Abstract
MAP2 is a neuron-specific microtubule-associated protein that binds and stabilizes dendritic microtubules. Previously, we showed that MAP2 expression is (a) activated in cutaneous primary melanoma and (b) inversely associated with melanoma tumor progression. We also showed that ectopic expression of MAP2 in metastatic melanoma cells inhibits cell growth by inducing mitotic spindle defects and apoptosis. However, molecular mechanisms of regulation of MAP2 gene expression in melanoma are not understood. Here, we show that in melanoma cells MAP2 expression is induced by the demethylating agent 5-aza-2′-cytidine, and MAP2 promoter is progressively methylated during melanoma progression, indicating that epigenetic mechanisms are involved in silencing of MAP2 in melanoma. In support of this, methylation of MAP2 promoter DNA in vitro inhibits its activity. Because MAP2 promoter activity levels in melanoma cell lines also correlated with activating mutation in BRAF, a gene that is highly expressed in neurons, we hypothesized that BRAF signaling is involved in MAP2 expression. We show that hyperactivation of BRAF-MEK signaling activates MAP2 expression in melanoma cells by two independent mechanisms, promoter demethylation or down-regulation of neuronal transcription repressor HES1. Our data suggest that BRAF oncogene levels can regulate melanoma neuronal differentiation and tumor progression.
Keywords: Cell/Differentiation, DNA/Methylation, DNA/Transcription, Gene/Regulation, Oncogene/Raf, Transcription, Transcription/Repressor
Introduction
MAP2 is a microtubule-associated protein that is expressed primarily in the mammalian neurons. MAP2 expression is a hallmark of postmitotic, terminally differentiated neurons, where it is preferentially localized to the dendrites. MAP2 stabilizes microtubules and is required for dendrite elongation, anchoring signaling molecules and organelle transport (1). Ectopic expression of MAP2 in heterologous cells induces rapid formation of stable microtubule bundles and dendritic extensions (2, 3). Because dynamic instability of microtubules is critical for mitotic spindle formation and segregation of chromosomes, ectopic expression of MAP2 in non-neuronal cells, especially in rapidly dividing cancer cells, can lead to disruption of dynamic instability of microtubules, mitotic spindle defects, cell cycle arrest, and apoptosis (4).
In previous studies, we showed that MAP2 expression is activated in cutaneous melanocytic lesions, specifically benign nevi and primary melanomas, but not metastatic melanomas (5). In a 5-year clinical follow-up, patients whose primary tumors expressed abundant MAP2 showed better disease-free survival compared with patients with tumors with weak or no MAP2 expression, suggesting that MAP2 expression is a good predictor of melanoma aggressiveness (6). In metastatic melanoma cells, MAP2 expression can be induced by treatment with a pharmacological agent (5). Ectopic expression of MAP2 by adenovirus-mediated gene transfer leads to mitotic spindle defects, cell cycle arrest, growth inhibition, and apoptosis (6). Thus, expression of MAP2, a neuronal marker, in melanoma appears to be not only inversely associated with tumor progression but also a potential therapeutic strategy similar to treatment with microtubule-stabilizing drugs, such as taxol (7–9).
Mechanisms of neural stem/progenitor cell differentiation have been extensively investigated. Epigenetic modification, including DNA methylation, and intracellular signaling mechanisms have been shown to be involved in neuronal differentiation and neural cell type-specific gene expression (10, 11). Although MAP2 expression is used as a hallmark of neuronal differentiation, the mechanism of MAP2 regulation is not well understood. We cloned and characterized the human MAP2 promoter. We identified several regulatory elements (NeuroD-binding E boxes and HES1 (Hairy and Enhancer of Split homolog-1)-binding N boxes) within the 3-kb region upstream of the MAP2 transcription start site. We also showed that HES1, a transcriptional repressor, is a critical regulator of MAP2 promoter activity in melanoma cells (12).
BRAF (v-Raf murine sarcoma viral oncogene homolog B1)-MEK3 -ERK signaling is known to play a role in neuronal differentiation. Although BRAF is expressed ubiquitously, the highest levels of BRAF mRNA are found in neuronal tissues (13–16). Because MAP2 is expressed in the majority of nevi (5) that also harbor a mutation in BRAF, we hypothesized that BRAF plays a role in MAP2 gene regulation in melanoma.
To understand the mechanisms involved in regulation of MAP2 gene expression, we studied the role of DNA methylation and BRAF signaling in activation of MAP2 in melanoma. Our results show that during melanoma tumor progression, the MAP2 promoter is progressively hypermethylated, and MAP2 gene expression can be activated by the DNA-demethylating agent 5-aza-2-deoxycytidine. Our data also show that overexpression of oncogenic BRAF activates MAP2 expression by two independent mechanisms, promoter demethylation or down-regulation of transcriptional repressor HES1.
EXPERIMENTAL PROCEDURES
Cell Culture
Melanoma cell lines WM115 and SK-MEL-2, -19, -28, and -31; human embryonal carcinoma cell line (NT2/D1); HeLa; and HEK293T were purchased from the American Type Culture Collection (Manassas, VA). WM35 and 451Lu melanoma cells were provided by Dr. M. Herlyn (Wistar Institute, Philadelphia, PA) and grown as described (5). Neonatal foreskin melanocytes were isolated and cultured as described (5).
Plasmids
BRAF expression plasmids pMCEFplink, pMCEFBRAFV600E, pEFBRAFV600E, wild type pEFBRAF, and pEFplink were from Dr. R. Marais (Institute of Cancer Research, London, UK), and mouse HES1 expression plasmid pCI-HES1 and HES1 antibody were gifts from Dr. R. Kageyama (Institute for Virus Research, Kyoto, Japan). Human MAP2 promoter-luciferase plasmids were constructed as described previously (12).
Antibodies
Anti-Raf-B, (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-p44/42 MAPK, anti-phospho-p44/42 MAPK (Thr202/Tyr204), anti-Notch1 (Cell Signaling Technology, Beverly, MA), anti-activated Notch1 (Abcam, Cambridge, MA), anti-MAP2, anti-neurofilament 70 kDa, anti-synaptophysin (Chemicon, Temecula, CA), anti-β-tubulin-III, anti-β-actin, and 4′,6-diamidino-2-phenylindole (Sigma) were used. Horseradish peroxidase-conjugated goat anti-mouse IgG and horseradish peroxidase-conjugated donkey anti-rabbit IgG were from GE Healthcare, and goat anti-mouse IgG Alexa 488 were from Molecular Probes (Carlsbad, CA).
Transfection
Transient transfection was performed using Lipofectamine Plus (Invitrogen) or the NHEM-Neo NucleofectorTM kit (Amaxa, Gaithersburg, MD). For stable clones, transfected 451Lu and SK-MEL-2 melanoma cells were selected and maintained in G418 (1 mg/ml). 451Lu stable clones 1 and 2 were established from two independent transfections that produced only a single clone each. SK-MEL-2 mBRAF stable cells represent a mixture of 15–20 separate clones.
Luciferase Promoter Assay
Cells cultured in 24-well tissue culture dishes, in triplicates, were transfected with either 650 ng of promoter reporter plasmid or control empty vector (pGL3). Normalization was done by cotransfection with the Renilla luciferase (pRL) plasmid. For BRAF co-transfection experiments, cells were transfected (Lipofectamine Plus) with 650 ng each of promoter reporter plasmid and pEFBRAFV600E or pEFBRAFwt. For HES1 co-transfection experiments, cells were transfected with 650 ng of promoter reporter plasmid, BRAF expression plasmid, and varying amounts of pCI-HES1 expression plasmid. Cells co-transfected with empty vector pGL3, pEFplink, and pcDNA served as controls, respectively. Forty-eight hours after transfection, cells were washed gently with 1× PBS and lysed in passive lysis buffer (Dual Luciferase Assay Kit, Promega). Firefly and Renilla luciferase activities were measured using a TD-20/20-luminometer (Turner Biosystems, Sunnyvale, CA). Firefly luciferase activity was normalized to Renilla luciferase activity, and the promoter activity was calculated as relative luciferase activity using enzyme activity in promoterless pGL3-transfected cells as 1.
Cell Proliferation Assays
Cell growth was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays using 1 × 104 cells plated in a 96-well plate. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide dye (5 mg/ml, Sigma) was added, and viable cell number (A540) was measured.
shRNA Plasmids and Lentiviruses
BRAF knockdown was performed using human pLKO.1 lentiviral shRNA plasmids from the RNAi Consortium collection (Open Biosystems, Inc., Huntsville, AL). Lentiviruses were produced by transfecting scrambled control or BRAF and HES1 shRNA with viral packing (pSVG) and envelope (pCMV ΔR8.2) plasmids into HEK293T cell lines. Supernatants were harvested and concentrated, and viral copy number was calculated using a quantitative RT-PCR titration kit (Clontech, Mountain View, CA). Cells were infected with an equal number of scrambled or BRAF viral particles.
Immunoblotting
Cells cultured under the indicated conditions were lysed in 1% Triton X-100 in phosphate-buffered saline or radioimmune precipitation buffer containing sodium vanadate (Sigma) and a mixture of protease inhibitors (Roche Applied Science). Protein was estimated using the BCA protein assay kit (Pierce), separated by SDS-PAGE, and transferred to a Polyscreen membrane (PerkinElmer Life Sciences). Membranes were blocked with 5% nonfat dry milk and incubated overnight at 4 °C with primary antibodies. The blots were incubated in appropriate horseradish peroxidase-conjugated secondary antibody, and proteins were detected by chemiluminescence (Amersham Biosciences). β-Actin was used as a control to monitor protein loading variability.
Immunofluorescence
Cells on coverslips were fixed in paraformaldehyde, permeabilized with cold methanol, incubated with antibody, and washed, and bound antibody was visualized using Alexa Fluor 488 (green)-conjugated secondary antibody. Nuclei were stained with propidium iodide or 4′,6-diamidino-2-phenylindole, and images were acquired using a Zeiss 510 confocal microscope.
RT-PCR and TaqMan Multiplex qPCR
Total RNA was isolated using the RNeasy minikit (Qiagen, CA), and cDNA was synthesized with reverse transcriptase SuperScript III (Invitrogen) according to the manufacturer's protocol. Total RNA was used to amplify MAP2 (200 ng) and HES1 and others (50 ng) using the following primers: MAP2, sense (5′-GCAGTTCTCAAAGGCTAGAC-3′) and antisense (5′-TGATCGTGGAACTCCATCT-3′); HES1, sense (5′-CACGACACCGGACAAACCAA-3′) and antisense (5′-TTCATGCACTCGCTGAAGC-3′); Olig2, sense (5′-AAGGAGGCAGTGGCTTCAAGTC-3′) and antisense (5′-CGCTCACCAGTCGCTTCATC-3′); Nkx2.2, sense (5′-TGCCTCTCCTTCTGAACCTTGG-3′) and antisense (5′-GCGAAATCTGCCACCAGTTG-3′); Pax6, sense (5′-GGCAACCTACGCAAGATGGC-3′) and antisense (5′-TGAGGGCTGTGTCTGTTCGG-3′); Nkx6.1, sense (5′-ACACGAGACCCACTTTTTCCG-3′) and antisense (5′- TGCTGGACTTGTGCTTCTTCAAC-3′); N-CAM, sense (5′-ATTGCGGTCAACCTGTGTGG-3′) and antisense (5′-TGGCACTCTGGCTTTGCTTC-3′); β-actin, sense (5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3′) and antisense (5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′). MAP2 expression was quantified by multiplex quantitative PCR using TaqMan® gene expression assays (Applied Biosystems, Foster City, CA). Briefly, cDNA was synthesized by two-step reverse transcriptase Superscript III kit (Invitrogen), and 50 ng of cDNA was used for multiplex qPCR with MAP2 TaqMan® minor groove binder probe with 6-carboxyfluorescein dye (Hs01103234-g1MAP2) and huGAPDH TaqMan® MGB with VIC dye (Applied Biosystems) using the StepOnePlus real-time PCR system (Applied Biosystems).
Bisulfite Modification of Genomic DNA and Sequencing
Genomic DNA was isolated using the Genelute mammalian genomic DNA isolation kit (Sigma). Human brain genomic DNA was purchased from the BioChain Institute (Hayward, CA). Bisulfite modification of genomic DNA was done as described (17, 18). Briefly, 1 μg of genomic DNA in a 50-μl volume was denatured by using mild heat at an alkaline pH by adding 3.5 μl of 3 m NaOH and incubating for 10 min at 37 °C. Immediately, 10 μl of 10 mm hydroquinone (Sigma) and 520 μl of 3 m sodium bisulfite (Sigma) were added and incubated for 12–16 h at 50 °C. Bisulfite-treated DNA was purified using the (Qiagen, Valencia, CA) and eluted in 50 μl of H2O. The conversion of uracil was completed by alkaline desulfonation, by adding 5 μl of 3 m NaOH, and incubated at 37 °C. The treated DNA was purified by using Qiaquick gel extraction kit and eluted in 30 μl of H2O. Sequencing primers for bisulfite-modified DNA were designed by using the online program MethPrimer (available at the University of California San Diego Web site). Bisulfite-modified DNA was amplified using sequencing primers for the MAP2 promoter: 3′-proximal CpG island primers, MAP2 P(F), 5′-GGAGTTGAGTGGTTTGTTATTATTTT-3′; MAP2 P(R), 5′-CATTATCCCCACAAACTAACTCTCTA-3′; 5′-distal CpG island primers, MAP2 D(F), 5′-TGTTTGAAAATTGTTTTTTTATTTAAG-3′; MAP2 D(R), 5′-TTAATTACCTCCATCCAAATATATCC-3′. PCR conditions were as follows: 10 min at 95 °C, 40 cycles of 40 s at 95 °C, 40 s at 55 °C, and 40 s at 72 °C, followed by 72 °C extension for 7 min. The amplified product was confirmed by electrophoresis on agarose gel. PCR products were sequenced directly with MAP2 sequencing primers or cloned into pCR4-TOPO vector using the TOPO TA cloning kit according to the manufacturer's manual (Invitrogen). Single clones were selected and cultured, and plasmid DNA was isolated using GeneElute plasmid miniprep kit (Sigma) and sequenced using M13 primers (Invitrogen) at the UW Biotech sequencing facility (University of Wisconsin). 5–10 clones were sequenced for each sample.
In Vitro Methylation of MAP2 Promoter
A 2.2-kb region of MAP2 promoter corresponding to −1854 to +368 was cloned into pGL3 luciferase plasmid (phMAP2-2) as described (12). phMAP2–2 promoter-luciferase plasmid (20 μg) was digested with PstI (New England Biolabs, Ipswich, MA) followed by ApaI (New England Biolabs). pGL3 vector and MAP2 insert were separated on a 0.8% agarose gel, bands were extracted separately using a gel extraction kit (Qiagen), and both vector (pGL3) and insert (MAP2 promoter) were eluted in 50 μl of H2O. In vitro methylation was performed as described (19). Briefly, insert DNA was separated into two equal halves, 24 μl (one half) was incubated overnight at 37 °C with methyl donor S-adenosylmethionine (AdoMet; Sigma) and M.SssI (30 unit), whereas the other half (24 μl) was incubated without SssI enzyme. pGL3 vector fragment was kept free of additional modification. DNA was purified by using a gel extraction kit and eluted in 30 μl of H2O. Completeness of the in vitro methylation reaction was confirmed by digesting with methylation-sensitive (HpaII, New England Biolabs) and methylation-insensitive (MspI, New England Biolabs) enzyme. Methylated and unmethylated inserts were ligated into linearized pGL3 vector separately. The ligation product was purified by a PCR purification kit (Qiagen), and purified plasmids were transfected using Lipofectamine Plus. Luciferase activity of methylated and unmethylated MAP2 promoter was normalized by cotransfection of Renilla luciferase (pRL) plasmid and represented as relative luciferase activity.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation assay was performed using the ChIP-IT Express kit (Active Motif, Carlsbad, CA). Formaldehyde-treated nuclear lysates were prepared and immunoprecipitated with anti-Hes1 antibody (Santa Cruz Biotechnology, Inc.) or control IgG. Immunoprecipitated DNA fragments were eluted and amplified using primers for the N2 box region in the MAP2 promoter as described earlier (12) and SYBR® Green in StepOnePlus PCR systems (Applied Biosytems). Differences in N2 box DNA specifically bound to HES1 were calculated by the 2−ΔΔCT method (20) using control IgG input DNA for normalization (21).
Statistical Analysis
All statistical analyses of data were performed using Prism 4.0 (GraphPad Software, La Jolla, CA).
RESULTS
MAP2 Expression Is Activated by Treatment with 5-Aza-2′-deoxycytidine (5-Aza)
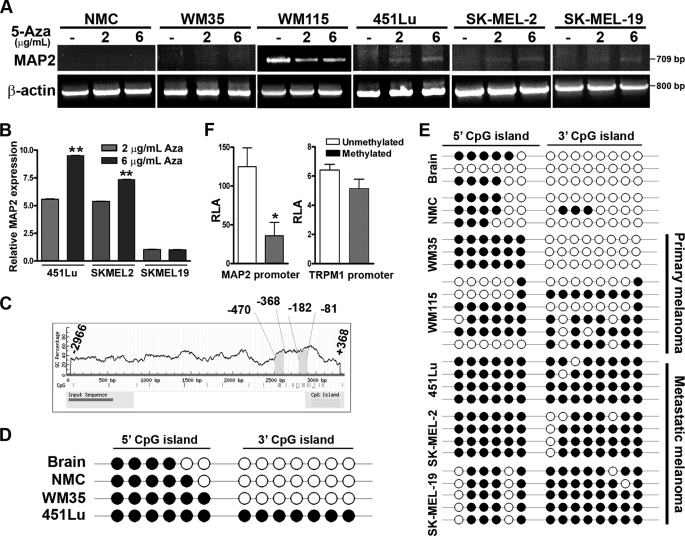
To investigate the role of DNA methylation in regulation of MAP2 gene expression, we treated cultured human melanocytes and melanoma cell lines (non-tumorigenic primary melanoma WM35, invasive primary melanoma WM115, and metastatic cell lines SK-MEL-2 and -19 and 451Lu) with DNA-demethylating agent 5-aza. In primary melanocytes and WM35 melanoma cells, MAP2 mRNA was not detectable, and treatment with 5-aza did not induce MAP2 expression. WM115, a cell line derived from invasive primary melanoma, showed constitutive expression of MAP2 mRNA that appeared to be slightly decreased by 5-aza. In metastatic melanoma cell lines, SK-MEL-2 and 451Lu, which show no detectable MAP2 expression, there was a dose-dependent induction of MAP2 mRNA by 5-aza (Fig. 1, A and B). Direct sequencing of the amplicons from 451Lu cells confirmed amplification of the expected 709-bp region of human MAP2 mRNA corresponding to exons 3–9 (12). Immunofluorescence staining of 451Lu cells with a MAP2-specific monoclonal antibody showed that MAP2 protein was also expressed after a 72-h treatment with 5-aza (supplemental Fig. S1). These data show that MAP2 expression can be activated in some metastatic melanoma cells by treatment with demthylating agent 5-aza.
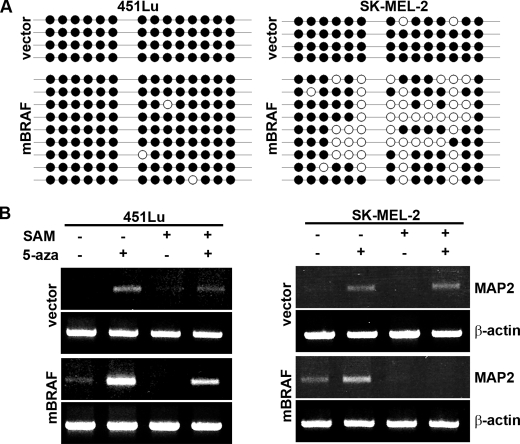
FIGURE 1.
Demethylation induces MAP2 expression in metastatic melanoma cells. A, MAP2 expression is induced by the DNA-demethylating agent 5-aza. Normal neonatal melanocytes (NMC) and a panel of melanoma cell lines were treated with or without 5-aza for 72 h and analyzed for MAP2 expression by RT-PCR using primers that amplify a 709-bp fragment that spans human MAP2 exons 3–9. Actin as a control for RNA loading is shown. B, MAP2 expression in melanoma cells treated with or without 5-aza was analyzed by multiplex qPCR using the TaqMan® expression assay. Data are represented as relative expression by calculating 2−ΔΔCT normalized to its untreated control. Representative data from three experiments are shown (mean ± S.E.; *, p < 0.001). C, computational prediction of CpG islands in the MAP2 promoter region using MethPrime software. Two CpG islands were identified in the 3.3-kb (upstream from transcription start site to +368 to −2966) MAP2 promoter region, a promoter-proximal 3′ (−81 to −182) CpG island, and a distal 5′ (−368 to −470) CpG island. Criteria used for prediction were island size >100 bp, GC percentage >30%, and observed/expected CpG ratio >0.6). CpG islands are indicated by a gray box, and the numbers indicate nucleotides in the promoter region upstream from the transcription start site. D, the MAP2 promoter is hypermethylated in metastatic melanoma. The methylation status of each CpG site was determined by direct bisulfite DNA sequencing of PCR products of MAP2 promoter CpG islands in human brain DNA, human melanocyte DNA, and melanoma cell line DNA. The black circles indicate methylated CpG sites, and white circles indicate unmethylated CpG sites. E, sequencing of the individual clones generated by cloning PCR products of bisulfite DNA from brain, melanocyte, and human primary and metastatic melanoma cell lines. F, in vitro methylation of MAP2 promoter inhibits its activity. MAP2 and TRPM1 promoters were methylated in vitro using MsssI enzyme, ligated to pGL3-luciferase reporter plasmid, and transfected to 451Lu cells, and luciferase activity of methylated and unmethylated promoter was measured. Representative data from three independent experiments are shown (mean ± S.E.; *, p < 0.001). RLA, relative luciferase activity.
Identification and Analysis of MAP2 Promoter CpG Islands
Using MethPrimer, a CpG island prediction program, we identified two CpG-rich regions, a proximal island (−81 to −182) and a distal island (−368 to −470), in the MAP2 promoter (12) (Fig. 1C). We investigated the methylation status of individual cytosine residues in these CpG islands by direct sequencing of PCR products amplified using bisulfite-modified genomic DNA templates. In human brain tissue, where MAP2 is expressed, all CpGs in the 3′ island in MAP2 promoter were unmethylated, whereas several CpGs in the 5′ island were methylated. Cultured neonatal foreskin melanocytes and a non-tumorigenic primary melanoma cell line, WM35, also showed a methylation profile similar to that of brain. In metastatic melanoma cell lines (SK-MEL-2 and -19 and 451Lu), both 5′ and 3′ CpG islands were hypermethylated (Fig. 1D). This methylation profile was confirmed by cloning the PCR products (obtained from independently bisulfite-modified genomic DNA templates) and sequencing multiple clones (Fig. 1E and supplemental Fig. S2). These data show that MAP2 promoter is highly methylated in metastatic melanoma cells, whereas it retains a brainlike methylation profile in melanocytes and early primary melanoma cells. WM115, a cell line from invasive vertical growth phase (VGP) melanoma showed an intermediate profile suggesting that the MAP2 promoter is progressively hypermethylated during melanoma progression. Although melanocytes and primary melanoma WM35 show a brainlike MAP2 promoter methylation profile, they do not express MAP2 either constitutively or after 5-aza treatment (Fig. 1A), suggesting that promoter methylation is not the primary mechanism of silencing of MAP2 gene in melanocytes.
In Vitro Methylation Inhibits MAP2 Promoter Activity
To test directly whether methylation inhibits MAP2 promoter activity, we methylated a 2.2-kb MAP2 promoter fragment (+368/−1115) in vitro using CpG methylase M.SssI. Methylation was confirmed by digestion with methylation-sensitive restriction enzymes HpaII and MspI (supplemental Fig. S3). We ligated mock-treated and methylated MAP2 promoter DNA fragments to luciferase reporter plasmid pGL3 and measured MAP2 promoter activity in transiently transfected 451Lu melanoma cells. Methylated MAP2 promoter showed >75% lower luciferase activity compared with the mock-treated promoter (Fig. 1F). Similar in vitro methylation of TRPM1 promoter (22) showed no effect on its activity. These data support a role for promoter methylation in regulation of MAP2 expression in melanoma cells.
MAP2 Promoter Activity in Melanoma Cell Lines
Although the above experiments showed that MAP2 expression can be induced in melanoma cells by promoter demethylation, our observation that MAP2 is not expressed in melanocytes and a radial growth phase melanoma WM35 cells in which MAP2 promoter is hypomethylated suggests that additional mechanisms are involved in MAP2 gene regulation.
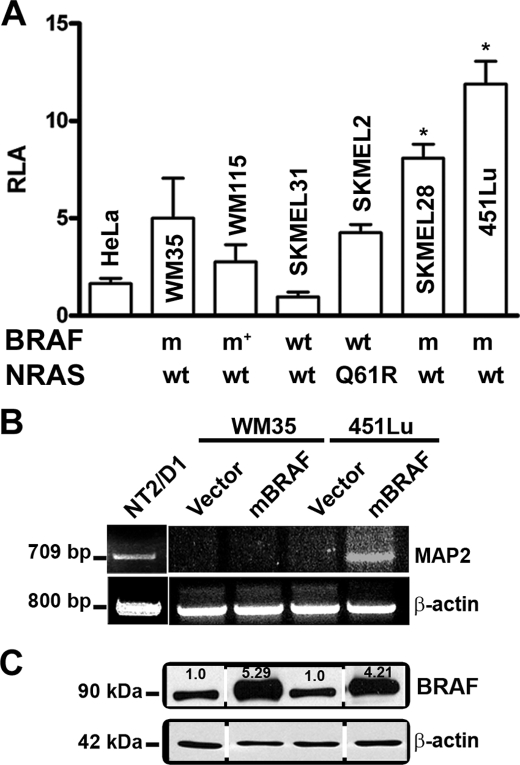
Since BRAF, which is frequently activated in melanomas, is also known to be highly expressed in neurons, we asked whether the ability of melanoma cells to support MAP2 promoter activity has a relationship to BRAF activity. We first compared the activity of the MAP2 promoter in a panel of melanoma cell lines with known RAS and BRAF mutations (23–26). All melanoma cell lines, with the exception of SK-MEL-31 (which harbors wild type RAS and BRAF genes), showed 2–6-fold greater MAP2 promoter activity than in non-melanoma HeLa cells (Fig. 2A). Melanoma cells lines (SK-MEL-28 and 451Lu) that harbor activating mutation BRAF (V600E) showed higher MAP2 promoter activity compared with melanoma cell lines that express wild-type BRAF and RAS (SK-MEL-31), V600D mutant BRAF (WM115), and mutant RAS (SK-MEL-2). Thus, MAP2 promoter activity appears to correlate with BRAFV600E mutation status.
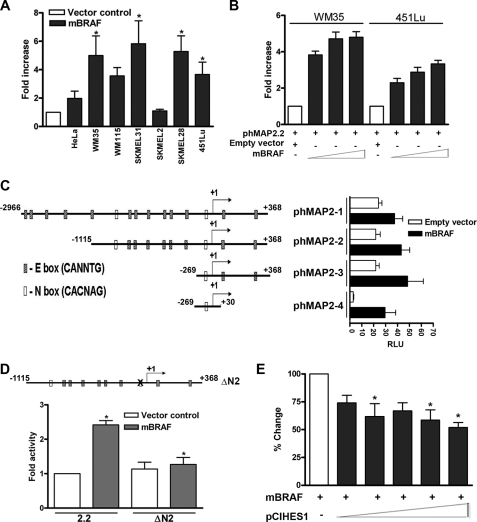
FIGURE 2.
Mutant BRAF up-regulates MAP2 promoter activity in melanoma cells. A, MAP2 promoter activity in melanoma cell lines compared with non- melanoma HeLa cells. Data shown are relative luciferase activity (RLA; mean ± S.E.; *, p < 0.001) from triplicate wells in three independent experiments. BRAF and RAS genotypes of melanoma cell lines are shown below the x axis. Q61R indicates a RAS mutation; m indicates a BRAFV600E mutation, m* indicates a BRAFV600D mutation; wt indicates the absence of NRAS or BRAF mutation in melanoma cell lines. B, mBRAF induces MAP2 mRNA in metastatic melanoma cells. RT-PCR was performed using total RNA from WM35 and 451Lu cells transfected with mBRAF. MAP2 expressions of NT2/D1 cells are shown as positive control. Actin as a control for RNA loading is shown. C, Western blot analysis of total BRAF protein expression in transiently transfected melanoma cells.
Overexpression of mBRAF Activates MAP2
To investigate whether hyperstimulation of BRAF could activate MAP2 expression in melanoma cells, we transiently transfected melanoma cell lines WM35 and 451Lu with a BRAFV600E expression plasmid. RT-PCR analysis showed MAP2 mRNA expression only in the metastatic cell line 451Lu and not in the primary cell line WM35 (Fig. 2B), although Western blot analysis showed increased BRAF protein in both cell lines (Fig. 2C). Thus, in WM35 cells, MAP2 expression could not be activated by 5-aza or by overexpression of mBRAF. These data suggest that MAP2 is silenced by highly efficient transcriptional repressor mechanisms.
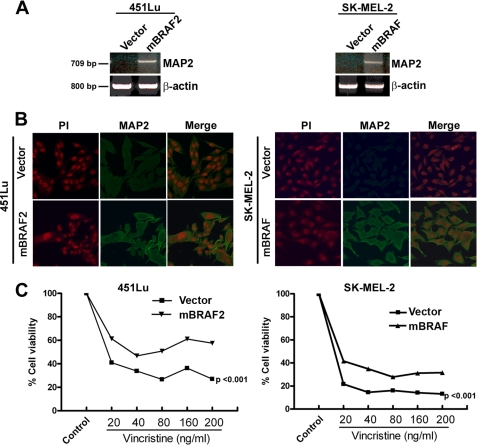
To further investigate the role of hyperactivation of BRAF in regulation of MAP2 gene expression, we transfected 451Lu (which harbors the BRAFV600E mutation) and SK-MEL-2 (wild-type BRAF) with mBRAFV600E expression plasmid and generated stable clones. These clones express up to 5-fold more BRAF protein compared with vector-transfected cells (data not shown). RT-PCR showed amplification of a PCR product of the expected size and sequence in high mBRAF stable clones but not in the vector control clones (Fig. 3A). Immunostaining with anti-MAP2 antibody showed MAP2 expression in mBRAF clones of both cell lines (Fig. 3B). Induction of MAP2 mRNA in transiently transfected 451Lu melanoma cells indicated that it is unlikely that the stable mBRAF clones represent clonally expanded rare G418-resistant MAP2-positive melanoma cells (Fig. 2B).
FIGURE 3.
High levels of oncogenic BRAF induce MAP2 expression. A, RT-PCR analysis of MAP2 mRNA expression in (BRAF mutant) 451Lu cells (left) and (RAS mutant) SK-MEL-2 cells stably expressing mBRAF or cells transfected with vector control. Actin served as control for RNA loading. B, immunostaining for MAP2 protein expression in 451Lu (left) and SK-MEL-2 (right). Cells on glass coverslips were stained using anti-MAP2 monoclonal antibody, followed by anti-mouse IgG-fluorescein isothiocyanate and propidium iodide (PI) and photographed using a Zeiss LSM 510 microscope. C, effects of stabilization of microtubules in MAP2-expressing melanoma cells. Vector control or 451Lu mBRAF2 and mBRAF-SK-MEL-2 cells were plated and treated with varying concentrations of vincristine, and after 3 days, cell growth was measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Data are represented as percentage of cell growth compared with untreated control (p < 0.001).
Since MAP2 is a microtubule-stabilizing protein, we predicted that MAP2 expression in melanoma cells could stabilize microtubules and as a consequence make these cells resistant or less sensitive to microtubule-destabilizing agents, such as vincristine. To test this, we treated vector control and mBRAF cells with a range of concentrations of vincristine (20–200 ng/ml) and estimated cell viability after 3 days. We found that over the entire concentration range of vincristine tested, mBRAF-expressing cells showed significantly higher (20–30%) cell viability compared with vector control cells (Fig. 3C). These data suggest that high BRAF levels can induce MAP2 expression, leading to stabilization of microtubules in melanoma cells.
Role of BRAF-MEK Signaling in MAP2 Expression
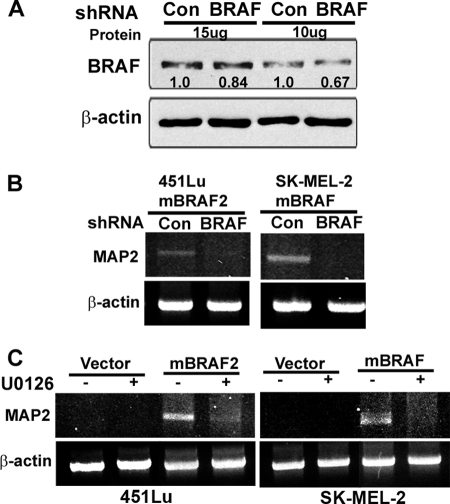
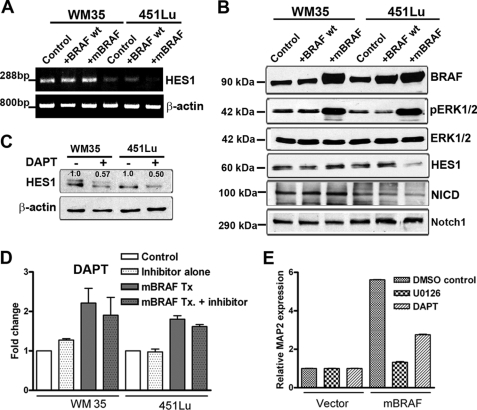
To understand whether activation of MAP2 expression in melanoma cells requires continued high mBRAF levels and increased MAPK signaling, we tested the effect of down-regulation of BRAF and inhibition of MEK kinase (a downstream effector of BRAF) on MAP2 expression in mBRAF-stable clones. Knockdown of BRAF using BRAF shRNA lentivirus (Fig. 4A) or treatment of cells with the MEK inhibitor U0126 resulted in silencing of MAP2 transcription in both 451Lu mBRAF- and SK-MEL-2 mBRAF-expressing cells (Fig. 4, B and C), suggesting that hyperactivation of BRAF-MEK signaling is required for maintaining MAP2 expression in melanoma cells.
FIGURE 4.
Down-regulation of BRAF or inhibition of MEK kinase inhibits MAP2 expression. A, knockdown of BRAF in 451Lu mBRAF-stable clones by BRAF shRNA lentivirus. Western blot analysis of total BRAF in cells infected with an equal multiplicity of infection of control and BRAF shRNA lentivirus. Actin was used as a control for protein loading. B, BRAF-induced MAP2 expression requires high levels of BRAF protein. RT-PCR analysis of 451Lu mBRAF and SK-MEL-2 mBRAF cells infected for 48 h with an equal multiplicity of infection of control or BRAF shRNA lentivirus. Actin served as control for RNA loading. C, MAPK signaling pathway is required for BRAF-induced MAP2 expression. RT-PCR analyses were performed in control cells and U0126 treated (10 μm for 24 h) vector control and 451Lu mBRAF and SK-MEL2 mBRAF cells. Actin served as loading control.
Effect of mBRAF Overexpression on MAP2 Promoter Methylation
Since we showed that MAP2 promoter is highly methylated in metastatic melanoma cells and demethylation by treatment with 5-aza can activate MAP2 expression, we asked whether high mBRAF expression activates MAP2 expression by MAP2 promoter demethylation. We cloned and sequenced MAP2 promoter region PCR products of bisulfite-modified genomic DNA of vector control and mBRAF cell lines. Methylation of CpG dinucleotides was determined by sequencing multiple clones. In SK-MEL-2 mBRAF cells, the majority of the CpGs in the promoter-proximal 3′ CpG island were demethylated compared with the vector control, whereas in 451Lu mBRAF cells, there was little or no change in the methylation profile of the MAP2 promoter compared with its vector control cells (Fig. 5A). Because mBRAF overexpression induces MAP2 expression in both SK-MEL-2 and 451Lu mBRAF cells, these data suggest promoter methylation-dependent and methylation-independent mechanisms of regulation of MAP2 expression in melanoma cells.
FIGURE 5.
Demethylation of MAP2 promoter by mBRAF overexpression. A, CpG methylation profiles of MAP2 promoter in bisulfite DNA of vector and mBRAF 451Lu and SK-MEL-2 stable clones. CpG methylation was determined by sequencing individual clones isolated by cloning MAP2 promoter PCR product of bisulfite DNA amplified. The black circles indicate methylated CpG sites, and white circles indicate unmethylated CpG sites. B, vector control and mBRAF 451Lu- and SK-MEL-2-stable clones were treated with or without AdoMet (SAM; 100 μg/ml) or 5-aza (6 μg/ml) and with both AdoMet and 5-aza. After 72 h, total RNA was isolated, and RT-PCR was performed for MAP2 transcript expression. Actin is used for equal loading of RNA.
To further understand the role of promoter demethylation in activation of MAP2 expression by mBRAF, we tested the effect of AdoMet, which when present in excess not only reverses/inhibits demethylation caused by 5-aza but also methylates all cytosine residues (27, 28). We treated 451Lu and SK-MEL-2 and vector control and mBRAF clones with 5-aza or AdoMet alone or AdoMet plus 5-aza. In vector control cell lines, 5-aza induced MAP2 expression. Treatment with AdoMet alone did not result in induction of MAP2 mRNA. As expected, induction of MAP2 by 5-aza was attenuated in the presence of AdoMet (Fig. 5B, top panels, lanes 2 and 4).
Although treatment with 5-aza increased MAP2 gene expression in both mBRAF cell lines (compare lanes 1 and 2 in the third panel of Fig. 5B), the effect of 5-aza was more pronounced in 451Lu mBRAF cells than in SK-MEL-2 mBRAF cells. Treatment with AdoMet alone inhibited MAP2 expression in both cell lines, although the MAP2 promoter in 451Lu mBRAF cells is already hypermethylated. These data suggest involvement of an indirect mechanism(s), such as methylation of promoter(s) for trans-acting genes, in silencing MAP2 by AdoMet in 451Lu mBRAF cells. This is in contrast to the direct effect of AdoMet in SK-MEL-2 mBRAF cells, where remethylation of the hypomethylated promoter silenced MAP2 expression (Fig. 5A).
Treatment of SK-MEL-2 mBRAF cells with the methylating agent AdoMet not only inhibited up-regulation of MAP2 expression by 5-aza but also completely silenced mBRAF-induced MAP2 expression (compare lanes 1, 2, and 4 in row 3, right panel). However, in 451Lu mBRAF cells, although treatment with AdoMet appeared to decrease 5-aza-induced MAP2 expression, it did not completely suppress mBRAF-induced constitutive MAP2 expression (compare lanes 1, 2, and 4 in row 3, left panel). These data are consistent with the methylation profiles of the MAP2 promoter in these two cell lines (Fig. 5A) and suggest that in melanoma, MAP2 can be activated by promoter demethylation-dependent (in SK-MEL-2) and methylation-independent (in 451Lu) mechanisms.
Mutant BRAF Stimulates MAP2 Promoter
Next, we tested the effect of mBRAF on MAP2 promoter activity. Co-transfection of BRAF increased MAP2 promoter activity in all melanoma cell lines, with the exception of SK-MEL-2 melanoma cells (that harbor a mutation in RAS) and non-melanoma HeLa cells (Fig. 6A). Co-transfection with increasing amounts of mBRAF together with the MAP2 promoter-luciferase plasmid resulted in an up to 6-fold increase in MAP2 promoter activity (Fig. 6B). We verified the specificity of MAP2 promoter up-regulation by mBRAF using catalase and human TYRP1 promoters as controls (supplemental Fig. S5A).
FIGURE 6.
Role of transcriptional repressor HES1 in up-regulation of MAP2 promoter activity by mBRAF. A, mBRAF increases MAP2 promoter activity in melanoma cells. Luciferase activity in cells co-transfected, in triplicate wells, with phMAP2.2 and vector control or mBRAF plasmid was measured, and representative data of three independent experiments are shown as the -fold increase (mean ± S.E.; *, p < 0.001) compared with the activity in empty vector-transfected cells. HeLa cells were used as a non-melanoma control. B, transfection with increasing amounts of mBRAF plasmid increases MAP2 promoter activity. Primary (WM35) and metastatic (451Lu) melanoma were co-transfected with phMAP2.2 promoter luciferase plasmid alone or with increasing amounts (25, 125, and 625 ng) of mBRAF plasmid in triplicate wells, luciferase activity was measured, and -fold increase in promoter activity (mean ± S.E.) in cells transfected with mBRAF over empty vector-transfected cells is shown. C, presence of HES1 binding box is sufficient for up-regulation of MAP2 promoter activity by mBRAF. Shown is a schematic diagram of various human genomic DNA fragments consisting of the MAP2 5′ regulatory region (phMAP2.1–2.4) showing E and N boxes and an N box mutant (ΔN2). Nucleotides are numbered using the transcription start site as +1. 451Lu melanoma cells were transfected with MAP2 promoter constructs and mBRAF expression plasmid, and luciferase activity was measured. Promoterless pGL3 and empty vector (pEFplink) were used as controls. Results are represented as relative luciferase units (RLU) (mean ± S.E.; *, p < 0.01). D, mutation of the HES1-binding N box abolishes BRAF-induced up-regulation of MAP2 promoter activity. The HES1-binding N2 box (GCCGCC) was mutated (mutated nucleotides are shown underlined) as described (12), and wild type and ΔN2 mutant promoter plasmids were co-transfected with mBRAF plasmid. Luciferase activity was measured, and results are shown as -fold change in activity as in A. *, p < 0.05. E, HES1 represses BRAF-induced MAP2 promoter up-regulation. 451Lu melanoma cells were co-transfected with MAP2.4 promoter plasmid, mBRAF expression plasmid, and increasing amounts (10, 20, 30, 40, and 50 ng) of mouse Hes1 expression plasmid pCI-Hes1. Forty-eight hours after transfection, luciferase activity was measured, and data (mean ± S.E.; *, p < 0.001) are represented as percentage change by normalizing with empty vector control.
BRAF-responsive Element(s) in MAP2 Promoter
Human MAP2 promoter contains several E (CANNTG) and two N (CACNAG) boxes that bind basic helix-loop-helix transcriptional factors (12). To identify the regulatory region(s) of MAP2 promoter that is responsive to BRAF signaling, we tested the effect of co-expression of mBRAF on a set of MAP2 promoter constructs described earlier (12). Luciferase activity driven by all MAP2 promoters tested was up-regulated by mBRAF (Fig. 6C). Activity of phMAP2.4 construct that contains only the N2 box and no other identifiable regulatory elements showed the largest (∼12-fold) increase in cells co-transfected with mBRAF. These data raised the possibility that the N2 box is necessary and sufficient for BRAF-induced up-regulation of the MAP2 promoter. To test this, we mutated the core sequence (CACNAG) within the N2 box and tested its activity with or without co-expressed mBRAF. As shown in Fig. 6D, mBRAF co-expression did not increase the activity of the N2 box mutant promoter, whereas the activity of the wild type promoter was up-regulated by mBRAF. This suggested that mBRAF activates MAP2 by attenuating N box-mediated transcription repression. However, since the wild type and the N box mutant promoters showed similar activity in the absence of overexpressed mBRAF, it is possible that pathways other than those that target the N2 box are also involved in MAP2 up-regulation in melanocytes.
To investigate the role of N box-binding neuronal repressor HES1 in mBRAF-induced MAP2 promoter activity, we asked whether overexpression of HES1 can inhibit mBRAF-mediated up-regulation of the MAP2 promoter. As shown in Fig. 6E, cotransfection with increasing amounts of HES1 expression plasmid pCI-Hes1 resulted in a significant (up to 50% with 50 ng of Hes1 plasmid) inhibition of mBRAF-induced MAP2 promoter activity. Further increase in the amount of pCIHes1 almost completely inhibited MAP2 promoter activity (supplemental Fig. S5B). These data show that the transcriptional repressor HES1 and its cognate N box play a role in regulation of MAP2 promoter activity by mBRAF.
mBRAF Down-regulates HES1 Expression
Since we previously showed that HES1 is a critical regulator of MAP2 promoter activity, we asked whether induction of MAP2 expression by mBRAF is due to down-regulation of HES1 expression. RT-PCR and Western blot analysis showed that transfection of mBRAF, but not wild-type BRAF, resulted in a decrease in HES1 mRNA and protein in 451Lu metastatic melanoma cells but not WM35 primary melanoma cells. Overexpression of wild-type BRAF did not increase ERK1/2 phosphorylation and also did not affect HES1 expression (Fig. 7, A and B). In WM35, overexpression of mBRAF appears to increase both HES1 mRNA and protein levels. Because the MAP2 promoter is unmethylated and MAP2 expression could not be activated in WM35 cells by either 5-aza or by mBRAF overexpression (Fig. 7, A and B), these data suggest that MAP2 gene in this cell line is either deleted or irreversibly silenced by mechanisms yet to be identified.
FIGURE 7.
BRAF activates the MAP2 promoter by attenuating Notch signaling and HES1 expression. A, mBRAF decreases HES1 mRNA in metastatic melanoma cells. RT-PCR analysis was performed using total RNA from WM35 and 451Lu cells transfected with mBRAF. Amplification of a 288-bp HES1 mRNA and actin control is shown. B, overexpression of mBRAF inhibits Notch cleavage and decreases HES1 protein expression in metastatic melanoma cells. Total cell lysates were analyzed by Western blots using the indicated antibodies. Actin levels indicate loading of equal protein. C, Western blot analysis confirmed inhibition of Notch downstream target HES1 by DAPT. The upper band in lanes 1 and 2 appears to be a nonspecific band. The numbers below the bands indicate relative protein levels based on densitometric quantitation and normalization to actin. D, effect of γ-secretase inhibitor DAPT has no effect on MAP2 promoter activity. Melanoma cell lines were transfected in triplicate wells with phMAP2.2 promoter-luciferase plasmid alone or co-transfected with mBRAF and treated with 10 μm DAPT. Forty-eight hours after transfection, cells were harvested, and luciferase activity was measured, and data are represented as -fold change in luciferase activity. Control, MAP2.2 promoter plasmid alone; Inhibitor alone, promoter with DAPT; mBRAF Tx, co-transfection of mBRAF with the promoter; mBRAF Tx + inhibitor, DAPT treatment after co-transfection with MAP2 promoter and mBRAF plasmids. E, γ-secretase inhibition does not enhance mBRAF-induced MAP2 expression. Vector and 451Lu mBRAF cells were treated with 10 μm DAPT or 10 μm U0126, and after 72 h, total RNA was isolated, and qRT-PCR was performed using MAP2 TaqMan® probes and normalized with glyceraldehyde-3-phosphate dehydrogenase VIC-labeled TaqMan® probes.
HES1 is a downstream target of Notch signaling. Therefore, we tested whether mBRAF down-regulates HES1 expression by down-regulating Notch signaling. Activation of Notch signaling can be monitored by the intracellular level of cleaved notch (Notch intracellular domain; NICD). Similar to its effects on HES1 expression in metastatic melanoma 451Lu cells, mBRAF overexpression resulted in a reduction of NICD protein levels, whereas in WM35 cells, the amount of NICD appeared to increase (Fig. 7B).
Notch signaling can be inhibited pharmacologically by γ-secretase inhibitor N-(N-3,5-difluorophenacetyl)-l-alanyl)-(S)-phenylglycine t-butyl ester (DAPT). DAPT inhibits cleavage of membrane-bound Notch to NICD. We tested whether inhibition of Notch signaling by treatment of melanoma cells with DAPT enhances the effect of mBRAF on MAP2 expression. Although DAPT treatment, resulted in a decrease of HES1 protein, as expected (Fig. 7C), similar treatment of cells transfected with MAP2 promoter alone or cells co-transfected with mBRAF did not cause a significant increase in MAP2 promoter-luciferase activity (Fig. 7D). TaqMan qPCR analysis of MAP2 expression showed that inhibition of the Notch-HES1 pathway by DAPT neither induced MAP2 expression in vector control cells nor enhanced MAP2 expression in 451Lu mBRAF cells. DAPT treatment, on the other hand, resulted in a modest decrease in MAP2 expression, presumably due to its off-target effects (Fig. 7E). These data show that although down-regulation of HES1 by mBRAF may be required, it is not sufficient to activate MAP2 expression. Thus, the neuronal marker MAP2 appears to be regulated in melanoma by both epigenetic mechanisms and modulation of transcription factor(s).
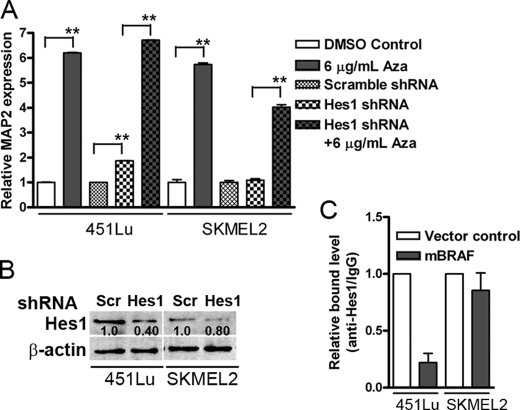
Next, to understand the relative contribution of epigenetic and HES1-dependent mechanisms, we studied MAP2 expression after 5-aza treatment or HES1 knockdown alone (by HES1 shRNA lentivirus) or HES1 knockdown plus 5-aza treatment using multiplex TaqMan qPCR. Treatment with 5-aza induced nearly 6-fold increase in MAP2 mRNA in both cell lines (Fig. 8A). HES1 knockdown (by ∼60%; Fig. 8B) induced nearly 2-fold increase in MAP2 mRNA expression in 451Lu cells, but not in SK-MEL-2 cells (consistent with the difference in the role of HES1 in MAP2 regulation between these two cell lines). HES1 knockdown together with 5-aza treatment did not result in significant further increase over 5-aza treatment alone in 451Lu cells. In SK-MEL-2 cells, although the combination of HES1 knockdown and 5-aza treatment also resulted in significant up-regulation of MAP2 mRNA compared with HES1 knockdown alone, this increase was not equal to that of 5-aza treatment (Fig. 8A). These data suggest that DNA demethylation and transcriptional repressor-mediated up-regulation of MAP2 expression operate independently with no evidence of positive cooperativity between these mechanisms.
FIGURE 8.
Knockdown of HES1 up-regulates MAP2 expression. A, 451Lu and SK-MEL-2 cells were transduced with scrambled (control) or HES1 shRNA lentivirus and treated with 5-aza for 72 h. MAP2 expression was assayed by RT-PCR using TaqMan® probes. Representative data from three experiments are shown as relative MAP2 expression. *, p < 0.001. B, Western blot analysis of cells transduced with control or HES1 shRNA virus in melanoma cells. C, chromatin immunoprecipitation analysis of MAP2 promoter occupancy by HES1. Formaldehyde-treated nuclear lysates from 451Lu and SK-MEL-2 vector and mBRAF-stable clones were immunoprecipitated with anti-HES1 antibody or control IgG, and antibody-bound DNA was amplified with N2 box primes and analyzed by real-time PCR.
To address whether mBRAF-induced activation of MAP2 expression is due to reduced occupancy of N2 box by HES1, we performed chromatin immunoprecipitation using anti-HES1 antibody and qPCR. As shown in Fig. 8C, in MAP2-positive 451Lu mBRAF cells, there was nearly 80% reduction in bound HES1. In contrast, in SK-MEL-2 mBRAF cells, there was no significant change in occupancy of N2 by HES1 compared with control cells. Since the methylation status of the MAP2 promoter in these two cells lines is different, these data suggest that methylation does not influence HES1 binding to N2. Taken together with the data on differences in HES1 regulation by mBRAF, these data support our conclusion that in melanoma cells MAP2 can be regulated by promoter demethylation and/or down-regulation of transcription repressor HES1.
DISCUSSION
The main findings of our study are as follows: (a) in melanoma cell expression, MAP2, a neuronal terminal differentiation marker and a prognostic marker for melanoma aggressiveness, is regulated by both epigenetic and transcriptional regulatory mechanisms, and (b) MAP2 expression can be activated by promoter demethylation and/or down-regulation of transcriptional repressor HES1, which appear to be regulated by activation levels of oncogenic BRAF. These observations point to a potential relationship between BRAF activation levels and neuronal differentiation, a prognostic feature of melanoma.
Neuronal Differentiation of Melanoma
Although it is widely accepted that neoplastic melanocytes exhibit plasticity, including a tendency to differentiate along neural cell pathways, the clinical significance of and mechanisms involved in melanoma neuronal differentiation are not completely understood (29, 30). For example, it is known that dermal nevus cells morphologically resemble Schwann cells of the peripheral nervous system and express Schwann cell-related antigens (29). Invasive primary melanoma cells, on the other hand, adopt a phenotype that includes the expression of certain proteins typical of neurons rather than Schwann cells (31–34). Consistent with this notion, we showed that MAP2 is expressed more frequently in invasive primary melanomas than in early non-invasive primary melanomas both in vivo (5) and in vitro (WM35 versus WM115 shown in this study). Melanoma cell lines, but not melanocytes in culture, show variable expression of neuronal differentiation marker proteins (e.g. class III β-tubulin) and transcription factors (e.g. Olig2 and Nkx6.1) (35–39), respectively (supplemental Fig. S4).
In previous published studies, we showed that MAP2, a marker of neuronal terminal differentiation, is expressed in benign melanocytic nevi and primary melanomas, but not metastatic lesions, and that MAP2 expression has clinical significance. Whereas MAP2 expression in primary lesions serves as a prognostic indicator of aggressiveness, forced expression of MAP2 in metastatic melanoma cells in vitro induces mitotic spindle defects and apoptosis and inhibits cell growth. Therefore, a detailed understanding of the regulation of MAP2 expression may help in designing novel treatments for melanoma. In this study, we show that MAP2 expression can be activated in melanoma cells by either treatment with 5-aza or hyperactivation of BRAF signaling, which induces MAP2 expression in different cell lines by promoter demethylation or down-regulation of transcriptional repressor HES1.
MAP2 Promoter Methylation
The role of epigenetic mechanisms in differentiation of neural stem/progenitor cells during development and in the adult brain has been extensively investigated (10, 11). Although MAP2 expression has been used as a marker of neuronal differentiation in wide ranging studies, including those investigating the effect of 5-aza treatment on neuronal differentiation (40), methylation status of the MAP2 promoter has not been investigated. We identified two CpG islands within 500 bp of the MAP2 3-kb promoter. In adult human brain, where MAP2 is expressed, the 3′ CpG cluster proximal to the transcription start site is mostly unmethylated compared with the distal cluster. Here, we show that cultured skin melanocytes, which do not normally express MAP2, also have a similar methylation pattern, suggesting that silencing of transcriptional machinery, but not promoter methylation, is a primary mechanism of silencing MAP2 expression in non-neural cells. Recently, in their studies on MAP2 gene expression in mantle cell lymphoma, Vater et al. (41) also showed that cytosine residues between −16 and −368 nucleotides upstream of MAP2 are predominantly non-methylated in human peripheral blood lymphocytes and tonsil tissue. Progressive methylation of the MAP2 promoter in transformed melanocytes, which express various neuronal transcription factors, seems to contribute to maintain transcriptional repression of MAP2 in melanoma. Activation of MAP2 expression in most melanoma cells, but not melanocytes, upon demethylation by 5-aza is consistent with this. Interestingly, a non-tumorigenic, radial growth phase early primary melanoma cell line WM35, in which 5-aza failed to induce MAP2 expression, also showed a CpG methylation pattern similar to melanocytes. These data suggest that whereas melanoma cells acquire plasticity, the ability to express genes such as MAP2 that could inhibit tumor progression may be reversed/silenced by epigenetic modifications.
BRAF and Melanoma Differentiation
Activating mutations in BRAF are frequently found in cutaneous melanoma (23, 42). Because benign melanocytic nevi also harbor BRAF mutations, BRAF is believed to elicit biphasic response. Activation of BRAF in melanocytes is thought to initiate a proliferative response, following which the cells withdraw from the cell cycle and become senescent (43). These benign nevus cells also exhibit characteristics of differentiation along neural pathways (29, 30). Therefore, we considered the possibility that activated BRAF might be involved in neuronal differentiation of malignant melanocytes, which express neuronal transcription factors (such as Olig2 and Nkx6.1 involved in commitment and differentiation of neural precursors). A role for BRAF in neuronal differentiation is supported by the observation that although BRAF is expressed in a wide range of tissues, the highest levels of its expression are found in neuronal tissues (15). Accordingly, B-raf−/− null mouse embryos die in utero due to growth retardation and vascular and neuronal defects (44). The BRAF gene is essential for survival of embryonic neurons in culture (16). It is also known that NGF-induced differentiation of PC12 cells is mediated by BRAF, and in neuronal cells NGF preferentially activates BRAF (45). Moreover, it is the regulated RAF (mainly B-RAF)-driven MAPK activity that seems to be involved in neuronal survival and differentiation (46, 47). It has been shown that whereas transient ERK activity leads to proliferation of neuronal precursors, sustained activity leads to their differentiation (48). Our data showing that overexpression of mBRAF and enhanced MAPK activity induce MAP2 expression in melanoma cells are consistent with this notion. A role for BRAF for activation of neuronal differentiation in melanoma cells is also supported by our observations that (a) MAP2 promoter activity levels correlate with BRAF mutation status (i.e. highest basal promoter activity in mutant BRAF melanoma cells compared with wild-type BRAF-expressing melanoma cells), (b) MAP2 promoter is activated by mBRAF preferentially in melanoma cells, and (c) high mBRAF induces MAP2 expression.
BRAF and Promoter Methylation
The CpG island methylator phenotype that is characterized by widespread methylation of gene promoters is known to be associated with BRAF mutation, specifically in colorectal cancers (49). Although epigenetic changes in melanoma have been extensively documented, association of this phenotype with BRAF mutation has not been investigated (50, 51). Our data on MAP2 promoter methylation in melanoma cells lines did not reveal any association between MAP2 promoter methylation and mutation in BRAF. On the other hand, hyperactivation of BRAF in SK-MEL-2 melanoma cells that carry wild type BRAF, but not in 451Lu melanoma cells that harbor mutant BRAF, resulted in demethylation of the MAP2 promoter, although MAP2 expression was activated in both cell lines. The exact role and mechanisms of MAP2 promoter demethylation by BRAF remain to be investigated.
Role of Transcriptional Repressor HES1
Using promoter deletion constructs, we identified the N box within the MAP2 proximal promoter as the sequence involved in mBRAF-induced up-regulation of the promoter activity. This suggests a role for the N box-binding transcriptional repressor HES1, a downstream target of Notch signaling, in regulation of MAP2 transcription by mBRAF. However, the observation that phMAP2.2 and phMAP2ΔN2 exhibit similar activity in the absence of co-expressed mBRAF (Fig. 6D) shows that although Notch-HES1/BRAF-MAPK signaling is necessary, it is not sufficient, for MAP2 up-regulation in melanocytes.
The Notch family consists of highly conserved receptors, which are activated by their ligands expressed on neighboring cells. Activation of Notch receptor results in its cleavage by γ-secretase, producing an NICD, which then translocates into the nucleus to initiate transcription of target genes, including HES transcriptional repressors (52, 53).
A functional role for Notch signaling and HES1 transcription factor in maintenance and survival of melanoblasts and melanocytes has been described (54). Dysregulated Notch signaling has been implicated in a wide variety of cancers, including melanoma (55, 56). Balint et al. (57) showed a stage-specific role for Notch1 signaling in promoting progression of primary melanoma. Thus, Notch signaling and HES1 appear to be involved in multiple aspects of melanocyte and melanoma biology, including neuronal differentiation. Our data suggest that mBRAF attenuates Notch signaling, as indicated by the HES1 expression, in metastatic but not primary melanoma cells. In metastatic melanoma cells, overexpression of mBRAF decreased the level of NICD (presumably by inhibiting cleavage of Notch receptor) and HES1 expression. Although HES1 co-transfection inhibited mBRAF-induced MAP2 promoter activity in both primary (data not shown) and metastatic melanoma cells, overexpression of mBRAF decreases NICD and HES1 levels only in metastatic cells. These data suggest a stage-specific role for Notch signaling as proposed earlier (57).
Role of Notch-HES1 in MAP2 Expression
Notch signaling maintains multipotency in some neural stem cells but promotes glial differentiation in others or at other times during development. Notch probably functions as a switch that terminates neurogenesis and causes glial lineage determination, in part by inhibiting expression of proneural bHLH factors and/or promoting expression of inhibitory bHLH factors, primarily HES and HEY family repressors (53, 58). Previously, we showed that although both proneural NeuroD1 and repressor HES1 are expressed in all melanoma cells, MAP2 promoter activity is regulated predominantly by the relative levels of transcriptional repressor HES1 (12). In this study, we show that activation levels of BRAF, a RAF kinase that is enriched in neuronal cells, play a role in regulation of expression of this important neuronal transcription repressor. The ability of mBRAF to induce endogenous MAP2 expression correlates with constitutive levels of HES1 and its down-regulation by mBRAF (Fig. 7, A and B). This potential relationship between mBRAF and neuronal gene expression in melanoma cells could explain the frequent display of neural phenotype by melanocytic lesions and neurotropism of melanoma (59, 60).
Relationship between BRAF and Notch Signaling
Our studies on regulation of MAP2 gene expression and its promoter activity by mBRAF and its relation to expression of transcription repressor HES1 uncovered potential interactions between BRAF-MAPK signaling and Notch-HES1 pathway in regulation of melanoma neuronal differentiation (Fig. 6). First, our data show that BRAF regulates MAP2 gene expression by down-regulation of Notch target gene HES1, only partly by down-regulation of NICD, because pharmacological inhibition of Notch cleavage does not enhance either basal activity or mBRAF-induced MAP2 promoter activity. Therefore, down-regulation of Notch signaling alone does not appear to be sufficient for up-regulation of the MAP2 promoter by mBRAF. This is consistent with the possibility that although HES1 is a Notch target gene, it is regulated by other signaling pathways (61, 62). Because BRAF activates the downstream MEK-ERK pathway, it is reasonable to argue that sustained MAPK activation might be involved in down-regulation of HES1. Although ERK activity has been shown to be involved in expression of HES1 (63), it is also known that, whereas transient ERK activity leads to proliferation in neural precursors, sustained activity of this kinase leads to their differentiation (48). In melanoma cells, mBRAF overexpression and increased ERK activity could also contribute to neuronal differentiation by down-regulation of HES1 expression. However, our data do not rule out the possibility that oncogenic BRAF might also regulate HES1 expression by Notch- and MAPK-independent mechanisms.
In summary, our data on regulation of MAP2 expression show that (a) MAP2 promoter is progressively methylated during melanoma progression and (b) MAP2 expression can be activated by both promoter demethylation and down-regulation of transcriptional repressor HES1. Although changes in DNA methylation and Notch signaling pathway have been implicated in melanoma tumor progression (57, 64), our in vitro molecular studies do not permit us to assign the specific physiologic contribution of each of these mechanisms of MAP2 regulation during melanoma progression. In view of our earlier studies on the potential role of MAP2 in melanoma tumor progression and the ability of MAP2 to inhibit growth of metastatic melanoma cells, our data provide the molecular basis for development of therapeutic strategies targeted toward activation of MAP2 expression in melanoma.
Supplementary Material
Acknowledgments
We thank Dr. Richard Marais for providing pEFBRAF constructs and Dr. Kageyama for providing pCI-HES1 construct and anti-HES1 antibody. We also thank Dr. Dali Yang for immunohistochemical staining and Smita Ramprakash and Ashley Zander for technical help.
This work was supported, in whole or in part, by National Institutes of Health Grants R21CA125091 (to V. S.) and R01NS045926 (to S.-C. Z.). This work was also supported by the Wisconsin Alumni Research Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- shRNA
- short hairpin RNA
- RT
- reverse transcription
- qPCR
- quantitative PCR
- AdoMet
- S-adenosylmethionine
- 5-aza
- 5-aza-2′-cytidine
- NICD
- Notch intracellular domain
- DAPT
- N-(N-3,5-difluorophenacetyl)-l-alanyl)-(S)-phenylglycine t-butyl ester.
REFERENCES
- 1.Harada A., Teng J., Takei Y., Oguchi K., Hirokawa N. (2002) J. Cell Biol. 158, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalcheva N., Rockwood J. M., Kress Y., Steiner A., Shafit-Zagardo B. (1998) Cell Motil. Cytoskeleton 40, 272–285 [DOI] [PubMed] [Google Scholar]
- 3.Nguyen H. L., Chari S., Gruber D., Lue C. M., Chapin S. J., Bulinski J. C. (1997) J. Cell Sci. 110, 281–294 [DOI] [PubMed] [Google Scholar]
- 4.Wittmann T., Hyman A., Desai A. (2001) Nat. Cell Biol. 3, E28–E34 [DOI] [PubMed] [Google Scholar]
- 5.Fang D., Hallman J., Sangha N., Kute T. E., Hammarback J. A., White W. L., Setaluri V. (2001) Am. J. Pathol. 158, 2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soltani M. H., Pichardo R., Song Z., Sangha N., Camacho F., Satyamoorthy K., Sangueza O. P., Setaluri V. (2005) Am. J. Pathol. 166, 1841–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Checchi P. M., Nettles J. H., Zhou J., Snyder J. P., Joshi H. C. (2003) Trends Pharmacol. Sci. 24, 361–365 [DOI] [PubMed] [Google Scholar]
- 8.Wassmann K., Benezra R. (2001) Curr. Opin. Genet. Dev. 11, 83–90 [DOI] [PubMed] [Google Scholar]
- 9.Bhat K. M., Setaluri V. (2007) Clin. Cancer Res. 13, 2849–2854 [DOI] [PubMed] [Google Scholar]
- 10.Kohyama J., Kojima T., Takatsuka E., Yamashita T., Namiki J., Hsieh J., Gage F. H., Namihira M., Okano H., Sawamoto K., Nakashima K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18012–18017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh J., Gage F. H. (2004) Curr. Opin. Genet. Dev. 14, 461–469 [DOI] [PubMed] [Google Scholar]
- 12.Bhat K. M., Maddodi N., Shashikant C., Setaluri V. (2006) Nucleic Acids Res. 34, 3819–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnier J. V., Papin C., Eychène A., Lecoq O., Calothy G. (1995) J. Biol. Chem. 270, 23381–23389 [DOI] [PubMed] [Google Scholar]
- 14.Storm S. M., Cleveland J. L., Rapp U. R. (1990) Oncogene 5, 345–351 [PubMed] [Google Scholar]
- 15.Wellbrock C., Karasarides M., Marais R. (2004) Nature Reviews 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 16.Wiese S., Pei G., Karch C., Troppmair J., Holtmann B., Rapp U. R., Sendtner M. (2001) Nat. Neurosci. 4, 137–142 [DOI] [PubMed] [Google Scholar]
- 17.Dai Z., Popkie A. P., Zhu W. G., Timmers C. D., Raval A., Tannehill-Gregg S., Morrison C. D., Auer H., Kratzke R. A., Niehans G., Amatschek S., Sommergruber W., Leone G. W., Rosol T., Otterson G. A., Plass C. (2004) Oncogene 23, 3521–3529 [DOI] [PubMed] [Google Scholar]
- 18.Herman J. G., Graff J. R., Myöhänen S., Nelkin B. D., Baylin S. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith L. T., Lin M., Brena R. M., Lang J. C., Schuller D. E., Otterson G. A., Morrison C. D., Smiraglia D. J., Plass C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmittgen T. D., Livak K. J. (2008) Nature Protocols 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 21.Jang H., Choi D. E., Kim H., Cho E. J., Youn H. D. (2007) J. Biol. Chem. 282, 11172–11179 [DOI] [PubMed] [Google Scholar]
- 22.Zhiqi S., Soltani M. H., Bhat K. M., Sangha N., Fang D., Hunter J. J., Setaluri V. (2004) Melanoma Res. 14, 509–516 [DOI] [PubMed] [Google Scholar]
- 23.Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. (2002) Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 24.Tanami H., Imoto I., Hirasawa A., Yuki Y., Sonoda I., Inoue J., Yasui K., Misawa-Furihata A., Kawakami Y., Inazawa J. (2004) Oncogene 23, 8796–8804 [DOI] [PubMed] [Google Scholar]
- 25.Hinselwood D. C., Abrahamsen T. W., Ekstrøm P. O. (2005) Electrophoresis 26, 2553–2561 [DOI] [PubMed] [Google Scholar]
- 26.Satyamoorthy K., Li G., Gerrero M. R., Brose M. S., Volpe P., Weber B. L., Van Belle P., Elder D. E., Herlyn M. (2003) Cancer Res. 63, 756–759 [PubMed] [Google Scholar]
- 27.Nelson E. D., Kavalali E. T., Monteggia L. M. (2008) J. Neurosci. 28, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabbara S., Bhagwat A. S. (1995) Biochem. J. 307, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed J. A., Finnerty B., Albino A. P. (1999) Am. J. Pathol. 155, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goovaerts G., Buyssens N. (1988) Am. J. Dermatopathol. 10, 20–27 [DOI] [PubMed] [Google Scholar]
- 31.Prieto V. G., McNutt N. S., Lugo J., Reed J. A. (1997) J. Cutan. Pathol. 24, 145–150 [DOI] [PubMed] [Google Scholar]
- 32.Lammerding-Köppel M., Noda S., Blum A., Schaumburg-Lever G., Rassner G., Drews U. (1997) J. Cutan. Pathol. 24, 137–144 [DOI] [PubMed] [Google Scholar]
- 33.Khare V. K., Albino A. P., Reed J. A. (1998) J. Cutan. Pathol. 25, 2–10 [DOI] [PubMed] [Google Scholar]
- 34.Dhillon A. P., Rode J., Leathem A. (1982) Histopathology 6, 81–92 [DOI] [PubMed] [Google Scholar]
- 35.Smith T. W., Nikulasson S., De Girolami U., De Gennaro L. J. (1993) Clin. Neuropathol. 12, 335–342 [PubMed] [Google Scholar]
- 36.Qiu M., Shimamura K., Sussel L., Chen S., Rubenstein J. L. R. (1998) Mech. Dev. 72, 77–88 [DOI] [PubMed] [Google Scholar]
- 37.Lee H. J., Elliot G. J., Hammond D. N., Lee V. M., Wainer B. H. (1991) Brain Res. 558, 197–208 [DOI] [PubMed] [Google Scholar]
- 38.Laggner U., Pipp I., Budka H., Hainfellner J. A., Preusser M. (2007) Histopathology 50, 949–952 [DOI] [PubMed] [Google Scholar]
- 39.Simpson T. I., Price D. J. (2002) BioEssays 24, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 40.Schinstine M., Iacovitti L. (1997) Exp. Neurol. 144, 315–325 [DOI] [PubMed] [Google Scholar]
- 41.Vater I., Wagner F., Kreuz M., Berger H., Martín-Subero J. I., Pott C., Martinez-Climent J. A., Klapper W., Krause K., Dyer M. J., Gesk S., Harder L., Zamo A., Dreyling M., Hasenclever D., Arnold N., Siebert R. (2009) Br. J. Haematol. 144, 317–331 [DOI] [PubMed] [Google Scholar]
- 42.Wellbrock C., Ogilvie L., Hedley D., Karasarides M., Martin J., Niculescu-Duvaz D., Springer C. J., Marais R. (2004) Cancer Res. 64, 2338–2342 [DOI] [PubMed] [Google Scholar]
- 43.Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., Peeper D. S. (2005) Nature 436, 720–724 [DOI] [PubMed] [Google Scholar]
- 44.Wojnowski L., Zimmer A. M., Beck T. W., Hahn H., Bernal R., Rapp U. R., Zimmer A. (1997) Nat. Genet. 16, 293–297 [DOI] [PubMed] [Google Scholar]
- 45.Kao S., Jaiswal R. K., Kolch W., Landreth G. E. (2001) J. Biol. Chem. 276, 18169–18177 [DOI] [PubMed] [Google Scholar]
- 46.Dugan L. L., Kim J. S., Zhang Y., Bart R. D., Sun Y., Holtzman D. M., Gutmann D. H. (1999) J. Biol. Chem. 274, 25842–25848 [DOI] [PubMed] [Google Scholar]
- 47.Frebel K., Wiese S., Funk N., Pühringer D., Sendtner M. (2007) Neurodegener. Dis. 4, 261–269 [DOI] [PubMed] [Google Scholar]
- 48.Marshall C. J. (1995) Cell 80, 179–185 [DOI] [PubMed] [Google Scholar]
- 49.Minoo P., Moyer M. P., Jass J. R. (2007) J. Pathol. 212, 124–133 [DOI] [PubMed] [Google Scholar]
- 50.Hoon D. S., Spugnardi M., Kuo C., Huang S. K., Morton D. L., Taback B. (2004) Oncogene 23, 4014–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halaban R., Krauthammer M., Pelizzola M., Cheng E., Kovacs D., Sznol M., Ariyan S., Narayan D., Bacchiocchi A., Molinaro A., Kluger Y., Deng M., Tran N., Zhang W., Picardo M., Enghild J. J. (2009) PLoS ONE 4, e4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinmaster G. (2000) Curr. Opin. Genet. Dev. 10, 363–369 [DOI] [PubMed] [Google Scholar]
- 53.Kageyama R., Ohtsuka T., Tomita K. (2000) Mol. Cells 29, 1–7 [DOI] [PubMed] [Google Scholar]
- 54.Moriyama M., Osawa M., Mak S. S., Ohtsuka T., Yamamoto N., Han H., Delmas V., Kageyama R., Beermann F., Larue L., Nishikawa S. (2006) J. Cell Biol. 173, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickoloff B. J., Hendrix M. J., Pollock P. M., Trent J. M., Miele L., Qin J. Z. (2005) J. Invest. Dermatol. Symp. Proc. 10, 95–104 [DOI] [PubMed] [Google Scholar]
- 56.Miele L. (2006) Clin. Cancer Res. 12, 1074–1079 [DOI] [PubMed] [Google Scholar]
- 57.Balint K., Xiao M., Pinnix C. C., Soma A., Veres I., Juhasz I., Brown E. J., Capobianco A. J., Herlyn M., Liu Z. J. (2005) J. Clin. Invest. 115, 3166–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A. J., Nye J. S., Conlon R. A., Mak T. W., Bernstein A., van der Kooy D. (2002) Genes Dev. 16, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horrigan E., Evans A. (2006) Surgery 24, 5–8 [Google Scholar]
- 60.Busam K. J. (2005) Adv. Anat. Pathol. 12, 92–102 [DOI] [PubMed] [Google Scholar]
- 61.Ingram W. J., McCue K. I., Tran T. H., Hallahan A. R., Wainwright B. J. (2008) Oncogene 27, 1489–1500 [DOI] [PubMed] [Google Scholar]
- 62.Curry C. L., Reed L. L., Nickoloff B. J., Miele L., Foreman K. E. (2006) Lab. Invest. 86, 842–852 [DOI] [PubMed] [Google Scholar]
- 63.Stockhausen M. T., Sjölund J., Axelson H. (2005) Exp. Cell Res. 310, 218–228 [DOI] [PubMed] [Google Scholar]
- 64.Howell P. M., Jr., Liu S., Ren S., Behlen C., Fodstad O., Riker A. I. (2009) Cancer Control 16, 200–218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.