNA ID 119657) and BiP (siRNA ID 16038) was purchased from Ambion (Austin, TX). HEK293F cells were transfected with 100 nm siRNA or a generic negative control siRNA (Ambion) using Lipofectamine 2000 (Invitrogen). After 72 h of incubation, the medium was changed to 293 SFM-II medium (Invitrogen), and cells were incubated for another 3 h. The conditioned medium and cell lysates were analyzed by Western blotting with appropriate antibodies. Cell-surface biotinylation was done as above.
Statistical Analysis
Data represent the mean and S.D. of three experiments. Statistical analysis was performed using an unpaired Student's t test.
RESULTS
Identification of Cellular Proteins That Complex with Pro-ADAMTS9
Because full-length pro-ADAMTS9 is a large enzyme (220 kDa), which is not robustly expressed (3, 11), we used Pro-Cat, a C-terminally truncated construct (Fig. 1A), which we previously found to be efficiently expressed, and was demonstrated to represent a valid surrogate for full-length ADAMTS9 in analysis of cell-surface propeptide processing (3, 11, 12). We cross-linked molecules complexed with Pro-Cat in HEK293F cells using a cell permeable, reducible cross-linker, and immunoprecipitated the resulting complexes with anti-Myc, followed by analysis of complexed proteins by reducing SDS-PAGE. For this purpose, we compared complexes isolated from cells expressing Pro-Cat with complexes from untransfected cells, and undertook LC/MS/MS analysis of the molecular species that were uniquely present in complexes from Pro-Cat expressing cells (Fig. 1B). MS analysis of these unique bands identified UDP-glucose:glycoprotein glucosyltransferase I, heat shock protein gp96 (GRP94), BiP (GRP78), and ERdj3 (Hsp40 homolog) as molecules that co-purified with Pro-Cat (Fig. 1B). At least 5 distinct peptides from each protein were present in spectral analysis of the respective band. We subsequently confirmed the formation of a complex that included gp96, BiP, ERdj3, and Pro-Cat by binary analysis of cell lysate in which cross-linking and immunoprecipitation with polyclonal rabbit anti-Myc antibody was followed by Western blotting with anti-gp96, anti-BiP, anti-ERdj3, and monoclonal anti-Myc. gp96, BiP, and ERdj3 were thus identified to form a complex with Pro-Cat (Fig. 1C). To ensure that these interactions also occurred with full-length ADAMTS9, i.e. to validate the continued use of Pro-Cat, we undertook affinity purification of full-length FLAG-tagged pro-ADAMTS9 after cross-linking and immunoprecipitation with anti-FLAG® M2 Affinity Gel. Co-isolation of gp96, BiP, ERdj3, and full-length pro-ADAMTS9 was observed after cross-linking and immunoprecipitation with anti-FLAG® M2 Affinity Gel (Fig. 1D).
Cell-surface Localization of Heat Shock Protein gp96 and BIP in HEK293F Cells
Because the initial cross-linking experiments did not distinguish between intracellular and cell-surface populations, we next asked whether gp96 and BiP were localized at the cell surface in HEK293F cells. To examine this, cell-surface proteins were labeled with cell non-permeable biotinylation reagents. Biotinylated proteins were captured using streptavidin-agarose and analyzed by Western blotting with appropriate antibodies. As a control to demonstrate that the captured proteins indeed were derived from the cell surface, cells were treated with trypsin prior to analysis, which led to loss of biotinylated BiP and gp96 (Fig. 2A). We found that gp96 and BiP were present at the cell surface both in the presence or absence of Pro-Cat, indicating that their cell-surface localization was not dependent on the presence of Pro-Cat and that it was not a consequence of stress induced by overexpression of the transfected cDNA (Fig. 2A). Because biotinylation is very sensitive and may detect small amounts of intracellular proteins released from lysed cells, we used an antibody to the intracellular protein α-actinin-4 for Western blotting of the biotinylated proteins. As shown in Fig. 2B, α-actinin-4 was not detected in the biotinylated populations, but only in the total cell lysate, which indicated that gp96 and BiP in the biotinylated isolates were indeed derived from the cell surface of HEK293F cells. Because ERdj3 is not secreted, we did not seek it at the cell surface.
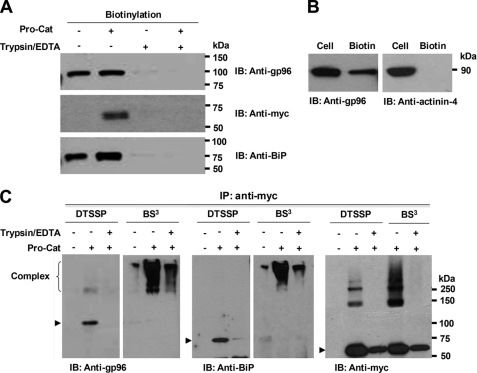
FIGURE 2.
Cell-surface localization of gp96 and BiP in HEK293F cells. Following cell-surface biotinylation (with (+) or without (−) prior trypsinization), Western blotting of streptavidin-agarose-purified proteins with the indicated antibodies demonstrates gp96, BiP, and ADAMTS9 Pro-Cat (A), but not α-actinin-4 (B) at the cell-surface. C, gp96 and BiP form complexes with Pro-Cat or furin at the cell surface. Cell-surface proteins were cross-linked with DTSSP or BS3 on ice. As a control, cell-surface proteins were eliminated by prior (DTSSP) or subsequent (BS3) treatment with trypsin. Cell lysates were immunoprecipitated with polyclonal anti-Myc, and eluted proteins were analyzed by Western blotting with anti-gp96, anti-BiP, or monoclonal anti-Myc. Note that similar high molecular weight forms (>250 kDa) with BS3 are recognized by all three antibodies, and the expected gp96, BiP, and Pro-Cat molecular species (arrowheads) disappear or are decreased when cross-linked with BS3.
Complex Formation of gp96 and BiP with pro-ADAMTS9 at the Cell Surface
To ask whether pro-ADAMTS9 existed in complex with gp96 and/or BiP at the cell surface, we next cross-linked cell-surface proteins using a thiol-cleavable cross-linker (DTSSP) or a thiol-uncleavable cross-linker (BS3), neither of which penetrates the cell membrane. The detergent-soluble proteins were immunoprecipitated using polyclonal anti-Myc, which recognizes the C-terminal tag on Pro-Cat. gp96 and BiP were co-immunoprecipitated with Pro-Cat in the presence of DTSSP, but these bands disappeared upon trypsin treatment of cells prior to cross-linking (Fig. 2C). When cells were cross-linked using BS3, high molecular weight forms (>250 kDa) were detected by anti-gp96, anti-BiP, or anti-Myc antibody, although the patterns were slightly different (Fig. 2C), suggesting that additional molecules could be part of the cell-surface complex containing ADAMTS9, BiP, and gp96. Furthermore, after trypsin treatment of BS3 cross-linked cells, these cross-linked bands were substantially diminished (Fig. 2C). These data indicate that pro-ADAMTS9 is present in a complex with gp96 and BiP, and possibly with additional proteins at the cell surface.
Furin Forms a Complex with gp96 and BiP at the Cell Surface
In a previous publication, we showed that furin was part of a cell-surface complex that contained ADAMTS9 (11). Moreover, we found here that ADAMTS9 was complexed with gp96 and BiP at the cell surface. To address whether furin could be part of a complex with gp96 and/or BiP at the cell surface, we cross-linked cell-surface proteins with DTSSP and immunoprecipitated cellular proteins using anti-gp96. Furin was co-immunoprecipitated with gp96 and BiP in the presence or absence of Pro-Cat (Fig. 3A). Furthermore, in the trypsin-treated cells, furin and BiP co-immunoprecipitation with gp96 was decreased, suggesting that furin existed as a complex with gp96 and BiP at the cell surface. We tried to detect high molecular weight complexes (as shown in Fig. 2C) on Western blot with anti-furin monoclonal antibody after treating the cells with BS3 and immunoprecipitation with anti-Myc (Pro-Cat), but these attempts were unsuccessful, possibly because of the alteration of the furin epitope by cross-linking with the thiol-noncleavable cross-linker. Nevertheless, these data, taken together with previous work (11, 12) suggested that furin, gp96, ADAMTS9, and BiP may together be part of a cell-surface complex.
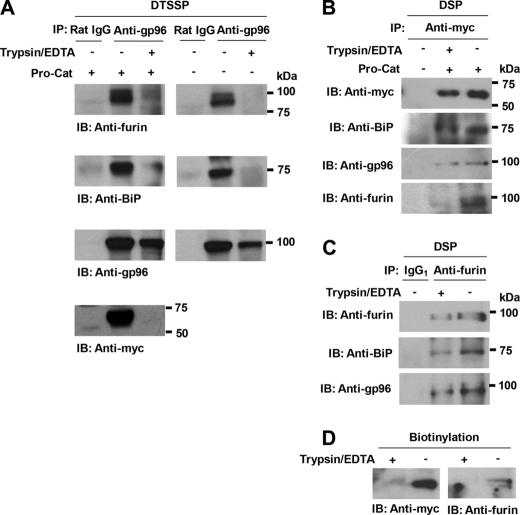
FIGURE 3.
Anti-gp96-precipitated, cross-linked cell-surface complexes contain furin, BiP, and Pro-Cat. A, cells were cross-linked with DTSSP as described. Immunoprecipitation was performed using rat anti-gp96 monoclonal antibody and protein G, followed by Western blotting with appropriate antibodies. As a control to eliminate cell-surface proteins, cells were treated with trypsin. Rat IgG was used as a negative control. Note that cell-surface BiP, Pro-Cat, and furin are substantially decreased in the trypsin-treated cells. B and C, HEK293F cells were treated with trypsin to remove cell-surface proteins. Cellular proteins were cross-linked with DSP on ice. Cell lysates were immunoprecipitated with polyclonal anti-Myc (B) or monoclonal anti-furin antibody (C), and the eluted proteins were analyzed by Western blotting with the indicated antibodies. Note that BiP and gp96 are co-immunoprecipitated with Pro-Cat or furin even if the cells are trypsin-treated, but furin is not co-immunoprecipitated with Pro-Cat in the trypsin-treated cells. D, removal of cell-surface Pro-Cat and furin was confirmed in the trypsin-treated cells after cell-surface biotinylation.
Pro-Cat and Furin Individually Form a Complex with gp96 and BiP Intracellularly
We considered the possibility that furin and ADAMTS9 could be independently transported to the cell surface, because furin was not isolated as one of proteins bound to ADAMTS9 in the secretory pathway (Fig. 1B). After treating the cells with trypsin to remove cell-surface protein, we cross-linked molecules complexed with Pro-Cat in HEK293F cells using a cell-permeable, reducible cross-linker (DSP), immunoprecipitated the complexes with polyclonal anti-Myc, and analyzed them by Western blotting with appropriate antibodies. Pro-Cat was co-immunoprecipitated with BiP and gp96, but not with furin, in the trypsin-treated cells (Fig. 3B). We did the same experiment as the above except using nontransfected cells and used anti-furin for immunoprecipitation. BiP and gp96 were also co-immunoprecipitated with furin in the trypsin-treated cells as well as in non-treated cells (Fig. 3C). As a control to clarify the removal of cell-surface Pro-Cat and furin, cells were treated with trypsin/EDTA before biotinylation. Biotinylated protein was streptavidin-captured and analyzed by Western blotting with anti-Myc and anti-furin, respectively. Pro-Cat and furin were detected in the biotinylated cell lysates, but not in the trypsin-treated cells, indicating that Pro-Cat and furin were completely removed from the cell surface (Fig. 3D). Together, these data suggest that pro-ADAMTS9 and furin form a complex with gp96 and BiP within the cell as well as on the cell surface. However, in contrast to their interaction on the cell surface, pro-ADAMTS9, and furin are not associated within the cell.
Furin Processing of ADAMTS9 Pro-Cat Is Significantly Reduced by Geldanamycin
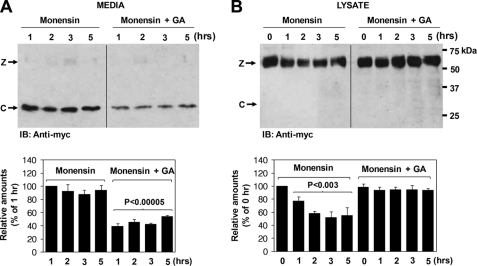
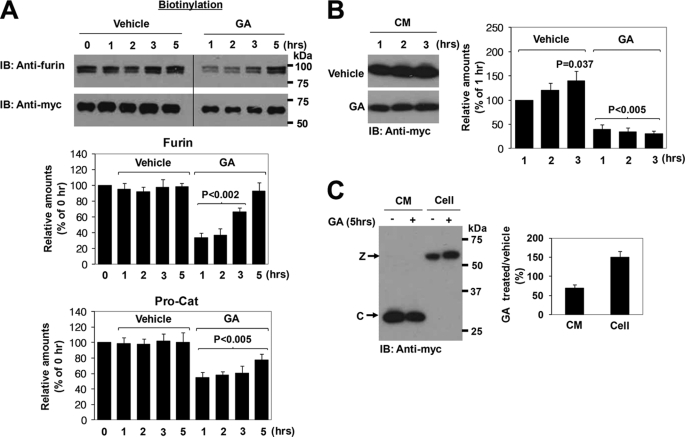
Geldanamycin is a benzoquinone ansamycin isolated from actinomyces (13). It binds with high affinity to the ATP binding site of molecules of the HSP90 family, which includes gp96, interfering with their function (14). To address whether extracellular HSP90 family proteins play a role in the processing of the ADAMTS9 propeptide at the cell surface, we first treated the cells with monensin to block trafficking in the cells. After cell-surface biotinylation, the processed biotinylated catalytic domain was chased in conditioned medium and cells treated with monensin alone or in the presence of monensin and geldanamycin. In the presence of geldanamycin, the processed catalytic domain was detectable at lower levels in the conditioned medium (Fig. 4A). Consistent with this, the level of cell-surface ADAMTS9 declined more rapidly in the absence of geldanamycin (Fig. 4B). As a control, to verify successful secretion blockade, monensin effectively blocked the secretion of fibronectin into the medium at the same concentration (data not shown).
FIGURE 4.
Geldanamycin prevents cell-surface processing of Pro-Cat by furin. A, monensin treatment was used to block protein trafficking. After cell-surface biotinylation, biotinylated ADAMTS9 Pro-Cat was chased in conditioned medium in the continuing presence of monensin (50 μg/ml) alone or in the presence of monensin (50 μg/ml) plus geldanamycin (GA) (10 μm). Western blotting was done with anti-Myc. Note that in the absence of geldanamycin, an increased amount of processed catalytic domain (C) is observed in the conditioned medium (left-hand panel). B, in the absence of geldanamycin, the zymogen (Z) declines more rapidly in the cell lysates (right-hand panel). Lower panels in A and B represent the mean and S.D. of three independent experiments. The statistical significance is shown.
The Trafficking of Furin and Pro-ADAMTS9 to the Cell Surface Is Retarded in the Presence of Geldanamycin
To ask if the HSP90 family was involved in the trafficking of furin and/or pro-ADAMTS9 to the cell surface, Pro-Cat-expressing cells were transfected with furin expression plasmid and incubated in the presence or absence of geldanamycin for up to 5 h. Conditioned media were collected, and cells were surface-biotinylated. Geldanamycin significantly inhibited the levels of both furin and pro-ADAMTS9 at the cell surface (Fig. 5A). However, the observed effects recovered somewhat on analysis after 5 h for reasons that are presently unclear. In addition, and consistent with the data in Fig. 4, a decreased amount of the processed catalytic domain in the conditioned medium was observed in the presence of geldanamycin (Fig. 5, B and C), whereas unprocessed zymogen accumulated in the geldanamycin-treated cells (Fig. 5C).
FIGURE 5.
Geldanamycin decreases levels of cell-surface furin. A, cells were incubated with vehicle alone or geldanamycin (GA) (10 μm) for up to 5 h. Conditioned medium was collected, and cells were biotinylated. Western blotting was performed for biotinylated proteins using anti-furin (upper panel) and anti-Myc (for Pro-Cat lower panel). Note that the levels of furin and Pro-Cat at the cell surface were decreased in the geldanamycin (GA)-treated cells. However, the furin doublet indicates that both the zymogen and mature furin are observed, suggesting that GA does not affect furin maturation. Data represent the mean and S.D. of three independent experiments. The statistical significance is shown. B, decreased amount of the processed catalytic domain recognized by anti-Myc in the conditioned medium was observed in the presence of geldanamycin. C, unprocessed zymogen (Z) accumulated in the geldanamycin-treated cells.
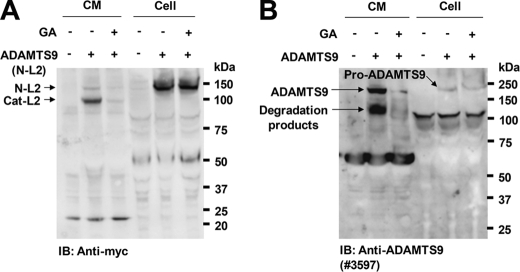
To seek evidence that geldanamycin also affected secretion of longer ADAMTS9 constructs, we transfected HEK293F cells using a full-length ADAMTS9 plasmid as well as a slightly shorter construct, ADAMTS9 N-L2, which is more robustly expressed than full-length ADAMTS9 (12). These experiments were done without furin co-transfection and relied on geldanamycin effects on endogenous furin. Because both constructs are weakly expressed relative to Pro-Cat, we could not reliably detect them on the cell surface. However, Western blotting showed that the conditioned medium of geldanamycin-treated cells had reduced levels of both constructs (Fig. 6, A and B).
FIGURE 6.
Geldanamycin decreases secretion of full-length ADAMTS9 and ADAMTS9 N-L2 constructs. A and B, HEK293F cells expressing ADAMTS9 (N-L2) (panel A) or full-length ADAMTS9 (panel B) were incubated with vehicle alone or geldanamycin (GA) (10 μm) for 6 h. Conditioned medium was collected, concentrated with trichloroacetic acid, and analyzed by Western blotting with the indicated antibodies.
Down-regulation of gp96 or BiP by siRNA Affects pro- ADAMTS9 and Furin
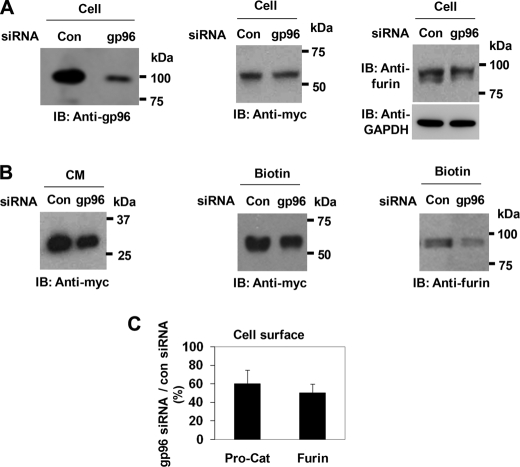
Because geldanamycin inhibits other HSP90 family members in addition to gp96, and because gp96 was the specific family member that was complexed with pro-ADAMTS9 and furin, we used gp96 siRNA to specifically test whether its down-regulation in HEK293F cells leads to reduction in pro-ADAMTS9 processing by furin. In cells transfected with gp96 siRNA, total cellular gp96 protein was down-regulated by about 70% (Fig. 7A, left panel). In these cells, the total expression level of Pro-Cat was not substantially altered by treatment with gp96 siRNA, although that of furin was slightly reduced (Fig. 7A, center and right-hand panel, respectively). However, compared with cells that were treated with control siRNA, a decreased amount of the processed ADAMTS9 was observed in the conditioned medium (63 + 8%) (Fig. 7B, left-hand panel). The levels of Pro-Cat and furin were both decreased at the cell surface of gp96 siRNA-treated cells (Fig. 7, B, center and right-hand panel and C).
FIGURE 7.
gp96 siRNA suppresses processing of ADAMTS9 Pro-Cat. A, gp96 siRNA suppresses the expression level of cellular gp96 (left panel). Transfection with gp96 siRNA does not significantly affect the total cellular levels of Pro-Cat (center panel) but decreases furin levels (right panel). B, expression of gp96 siRNA decreases levels of processed catalytic domain in conditioned medium (left panel) and the amounts of ADAMTS9 zymogen (center panel) and furin (right panel) at the cell surface. C, quantitation of cell-surface Pro-Cat and furin in gp96 siRNA-treated cells is expressed as a percentage of control siRNA-treated cells. Data represent the mean and S.D. of three independent experiments.
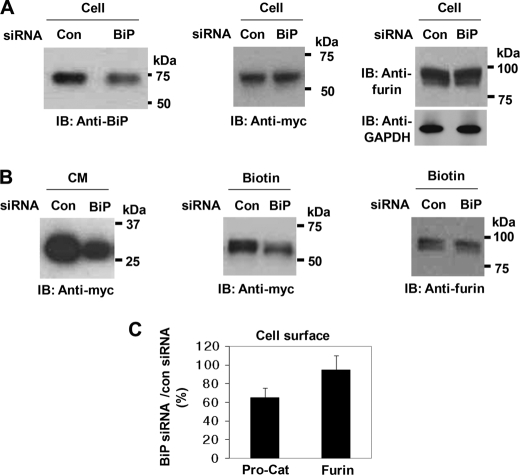
Similarly, to address the function of BiP in the processing of pro-ADAMTS9 by furin, we used BiP siRNA, which, however, reduced total cellular BiP protein by about 30% (Fig. 8A, left-hand panel). In BiP siRNA-treated cells, total expression levels of Pro-Cat and furin were not changed (Fig. 8A, center and right-hand panel). Decreased amounts of the processed catalytic domain was observed in the conditioned medium of BiP siRNA-treated cells (61 + 11%) (Fig. 8B, left-hand panel). In BiP siRNA-treated cells, the cell-surface level of Pro-Cat was decreased, whereas that of furin was unaltered (Fig. 7B, center and right panel, Fig. 8C).
FIGURE 8.
Processing of ADAMTS9 is decreased in BiP siRNA-transfected cells. A, BiP siRNA suppresses the levels of cellular BiP (left panel) but transfection with BiP siRNA does not affect either the total cellular levels of Pro-Cat (center panel) or furin (right-hand panel). B, expression of BiP siRNA markedly decreases processing (left-hand panel) and cell-surface levels (center panel) of Pro-Cat, but does not affect cell-surface level of furin (right panel). C, quantitation of cell-surface Pro-Cat and furin in BiP siRNA-treated cells is expressed as a percentage of control siRNA-treated cells. Data represent the mean and S.D. of three independent experiments.
Taken together with the preceding experiments using gp96 siRNA, these data indicate that both gp96 and BiP influence the net amount of the processed catalytic domain released in the medium, but do so via different effects, i.e. gp96 suppresses the cell-surface presentation of both furin and Pro-Cat, whereas BiP reduces Pro-Cat, but is without influence on furin.
DISCUSSION
Although ADAMTS9 has considerable biomedical significance, little is known about the mechanisms that influence its biosynthesis, cellular localization, and cell-surface processing. Through seeking cellular proteins that associated with pro-ADAMTS9 Pro-Cat, we sought to elucidate these mechanisms. Four well-known intracellular proteins that are associated with the secretory pathway were identified in a complex with ADAMTS9. UDP-glucose:glycoprotein glucosyltransferase I is a resident ER protein that binds to unfolded regions and adds a single glucose to the deglucosylated glycan (15). There are three N-glycosylation sites in the ADAMTS9 propeptide (3, 12), which might explain its complex formation with this ER resident protein. gp96 is the ER resident member of the HSP90 family and is constitutively expressed in all cell types (16). All gp96-interacting proteins are in the category of cell-surface ligands and receptors, or proteins localized in the secretory pathway (16). gp96 is transcriptionally co-regulated with BiP in many cell types and works cooperatively with it, although the association of gp96 with its substrates has been shown to be independent of BiP (17). BiP is an ER chaperone that binds to many newly synthesized proteins during the course of their maturation (18), with the help of Erdj3, which serves as a cofactor for BiP to interact with unfolded substrates (19). Thus, the association of these proteins with pro-ADAMTS9 suggests that they are likely involved in ensuring the proper folding of pro-ADAMTS9. Indeed, these are the first components of the secretory pathway that have been identified in association with an ADAMTS protease.
Although an association of BiP and gp96 with pro-ADAMTS9 at the cell surface was unexpected, their cell-surface localization has been previously reported (17, 20–23). Extracellular gp96 has been most extensively studied in regulation of immune function. Cell-surface expression of gp96 in tumor cells induced dendritic cell activation and anti-tumor immunity through increased cross-priming of CD8+ T cells (24, 25). Furthermore, significant dendritic cell activation and spontaneous lupus-like autoimmune disease was induced in a transgenic mouse expressing gp96 at the cell surface (23). A Listeria monocytogenes virulence factor utilized cell surface gp96 as a receptor for cell invasion (26).
Whereas BiP is generally thought to play a role in the structural maturation of nascent proteins in the ER, recent observations have shown that it may also have functions at the cell surface (23, 27, 28). BiP has been shown to associate with major histocompatibility complex class I protein (MHC class I) at the cell surface (29, 30). Furthermore, it acts as a co-receptor with MHC class I to facilitate the internalization of Coxsackievirus A9 by mammalian cells (29). It was also identified as a receptor for the dengue virus (31).
Because ADAMTS9 is part of a cell-surface complex that contains furin, we asked whether furin was also associated with gp96 and BiP. We demonstrated a complex containing furin, and these molecular chaperones at the cell surface in the presence as well as absence of ADAMTS9. Co-immunoprecipitation of furin with gp96 and BiP in the absence of ADAMTS9 Pro-Cat suggested that these chaperones were constitutively associated with and could potentially regulate cell-surface processing by furin. Indeed, the trafficking of furin and pro-ADAMTS9 to the cell surface was suppressed in the presence of geldanamycin and gp96 siRNA, with the net result of lowering ADAMTS9 catalytic domain released in the conditioned medium. BiP was involved in the trafficking and/or cell-surface localization of pro-ADAMTS9 but did not affect furin.
There is little existing literature on the role of molecular chaperones in the regulation of ADAMTS proteases or proprotein convertases. One such chaperone, 7B2, that is present in neuroendocrine cells, and thus specifically regulates PC2, has been previously identified (32–34), but this chaperone is not widely distributed and is thus unlikely to participate in furin regulation. The ubiquitous expression of BiP and gp96 is compatible with their observed association with furin, the most broadly expressed PC. It was previously shown that BiP was instrumental in preventing aggregation of lymphoma proprotein convertase (LPC/PC7) and that overexpression of BiP reduced intracellular LPC maturation (35). Because furin and other PCs were previously thought to act solely within the trans-Golgi, molecular chaperones may not have been previously felt to be relevant to their activity. The furin zymogen undergoes autocatalytic processing within the ER, but its propeptide dissociates to generate the active form within the trans-Golgi (36), where most of its known substrates are cleaved. However, furin is also located at the cell surface, and significant pools of furin exist on the surface of polarized intestinal and renal epithelial cells and endothelial cells of continuous, fenestrated, and discontinuous capillaries (37, 38). Specifically, furin is co-localized with integrin αV at the slit diaphragm of renal podocytes and around endothelial fenestrations (38). Furin can be shed from cells as a soluble protease (39, 40). The biological activities of cell surface and secreted furin are attracting increasing attention as a consequence of their role in regulating growth factors of the TGFβ superfamily such as Nodal (41) and TGFβ (42), proteases (such as ADAMTS7, ADAMTS4, ADAMTS9, and ADAMTS10) (3, 11, 43–45), the extracellular matrix glycoprotein profibrillin (46) and in facilitating pathogen invasion and toxicity (e.g. processing of anthrax protective antigen and diphtheria toxin) (39, 47). The observations regarding regulation of cell-surface furin may be relevant to the processing of these cell-surface substrates.
A gp96-related chaperone, HSP90α, was previously found to associate with the matrix metalloprotease (MMP) zymogen, proMMP-2, and to enable its proteolytic activation at the cell surface (48, 49). Pro-MMP2 is activated by cell-surface MT1-MMP (50), which is also associated with furin at the cell surface (38), and subsequently, the activated MMP2 mediates tumor invasion and angiogenesis (51). Pro-MMP2 activation was blocked by geldanamycin (49). In addition, antibodies to HSP90 have also been shown to block activation of pre-kallikrein on the endothelial cell surface (52). It has been suggested that in these situations, HSP90 may work less as a chaperone, and more as a scaffolding protein linking substrates to activating proteases (53). Our data suggest that the role of gp96 and BiP in folding, cell-surface transport and propeptide processing of ADAMTS9 is a constitutive and physiological one. However, in the context of known functions of ADAMTS9, pharmacologic agents, such as geldanamycin could be used to manipulate the cell-surface proteolysis of proteoglycans (3), and the role of ADAMTS9 in angiogenesis.3 The current observations support and further extend this paradigm and may be of broad significance to interference with activation of other ADAMTS proteases.
Acknowledgment
We thank Dr. Nabil Seidah for providing the furin expression plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants AR49930 and AR53890 (to S. A.).
Koo, B. H., Coe, D. M., Dixon, L. J., Somerville, R. P. T., Nelson, C. M., Wang, L. W., Young, M. E., Lindner, D. J., Apte, S. S. (2010) Am. J. Pathol., in press.
- ADAMTS9
- a disintegrin-like and metalloprotease domain with thrombospondin type 1 motifs 9
- Pro-Cat
- catalytic domain of pro-ADAMTS9
- DSP
- dithiobis[succinimidylpropionate]
- DTSSP
- 3,3′-dithiobis[sulfosuccinimidylpropionate]
- BS3
- bis(sulfosuccinimidyl)suberate
- siRNA
- short interfering RNA
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Porter S., Clark I. M., Kevorkian L., Edwards D. R. (2005) Biochem. J. 386, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apte S. S. (2009) J. Biol. Chem. 284, 31493–31497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somerville R. P., Longpre J. M., Jungers K. A., Engle J. M., Ross M., Evanko S., Wight T. N., Leduc R., Apte S. S. (2003) J. Biol. Chem. 278, 9503–9513 [DOI] [PubMed] [Google Scholar]
- 4.Silver D. L., Hou L., Somerville R., Young M. E., Apte S. S., Pavan W. J. (2008) PLoS Genet 4, e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCulloch D. R., Nelson C. M., Dixon L. J., Silver D. L., Wylie J. D., Lindner V., Sasaki T., Cooley M. A., W. S. A., Apte S. S. (2009) Dev. Cell 17, 697–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo P. H., Leung A. C., Kwok C. Y., Cheung W. S., Ko J. M., Yang L. C., Law S., Wang L. D., Li J., Stanbridge E. J., Srivastava G., Tang J. C., Tsao S. W., Lung M. L. (2007) Oncogene 26, 148–157 [DOI] [PubMed] [Google Scholar]
- 7.Lung H. L., Lo P. H., Xie D., Apte S. S., Cheung A. K., Cheng Y., Law E. W., Chua D., Zeng Y. X., Tsao S. W., Stanbridge E. J., Lung M. L. (2008) Int. J. Cancer 123, 401–408 [DOI] [PubMed] [Google Scholar]
- 8.Silver D. L., Hou L., Somerville R., Young M. E., Apte S. S., Pavan W. J. (2008) PLoS Genet. 4, e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demircan K., Hirohata S., Nishida K., Hatipoglu O. F., Oohashi T., Yonezawa T., Apte S. S., Ninomiya Y. (2005) Arthritis Rheum. 52, 1451–1460 [DOI] [PubMed] [Google Scholar]
- 10.Blelloch R., Kimble J. (1999) Nature 399, 586–590 [DOI] [PubMed] [Google Scholar]
- 11.Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., Apte S. S. (2006) J. Biol. Chem. 281, 12485–12494 [DOI] [PubMed] [Google Scholar]
- 12.Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., Apte S. S. (2007) J. Biol. Chem. 282, 16146–16154 [DOI] [PubMed] [Google Scholar]
- 13.DeBoer C., Meulman P. A., Wnuk R. J., Peterson D. H. (1970) J. Antibiot. 23, 442–447 [DOI] [PubMed] [Google Scholar]
- 14.Whitesell L., Mimnaugh E. G., De Costa B., Myers C. E., Neckers L. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8324–8328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trombetta S. E., Parodi A. J. (1992) J. Biol. Chem. 267, 9236–9240 [PubMed] [Google Scholar]
- 16.Yang Y., Li Z. (2005) Mol Cells 20, 173–182 [PubMed] [Google Scholar]
- 17.Melnick J., Dul J. L., Argon Y. (1994) Nature 370, 373–375 [DOI] [PubMed] [Google Scholar]
- 18.Kuznetsov G., Nigam S. K. (1998) N. Engl. J. Med. 339, 1688–1695 [DOI] [PubMed] [Google Scholar]
- 19.Shen Y., Hendershot L. M. (2005) Mol. Biol. Cell 16, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiest D. L., Bhandoola A., Punt J., Kreibich G., McKean D., Singer A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1884–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert J., Ménoret A., Cohen N. (1999) J. Immunol. 163, 4133–4139 [PubMed] [Google Scholar]
- 22.Shin B. K., Wang H., Yim A. M., Le Naour F., Brichory F., Jang J. H., Zhao R., Puravs E., Tra J., Michael C. W., Misek D. E., Hanash S. M. (2003) J. Biol. Chem. 278, 7607–7616 [DOI] [PubMed] [Google Scholar]
- 23.Liu B., Dai J., Zheng H., Stoilova D., Sun S., Li Z. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15824–15829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J., Liu B., Caudill M. M., Zheng H., Qiao Y., Podack E. R., Li Z. (2003) Cancer Immun. 3, 1. [PubMed] [Google Scholar]
- 25.Zheng H., Dai J., Stoilova D., Li Z. (2001) J. Immunol. 167, 6731–6735 [DOI] [PubMed] [Google Scholar]
- 26.Cabanes D., Sousa S., Cebriá A., Lecuit M., Garcia-del Portillo F., Cossart P. (2005) EMBO J. 24, 2827–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delpino A., Castelli M. (2002) Biosci. Rep. 22, 407–420 [DOI] [PubMed] [Google Scholar]
- 28.Jang J. H., Hanash S. (2003) Proteomics 3, 1947–1954 [DOI] [PubMed] [Google Scholar]
- 29.Triantafilou K., Fradelizi D., Wilson K., Triantafilou M. (2002) J. Virol. 76, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vabulas R. M., Braedel S., Hilf N., Singh-Jasuja H., Herter S., Ahmad-Nejad P., Kirschning C. J., Da Costa C., Rammensee H. G., Wagner H., Schild H. (2002) J. Biol. Chem. 277, 20847–20853 [DOI] [PubMed] [Google Scholar]
- 31.Jindadamrongwech S., Thepparit C., Smith D. R. (2004) Arch Virol 149, 915–927 [DOI] [PubMed] [Google Scholar]
- 32.Muller L., Zhu P., Juliano M. A., Juliano L., Lindberg I. (1999) J. Biol. Chem. 274, 21471–21477 [DOI] [PubMed] [Google Scholar]
- 33.Martens G. J., Braks J. A., Eib D. W., Zhou Y., Lindberg I. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5784–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braks J. A., Martens G. J. (1994) Cell 78, 263–273 [DOI] [PubMed] [Google Scholar]
- 35.Creemers J. W., van de Loo J. W., Plets E., Hendershot L. M., Van De Ven W. J. (2000) J. Biol. Chem. 275, 38842–38847 [DOI] [PubMed] [Google Scholar]
- 36.Leduc R., Molloy S. S., Thorne B. A., Thomas G. (1992) J. Biol. Chem. 267, 14304–14308 [PubMed] [Google Scholar]
- 37.Mayer G., Boileau G., Bendayan M. (2004) J. Histochem. Cytochem. 52, 567–579 [DOI] [PubMed] [Google Scholar]
- 38.Mayer G., Boileau G., Bendayan M. (2003) J. Cell Sci. 116, 1763–1773 [DOI] [PubMed] [Google Scholar]
- 39.Tsuneoka M., Nakayama K., Hatsuzawa K., Komada M., Kitamura N., Mekada E. (1993) J. Biol. Chem. 268, 26461–26465 [PubMed] [Google Scholar]
- 40.Thimon V., Belghazi M., Dacheux J. L., Gatti J. L. (2006) Reproduction 132, 899–908 [DOI] [PubMed] [Google Scholar]
- 41.Guzman-Ayala M., Ben-Haim N., Beck S., Constam D. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15656–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zacchigna L., Vecchione C., Notte A., Cordenonsi M., Dupont S., Maretto S., Cifelli G., Ferrari A., Maffei A., Fabbro C., Braghetta P., Marino G., Selvetella G., Aretini A., Colonnese C., Bettarini U., Russo G., Soligo S., Adorno M., Bonaldo P., Volpin D., Piccolo S., Lembo G., Bressan G. M. (2006) Cell 124, 929–942 [DOI] [PubMed] [Google Scholar]
- 43.Somerville R. P., Jungers K. A., Apte S. S. (2004) J. Biol. Chem. 279, 51208–51217 [DOI] [PubMed] [Google Scholar]
- 44.Somerville R. P., Longpré J. M., Apel E. D., Lewis R. M., Wang L. W., Sanes J. R., Leduc R., Apte S. S. (2004) J. Biol. Chem. 279, 35159–35175 [DOI] [PubMed] [Google Scholar]
- 45.Wang P., Tortorella M., England K., Malfait A. M., Thomas G., Arner E. C., Pei D. (2004) J. Biol. Chem. 279, 15434–15440 [DOI] [PubMed] [Google Scholar]
- 46.Wallis D. D., Putnam E. A., Cretoiu J. S., Carmical S. G., Cao S. N., Thomas G., Milewicz D. M. (2003) J. Cell. Biochem. 90, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klimpel K. R., Molloy S. S., Thomas G., Leppla S. H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10277–10281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eustace B. K., Jay D. G. (2004) Cell Cycle 3, 1098–1100 [PubMed] [Google Scholar]
- 49.Eustace B. K., Sakurai T., Stewart J. K., Yimlamai D., Unger C., Zehetmeier C., Lain B., Torella C., Henning S. W., Beste G., Scroggins B. T., Neckers L., Ilag L. L., Jay D. G. (2004) Nat. Cell Biol. 6, 507–514 [DOI] [PubMed] [Google Scholar]
- 50.Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 51.Nakamura H., Ueno H., Yamashita K., Shimada T., Yamamoto E., Noguchi M., Fujimoto N., Sato H., Seiki M., Okada Y. (1999) Cancer Res. 59, 467–473 [PubMed] [Google Scholar]
- 52.Joseph K., Tholanikunnel B. G., Kaplan A. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 896–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picard D. (2004) Nat. Cell Biol. 6, 479–480 [DOI] [PubMed] [Google Scholar]