Abstract
Lignin is a complex polymer derived from the oxidative coupling of three classical monolignols. Lignin precursors are methylated exclusively at the meta-positions (i.e. 3/5-OH) of their phenyl rings by native O-methyltransferases, and are precluded from substitution of the para-hydroxyl (4-OH) position. Ostensibly, the para-hydroxyls of phenolics are critically important for oxidative coupling of phenoxy radicals to form polymers. Therefore, creating a 4-O-methyltransferase to substitute the para-hydroxyl of monolignols might well interfere with the synthesis of lignin. The phylogeny of plant phenolic O-methyltransferases points to the existence of a batch of evolutionarily “plastic” amino acid residues. Following one amino acid at a time path of directed evolution, and using the strategy of structure-based iterative site-saturation mutagenesis, we created a novel monolignol 4-O-methyltransferase from the enzyme responsible for methylating phenylpropenes. We show that two plastic residues in the active site of the parental enzyme are vital in dominating substrate discrimination. Mutations at either one of these separate the evolutionarily tightly linked properties of substrate specificity and regioselective methylation of native O-methyltransferase, thereby conferring the ability for para-methylation of the lignin monomeric precursors, primarily monolignols. Beneficial mutations at both sites have an additive effect. By further optimizing enzyme activity, we generated a triple mutant variant that may structurally constitute a novel phenolic substrate binding pocket, leading to its high binding affinity and catalytic efficiency on monolignols. The 4-O-methoxylation of monolignol efficiently impairs oxidative radical coupling in vitro, highlighting the potential for applying this novel enzyme in managing lignin polymerization in planta.
Introduction
Lignin is a complex, irregular biopolymer composed of hydroxylated and methylated phenylpropane units. It is derived mainly from the oxidative coupling of three different hydroxycinnamyl alcohols (or monolignols, i.e. p-coumaryl, coniferyl, and sinapyl alcohols), differing only in their degree of methoxylation (supplemental Fig. S1A). These three monolignols, incorporated into the lignin polymer, respectively, produce p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) subunits. The G unit is methylated singly on the 3-hydroxyl group, whereas the S unit is methylated on both the 3- and 5-hydroxyl moieties. The ratio of S to G subunits dictates the degree of lignin condensation by allowing different types of polymeric linkages. A higher G content creates a highly condensed lignin composed of a greater portion of biphenyl and other carbon-carbon linkages, whereas S subunits commonly are linked through more labile ether bonds at the 4-hydroxyl position (1, 2). To ensure the efficient degradation of the cell-wall biomass in agriculture, paper making, and biofuel production, lignin content should be lowered, or the amount of the more chemically labile S lignin increased (2, 3).
Monolignols, synthesized in the cytosol, are sequestered into the cell wall, and subsequently polymerized to afford a wall-reinforcing biopolymer (1). Polymerization begins with the dehydrogenation (single-electron oxidation) of monolignols that produce phenoxy radicals (supplemental Fig. S1B). Supposedly, dehydrogenation is initiated by oxidative enzymes (peroxidases/laccases) at the para-hydroxyl (4-OH) site of the aromatic ring, and is followed by electron resonance to generate different p-quinoid species, i.e. the free radical intermediates (depicted with p-coniferyl alcohol in supplemental Fig. S1B) (4–6). Subsequent coupling of the phenoxy radicals to each other, or to the growing polymer during lignification forms the lignin polymer. The cross-coupling of these phenoxy radicals in planta generates different inter-unit bonds, including the most frequent β-O-4 (β-aryl ether) linkages, and less frequent 5-O-4 ether linkages (4, 7). Apparently, the generation of radical intermediates and formation of ether linkages between the phenylpropane units requires unsubstituted 4-hydroxyl (supplemental Fig. S1B).
Natural evolution and selection created numerous specialized enzymes that catalyze specific chemical reactions in cells. In monolignol biosynthesis, the O-methylation of lignin monomeric precursors is catalyzed by a caffeate/5-hydroxyferulate 3/5-O-methyltransferase (COMT)2 (EC. 2.1.1.6) (8–10), and a caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) (EC. 2.1.1.104) (11, 12). The latter is a prototypic methyltransferase whose catalysis require divalent cations, whereas COMT is a homodimer protein whose activity does not necessitate the presence of metal ions (13). COMT originally was recognized as responsible for methylating caffeic acid and 5-hydroxyferulic acid, and later was evaluated as predominating in the methylation of p-cinnamaldehyde and cinnamyl alcohol (monolignol) (14–16). Despite the broad substrate specificity of both enzymes in vitro, in nature the activities of lignin O-methyltransferases are confined to exclusive regiospecificity for meta (3 or 5)-hydroxyl methylation (11, 14–16) (see Fig. 1A). The crystal structure of alfalfa COMT has been determined (17). Its ternary structural complexes, with the methyl donor AdoMet/S-adenosylhomocysteine and the substrate/product 5-hydroxyconiferaldehyde/ferulic acid, clearly revealed the structural basis for its substrate specificity and selective methylation of meta-hydroxyl (3/5-OH). Consequently, upon exposure to the activity of the lignin O-methyltransferases, the monolignols and their monomeric precursors are methylated exclusively at the meta-positions (i.e. 3/5-OH) of their phenyl rings (supplemental Fig. 1A). The para-hydroxyl position of lignin precursors remains unmethylated, pointing to the importance of the free para-hydroxyl of monolignols in lignin biosynthesis and polymerization. In fact, all current lignin biosynthetic scenarios implicate the free para-hydroxyl of monolignol as critical for monolignol dehydrogenation, and for cross-coupling phenoxy radicals to form inter-unit linkages (4–6) (supplemental Fig. S1B). Therefore, obtaining an enzyme that enables the methylation of the para-hydroxyls (i.e. 4-OH) of monolignols may well lead to a new strategy for managing the biosynthesis and polymerization of lignin.
FIGURE 1.
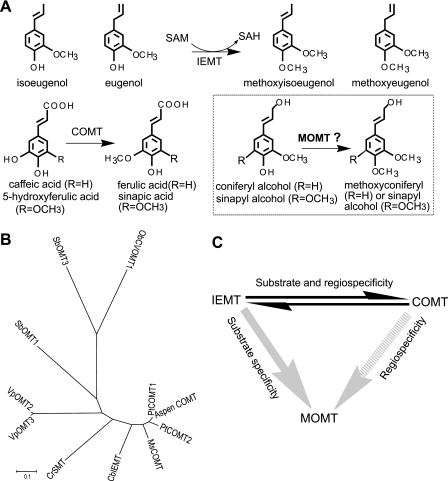
Reactions and phylogeny of phenolic O-methyltransferases. A, the biochemical reactions catalyzed by COMT, IEMT, and the desired monolignol 4-O-methyltransferase (MOMT) (in box); note the structural similarity of monolignol coniferyl alcohol and phenylpropene (iso)eugenols. B, phylogenetic tree of phenolic O-methyltransferases, showing the evolutionary relationship with COMT; PtCOMT1 (P. trichocarpa, accession No EU889124), PtCOMT2 (EU889125), Aspen COMT (P. tremuloides, Q00763), MsCOMT (M. sativa, P28002), CbIEMT (C. breweri, O04385), VanOMT2 (Vanilla planifolia, ABD61227), VanOMT3 (ABD61228), SbOMT3 (S. bicolor, No ABP01564), ObCVOMT1 (O. basilicum, AAL30423), and CrSMT (Catharanthus roseus, DQ084384). C, schematic strategy for engineering the desired monolignol 4-OMT.
Several phenolic O-methyltransferases of plants were characterized functionally. Sequence and phylogenic analyses indicated that some originated from COMT through a few amino acid substitutions (18–24) (Fig. 2B). In Clarkia breweri (fairy fans) and sweet basil (Ocimum basilicum), the identified O-methyltransferases catalyze the 4-O-methylation of a group of volatile compounds, viz. the phenylpropenes, isoeugenol, eugenol, and/or chavicol (23, 24). These allyl/propenylphenols are structural analogs of monolignols, differing only in their propanoid tail. Particularly, the (iso)eugenols structurally resemble p-coniferyl alcohol (see Fig. 1A). Early pioneering studies by Wang and Pichersky (19, 24) revealed that (iso)eugenol 4-O-methyltransferase (IEMT) (EC. 2.1.1.146) evolved from COMT by gene duplication and subsequent mutations. The IEMT from C. breweri shares more than 83% amino acid sequence identity with COMT from the same species, but exhibits distinct substrate preferences and regiospecificity for the 4-hydroxyl methylation of isoeugenol and eugenol (see Fig. 1A). Swapping sequence fragments of IEMT and COMT, or reversely converting the residues around their active sites inter-transformed their activities, although it greatly compromised the catalytic efficiency of the resulting enzyme variants (19, 24). Interestingly, these designed site-directed mutations invariably and simultaneously alter substrate preference and regioselective methylation, i.e. the mutations change the activity of IEMT from the 4-O-methylation of allyl/propenylphenols to the 3/5-O-methylation of caffeic acid, and vice versa (19, 24).
FIGURE 2.
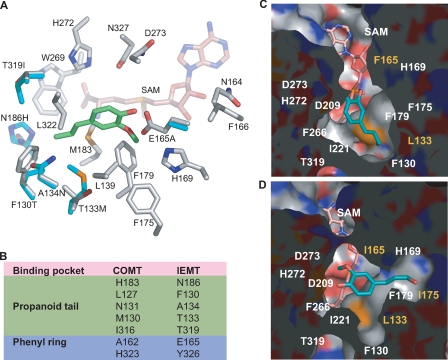
The putative active sites of IEMT and its mutant variants. A, close-up view of the modeled IEMT active site superimposed on that of COMT; the amino acid residues of IEMT are color coded, with carbon in gray, nitrogen in blue, oxygen in red, and sulfur in yellow. Only the distinct residues of COMT in the active site are shown (carbons in cyan). The docked isoeugenol in the IEMT active site is shown in green. For clarity, one distinct residue, Tyr-326, in IEMT (His-323 in COMT) is omitted. B, the seven distinct amino acid residues in IEMT and COMT that might directly interact with the accommodated phenolics. C and D, surface representations of the active site cavity of the mutant T133L/E165F (C), and T133L/E165I/F175I (D) from the same view, illustrating the architectural changes in the binding pocket and repositioning the docked coniferyl alcohol.
Directed evolution is a proven effective process for protein engineering and optimization (25, 26). Within the short time scale, a remarkable range of new enzymatic properties can be generated, based on creating different libraries and adopting different screening approaches. To create a novel enzyme exhibiting 4-OH methylation activity, preferentially for lignin monomeric precursors, primarily monolignols, i.e. a monolignol 4-O-methyltransferase (see Fig. 1C), we explored the structural basis of IEMT for its regioselective methylation and substrate discrimination by homology modeling. We identified seven putative “plasticity” amino acid residues in its active site that might contribute to the binding surface directly interacting with the accommodated phenolics. Thereafter, we established sets of enzyme-mutant libraries via site saturation and iterative saturation mutagenesis (27) primarily focusing on those identified plastic residues. Screening the activity of enzyme variants, we discovered that two evolutionarily plastic amino acid sites, Glu-165 and Thr-133, in the active site of IEMT dominate substrate specificity. Mutation at either site liberates the latent substrate flexibility of IEMT, enabling enzyme variants to accommodate lignin monomeric precursors, while retaining 4-O-methylation selectivity, i.e. the enzyme can methylate the para-hydroxyls of monolignols. The subsequent iterative saturation mutagenesis to optimize enzyme activity affirmed the additive effect of the beneficial mutations of both sites on the enzymes' catalytic efficiency; eventually, after three rounds of iterative mutagenesis, we obtained a triple mutant variant displaying more than a 70-fold increase in catalytic efficiency for the 4-O-methylation of monolignols compared with the parental enzyme. An in vitro polymerization assay confirmed that the 4-O-methoxy monolignol did not yield any type of oxidative coupling products, denoting that para-substitution of phenolics impairs the processes of dehydrogenation and oxidative coupling.
EXPERIMENTAL PROCEDURES
Chemicals
[methyl-14C]S-Adenosyl-l-methionine was purchased from Amersham Biosciences. BugBuster and Bradford solution, respectively, were bought from Novagen (Madison, WI) and Bio-Rad. We purchased all other chemicals, including the phenolics, from Sigma.
Site-saturation Mutagenesis
The cDNA encoding C. breweri IEMT was PCR amplified from the pET11-IEMT plasmid (24) and subcloned into a modified pET28a(+) vector compatible with Gateway cloning to fuse the IEMT with a His tag to facilitate Ni+-mediated affinity protein purification (28).
Saturation mutagenesis was performed at sites 130, 131, 133, 134,139,164,165, 175,186, 319, 326, and 327 of IEMT following the QuikChange site-directed mutagenesis strategy (Stratagene) using NNK degenerate primers (N represents a mixture of A, T, G, C, and K for G/T) (supplemental Table S1). The codon NNK has 32-fold degeneracy and encodes all 20 amino acids without rare codons.
Mutant Library Screening and Enzymatic Assay
We inoculated the Escherichia coli transformants in 96-well plates. Protein production was induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside, and the harvested cell cultures were lysed with 60 μl of BugBuster solution per well (Novagen). We either used the lysate directly or further purified the recombinant protein in a 96-well purification plate for screening enzymatic activity.
Screening was performed in a polypropylene microplate (Bio-Rad) containing 50 μl of the reaction in each well, 500 μm p-coniferyl-alcohol substrates, and 500 μm S-adenosyl-l-methionine augmented with (for detecting isotope activity) or without (for detecting by LC-MS) 0.005 μCi of [methyl-14C]S-adenosyl-l-Met, and 20 μl of lysate or purified eluent. We extracted the reaction product with ethyl acetate and moved the extracts into a new microplate. The radioactivity of the product was measured with a microplate scintillation counter (TopCount NXT, Packard), or the extracted product was analyzed by high pressure liquid chromatography using a reverse-phase C18 column (Gemini, 5-μm, 4.6 × 250-mm, Phenomenex). The samples were resolved in 0.2% acetic acid (A) with an increasing concentration gradient of acetonitrile containing 0.2% acetic acid (B) for 5 min, 30%; then to 12 min, 60%; and 14 min, 100%, at a flow rate of 1 ml/min. UV absorption was monitored at 254, 260, 280, 310, and 510 nm with a multiple wavelength photodiode array detector.
To examine the substrate specificity of the enzymes, the mutant enzymes obtained from the quick screen were purified as described previously (28). They were then assayed in 50 μl of 50 mm Tris-HCl, pH 7.5, containing 1 mm dithiothreitol, 500 μm S-adenosyl-l-methionine augmented with 0.005 μCi of [methyl-14C]S-adenosyl-l-Met, 500 μm phenolic substrates along with 5 μg of purified protein. The reactions proceeded for 5 min at 30 °C. We partitioned the products with ethyl acetate, and used aliquots of the extract for measuring radioactivity with a scintillation counter (Packard, 2500 TR).
To measure the steady-state kinetic constants of the IEMT wild-type and mutant variants, their enzyme activities were determined in different concentrations of coniferyl alcohol (10–960 μm) at a fixed concentration of 500 μm AdoMet, augmented with 0.005 μCi of [methyl-14C]S-adenosyl-l-Met. The reactions (50 μl) proceeded for 5 min at 30 °C. The activities were quantified by a liquid scintillation counter. The Vmax and Km were determined by nonlinear regression analysis via fitting the velocity concentration data to the Michaelis-Menten equation. The results represent the mean of three replicate assays.
In Vitro Polymerization
We produced 4-O-methoxyconiferyl alcohol in larger reactions using the purified mutant E165F. P-Coniferyl alcohol (6.0 μm) or purified 4-O-methoxy coniferyl alcohol (6.5 μm) were incubated with horseradish peroxidase (40 ng) (Sigma), and a final 1.14 mm H2O2 in 295 μl of 25 mm phosphate buffer, pH 7.0. The reaction was continued for 30 min at room temperature. The products were extracted twice with ethyl acetate, dried under N2 gas, re-dissolved in 50 μl of methanol, and examined by LC-MS (Agilent). For this analysis, the samples were resolved in 0.2% acetic acid (A) with an increasing concentration gradient of acetonitrile containing 0.2% acetic acid (B) for 0 to 2 min, 5%; 2 to 22 min, 5- to 73-%; and 22 to 24 min, 73 to 100%. Thereafter, they were maintained at 100% for 2 min at a flow rate of 1 ml/min. UV absorption was monitored at 254, 260, 280, 310, and 510 nm using a multiple wavelength photodiode array detector. For MS analysis, we coupled an HP 1100 series II LC system (Agilent) to a Bruker Esquire ion-trap mass spectrometer (MSD trap XCT system) equipped with an atmospheric pressure chemical ionization source. The flow rate to the MSD trap was 0.5 ml/min; positive ionization was attained using an ion source voltage of 3.5 kV, a corona of 4000 nA, and a skimmer at a voltage of 40 V. Nebulization was aided by a coaxial nitrogen sheath gas at 60 p.s.i. pressure. Desolvation was insured by a counter current nitrogen flow set at 7 p.s.i., with both the capillary and vaporizer temperature at 350 °C. Mass spectra were recorded over 50 to 1000 m/z in the positive mode.
Homology Modeling
The amino acid sequences of IEMT or its mutant variants were aligned to that of Medicago sativa COMT using CLUSTAL W version 1.83. The homology model of IEMT was built on the structure of M. sativa COMT (Protein Data Bank code 1KYW; 17) using the program MODELLER (29). The phenolics, isoeugenol, or p-coniferyl alcohol were docked into the built IEMT wild-type or mutant models via the GOLD program (CCDC, United Kingdom).
RESULTS
Re-evaluating the Substrate Specificity of Phenolic OMTs
Directed evolution of novel enzyme function on a laboratory time scale requires carefully selecting a suitable starting gene. To engineer a monolignol 4-O-methyltransferase, we first re-evaluated the substrate specificity and regioselective methylation of a few characterized plant phenolic O-methyltransferases (Fig. 1B), including the (iso)eugenol OMT (IEMT) from C. breweri (24), two COMT from Populus tricocharpa (28), and two phenolic/polyphenolic O-methyltransferases from Sorghum bicolor (20). LC-MS analysis showed that whereas the individual enzymes exhibited prominent regioselective transmethylation activities on their reported native substrates, C. breweri IEMT, poplar COMTs, and sorghum SbOMT1 showed almost negligible residual activity in methylating monolignol, p-coniferyl alcohol. Among them, C. breweri IEMT had the highest activity (Table 1), about ∼1% of that of the reported native substrate isoeugenol (19). Although such residual activity is kinetically meaningless (Table 1), this result indicates the intrinsic, yet highly constrained, substrate promiscuity of native IEMT.
TABLE 1.
Kinetic parameters of IEMT wild-type (WT) and the mutants for monolignols
The data represent mean ± S.D. of three replicates.
| Mutant | Km | Vmax | Kcat/Km |
|---|---|---|---|
| μm | nmol mg−1min−1 | m−1s−1 | |
| Coniferyl alcohol | |||
| IEMT WT | 1591 ± 182.0 | 42.0 ± 3.5 | 17.6 |
| E165F | 382.8 ± 27.0 | 165.1 ± 5.0 | 287.5 |
| E165I | 386.0 ± 77.8 | 110.0 ± 9.5 | 190.0 |
| E165V | 374.3 ± 95.7 | 66.0 ± 7.2 | 117.5 |
| E165M | 464.8 ± 73.7 | 53.7 ± 9.1 | 77.1 |
| E165L | 589.1 ± 81.1 | 62.2 ± 4.6 | 70.4 |
| E165S | 239.3 ± 24.2 | 19.1 ± 0.7 | 53.3 |
| E165P | 475.7 ± 45.9 | 15.0 ± 0.7 | 21.0 |
| E165Y | 884.3 ± 153.1 | 13.6 ± 1.4 | 10.3 |
| T133L | 534.0 ± 78.2 | 31.6 ± 2.3 | 39.5 |
| T133M | 344.8 ± 33.9 | 56.5 ± 2.3 | 109.5 |
| T133L/L139Q/E165F | 304.4 ± 21.0 | 363.0 ± 9.6 | 794.9 |
| T133L/E165I/F175I | 198.5 ± 23.7 | 371.0 ± 14.8 | 1245.8 |
| T133L/L139Q/E165I/F175I | 229.5 ± 19.5 | 330.9 ± 9.2 | 961.1 |
| Sinapyl alcohol | |||
| IEMT WT | 1495 ± 214.4 | 44.6 ± 3.9 | 19.9 |
| T133L/L139Q/E165F | 615.0 ± 57.9 | 424.9 ± 20.0 | 459.5 |
| T133L/E165I/F175I | 118.7 ± 11.0 | 260.6 ± 6.3 | 1463.4 |
| T133L/L139Q/E165I/F175I | 155.9 ± 13.7 | 169.3 ± 4.2 | 723.8 |
Identifying Amino Acid Residues of IEMT Potentially Responsible for Substrate Discrimination
The high sequence similarities between C. breweri IEMT and alfalfa COMT (∼84% at the amino acid level) allowed us to undertake homology modeling and substrate docking analyses, based on the crystal structure of the latter (PDB code 1KYW) (17). The homology model shows that IEMT exhibits obvious architectural changes in the putative active site in respect to the evolution of 4-O-methylation activity on phenylpropenes. About 18 amino acid residues of IEMT constellated to constitute the binding pocket for the accommodated phenolic substrate in the putative active site. Seven are distinct from COMT, representing key substitutions changing the architecture of the active site (Fig. 2, A and B). Among them, five substitutions take place in the binding pocket for the propanoid tail of the docked compound, and the other two, i.e. Glu-165 in IEMT versus Ala-162 in COMT, and Tyr-326 versus His-323 occur near the aryl ring of the bound phenylpropene (Fig. 2, A and B). The changes of these residues in the IEMT active site probably offer a more favorable binding environment for the allylphenols, and re-position the bound phenylpropene for transmethylating the 4-hydroxyl moiety. Particularly, changing Ala-162 in COMT to the bulky, acidic residue Glu-165 likely imposes a steric constraint on the phenyl ring of the compound. Meanwhile, the carboxylate of its side chain donates a hydrogen bond with the 4-hydroxyl of the bound compound, thus enabling the 4-hydroxyl proximal to the methyl donor AdoMet, and the putative catalytic base His-272 to facilitate a Sn2 reaction for 4-O-methylation (Fig. 2A).
Evolving IEMT for 4-O-Methylation of Monolignols
To promote the latent activity of IEMT (or COMT) for the 4-O-methylation of monolignols, we first carried out conventional site-directed mutagenesis by either converting the identified distinct active site residues from COMT to the corresponding one of IEMT (or vice visa) (supplemental Fig. 2A). Alternatively, we designed substitutes for the IEMT residues to afford potential H-bonds or eliminate steric hindrance for favorably accommodating monolignols in the active site. For example, we changed the IEMT Phe-130 to Thr, Asp, and Cys, or Thr-133 to Ser, and/or combining those mutations into the double and triple mutants (supplemental Fig. S2B). However, these activities did not alter the regioselectivity of COMT for the recognized lignin precursors, or the substrate preferences of IEMT that favor monolignols (supplemental Fig. S2).
To identify the residues that specifically entail the substrate preference of IEMT for monolignols, we next subjected the seven distinct active site residues (Fig. 2B), and a few adjacent ones (Leu-131, Leu-139, Phe-175, Asn-164, and Asn-327) to directed evolution by site-saturation mutagenesis. We adopted the NNK strategy with 32-fold degeneracy to construct mutant libraries (supplemental Table S1). We constituted a high throughput mutant-screening procedure (supplemental Fig. S3), in which we employed a 96-well plate-based isotope activity assay, and LC-MS to confirm the enzymatic product. About 30 active IEMT mutant clones were selected from the single site-saturation mutant libraries, based on their similar, or higher, methylation activity for coniferyl alcohol than that of the wild-type enzyme. These active clones represent 18 distinct codon mutations (supplemental Table S2). All the mutations that conferred better methylation activity for coniferyl alcohol occurred at two amino acid sites: Glu-165, the residue that potentially interacts with the phenyl ring of the bound substrate, and, Thr-133, the residue involved in binding the propanoid tail of the compound (Fig. 2A). The recombinant proteins of all 18 mutants were purified further, and their activities again confirmed by LC-MS. Incubating the purified enzymes with coniferyl or sinapyl alcohol and the methyl donor AdoMet efficiently converted the phenolic substrate to a novel product with a similar UV spectrum as that of the monolignol substrate. However, its atmospheric pressure chemical ionization mass spectrum shows a 14 increment mass to charge ratio (m/z) at the molecular ion and the major fragment ions, indicating the addition of a methyl group to the monolignol substrate (Fig. 3, A and B). The tandem MS analysis on the ion fragments suggest that methylation occurred on the para-hydroxyl of monolignols (supplemental Fig. S4).
FIGURE 3.
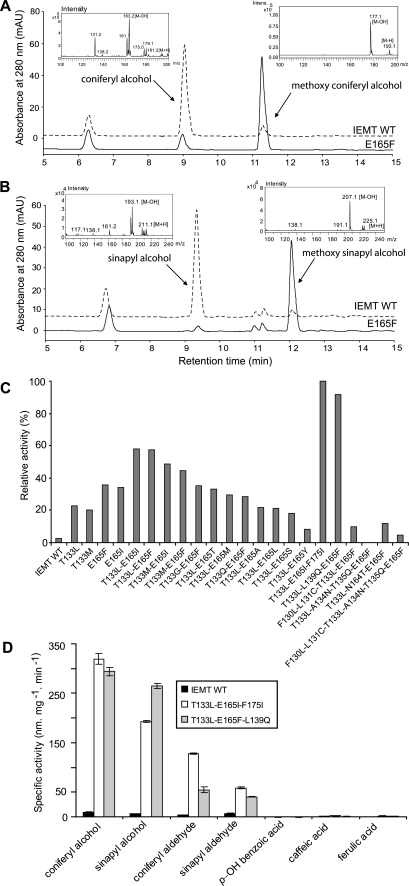
Catalytic activity and substrate specificity of the engineered IEMT mutants. A, LC-MS profiling of the reactions catalyzed by IEMT (dashed line), and the mutant E165F (solid line) incubated with coniferyl alcohol and the methyl donor AdoMet. B, LC-MS profiling of the reactions catalyzed by IEMT (dashed line) and mutant E165F (solid line) incubated with sinapyl alcohol and the methyl donor AdoMet. C, relative activity of the selected IEMT mutant variants in the 4-O-methylation of coniferyl alcohol. 100% activity represents 147 ± 6 nmol mg−1 min−1. D, substrate specificity of the engineered monolignol 4-O-methyltransfeases (IEMT triple mutants). The error bars represent S.D. of three replicate.
Kinetic analysis showed that substituting Glu-165 with hydrophobic residues, like Phe, Ile, Val, Met, and Leu increased the binding affinity (Km) and the turnover (Vmax) for 4-O-methylation of monolignols (Table 1). The improvement of the catalytic activity of these mutant variants, particularly, of their binding affinity for monolignol generally correlates with the hydrophobicity of the substituted residues; the higher that was, the better the catalytic activity (supplemental Fig. S5A). Structurally, those substitutions provided a more hydrophobic environment in the active site for binding aromatic substrates (supplemental Fig. S5B). The variant E165F with a substitution showing the highest hydrophobicity yielded a catalytic ratio (Kcat/Km) for coniferyl alcohol at 287 m−1 s−1, about 17-fold higher than that of IEMT wild-type. Substituting Glu-165 with the polar residue Ser also increased its binding affinity for coniferyl alcohol (Table 1); however, the catalytic activity of this variant did not rise proportionally. Presumably, this reflected the imposition by the side chain hydroxyl of Ser of an H-bond on the 4-O-methoxyl group of the product yielded, thus impairing its release. Enzyme activity was impaired by replacing Glu-165 with residues such as Tyr and Arg bearing a bulky side chain, or residue Pro that potentially entails severe changes in structural conformation (Table 1 and supplemental Fig. S5A). Particularly, changing Glu-165 to Arg totally abolished the activity for monolignol and isoeugenol (supplemental Fig. S5A). These results suggest that the hydrophobic interaction and steric effect imposed by residue 165 on the phenyl ring of the bound compound in IEMT mutants play a crucial role in governing substrate binding and transmethylation proficiency.
Similar to Glu-165, Thr-133, one of the identified distinct residues that potentially interact with the propanoid tail of the docked phenolics in IEMT model, affords beneficial mutations for methylating monolignols. Substituting Thr-133 with hydrophobic residues Leu and Met correspondingly improved the catalytic efficiency for coniferyl alcohol by 2.2- and 6.2-fold compared with that of IEMT wild-type (Table 1).
Improving the Catalytic Activity of Mutant Variants
To improve further the activity of the single mutant variants for 4-O-methylation of monolignols, we employed iterative saturation mutagenesis on Glu-165 and Thr-133 variants. We used the E165F mutant as the parental enzyme to introduce saturation mutations in all six remaining distinct residues (Fig. 2B); meanwhile, we employed T133L and T133M variants to mutate specifically Glu-165 with the 32-fold degenerative primers. A range of double mutants was obtained (Fig. 3C). Serendipitously, only those variants arising from the combination mutations of Glu-165 and Thr-133 exhibited activity for 4-O-methylation of monolignols. Among them, mutants of T133L/E165F, T133L/E165I, T133M/E165F, and T133M/E165I demonstrated 1.6–2.6-fold better activity for coniferyl alcohol compared with their respective single site mutant parents (Fig. 3C). Previous reports suggest that simultaneously converting the residues adjacent to Thr-133 and Glu-165 of IEMT (i.e. Phe-130, Leu-131, Ala-134, Thr-135, and Asn-164) to those of COMT confer on the variants the ability to methylate the lignin-precursor caffeic acid or 5-hydroxyferulic acid (at meta-position) (19). However, when we introduced similar mutations onto the T133L/E165F variant, the new enzymes did not display any beneficial effect on the activity in 4-O-methylating monolignols. In the mutant T133L/A134N/T135Q/E165F, all such activity was totally abolished (Fig. 3C). Furthermore, using T133L/E165F and T133M/E165F as templates to introduce the saturation mutations on that set of adjacent sites did not generate any valuable variants.
By modeling the double mutant variant and docking monolignol into its active site (Fig. 2C), we identified a range of additional amino acid sites potentially directly contacting, or proximal to the docked monolignol. They included Leu-139 and Phe-175 that might contribute to constructing a hydrophobic cavity to interact with the 3-methoxy group of the bound compound, as do the corresponding residues in COMT (17). Therefore, to optimize enzymatic activity, we imposed further saturation mutations on those recognized sites via T133L/E165F and T133L/E165I as parental templates. Screening the triple mutant libraries, we identified two mutants, T133L/E165I/F175I and T133L/L139Q/E165F, that exhibit a major increase in their specific activity on monolignols (Fig. 3C). Kinetic analysis revealed that the former displays the catalytic efficiency of 1245 and 1463 m−1 s−1, respectively, for coniferyl and sinapyl alcohols, namely, more than 70-fold increases compared with the wild-type enzyme (Table 1).
Substitutions in T133L/E165I/F175I, particularly the replacement of Phe-175 with Ile, greatly enlarge the original hydrophobic cavity that apparently contributes to constraining the 3-methoxy group of the bound phenolics in the IEMT and its single or double mutant models. This architectural change entails an artificial hydrophobic binding pocket that can accommodate snugly the propanoid tail of the compound (Fig. 2D). Consequently, the docked monolignol in the putative active site of this triple mutant variant displays drastic re-orientation compared with the compound in the wild type, single or double mutants (Fig. 2, C and D). Such repositioning of the substrate probably facilitates the efficient transmethylation of the 4-hydroxyl moiety.
Because substituting Leu-139 with Gln in the T133L/E165F variant increased enzyme activity on monolignols, we incorporated the mutation of L139Q into the triple mutant T133L/E165I/F175I. However, the resulting quadruple mutant variant kinetically did not show any improvement in binding affinity or catalytic efficiency (Table 1). This is probably because Leu/Gln-139 is relatively distant from the newly created binding pocket for monolignols in T133L/E165I/F175I; thus, it has little advantage on substrate repositioning or catalysis.
Substrate Specificity of Mutant Variants
The mutant variants, represented by T133L/E165I/F175I and T133L/L139Q/E165F, exhibited high activities for a range of lignin monomeric precursors, the p-hydroxycinnamyl alcohols and p-hydroxycinnamaldehydes (Fig. 3D). The activity of the wild-type IEMT, in contrast, was barely measurable for all of them. Kinetically, the mutant T133L/E165I/F175I equally preferred two major types of monolignols in angiosperms, viz. coniferyl and sinapyl alcohols, whereas the T133L/L139Q/E165F variant favored coniferyl slightly over the sinapyl alcohol as reflected in its better binding capacity for the former (Table 1). Like the wild-type enzyme, no activity was detected when the mutant variants were incubated with phenolics bearing the carboxylic function group, i.e. p-hydroxybenzoic, caffeic, and ferulic acids (Fig. 3D).
4-O-Methylation of Monolignol Impairs Its Dehydrogenative Polymerization in Vitro
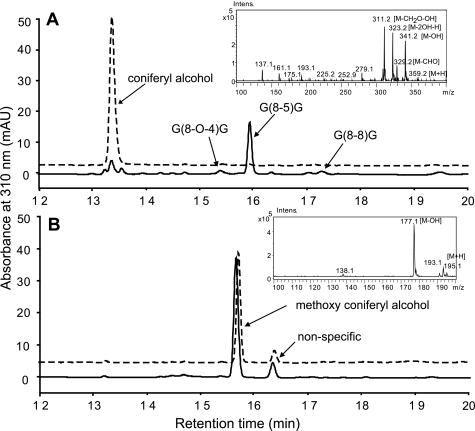
In vitro dehydrogenative polymerization is commonly used as a biomimetic model to explore in vivo lignin formation (30, 31). Using the novel mutant variants, we produced methoxylated monolignol, viz. the 4-O-methoxyconiferyl alcohol, and subjected it to peroxidase-catalyzed dehydrogenative polymerization, and compared with the classic p-coniferyl alcohol. After incubating these phenolics with horseradish peroxidase and H2O2, coniferyl alcohol was oxidized and conjugated, yielding several oligolignols, as previously documented (32). These oligomer products include a predominant dimer, the β-5 inter subunit linkage, G(8–5)G, and a few minor peaks of the G(8-O-4)G and G(8–8)G dimers (Figs. 4A and supplemental Fig. S6). However, after incubating 4-O-methylated coniferyl alcohol with horseradish peroxidase/H2O2, essentially there was no conversion and no oligomeric products were detected (Fig. 4B).
FIGURE 4.
In vitro polymerization of monolignols. A, LC-MS profiling of the reaction of coniferyl alcohol incubated with horseradish peroxidase and H2O2 (solid line), or with buffer alone as the control (dashed line). The profile shows a predominant dimer G(8–5)G and the minor products of G(8-O-4)G and G(8–8)G. The inset shows atmospheric pressure chemical ionization mass spectrum of the former. The mass and UV spectra for the other products are shown in supplemental Fig. S6. B, LC-MS profiling of the reaction of methoxyconiferyl alcohol incubated with horseradish peroxidase and H2O2 (solid line), or with buffer alone as the control (dashed line). No dimers/oligomers were detected.
DISCUSSION
Following a path of directed evolution with one amino acid substitution at a time, and employing iterative site-saturation mutagenesis, we created an efficient, novel monolignol 4-O-methyltransferase from phenylpropene O-methyltransferase. Phenylpropene O-methyltransferases and a few other phenolic O-methyltransferases were demonstrated to have evolved naturally in plants from the lignin biosynthetic enzyme, COMT (18–24). The evolution of these phenolic O-methyltransferases primarily was archived through gene duplication and subsequent substitutions of only a limited set of amino acid residues. A previous study (17) and our current homology modeling indicate that IEMT differs from COMT in its putative active site, primarily seven amino acid residues (Fig. 2). Therefore, these distinct active site residues in the two enzymes most likely represent evolutionarily plastic sites that dominate substrate discrimination and regioselective methylation. Further modulating these plastic sites might interrogate and engender the desired novel functionalities. By saturation mutagenesis, during which we introduced a full set of 20 amino acid substitutions at each of the seven active plastic sites, we demonstrated that two amino acid sites in IEMT, Glu-165 and Thr-133, are critical for substrate discrimination/binding. Substituting both sites with hydrophobic residues enables the resulting variants to effectively recognize and accommodate a monolignol substrate, while retaining the ability for 4-O-methylation (see Table 1 and Fig. 3). Other sites tested displayed a lesser effect, or none, in initiating the novel substrate preference of the enzyme. Subsequent iterative mutations using the single mutant variants from both sites created the double mutations with higher activity. Serendipitously, these double mutants combine the beneficial substitutions of Glu-165 and Thr-133. These apparent additive mutation effects support the notion that the targeted property in directed enzyme evolution can be acquired through a series of single beneficial mutations, and that combination of the single mutations would retain the desired properties (26). After three rounds of saturation mutations, variant T133L/E165I/F175I showed adequate catalytic capacity in the 4-O-methylation of monolignols; its catalytic efficiency and binding affinity to monolignols, respectively, were more than 70- and 13-fold higher than those of the wild-type enzyme. Interestingly, these three site mutations created an apparently novel substrate binding pocket for accommodating monolignols (Fig. 2D), pointing to the facile structural plasticity of phenolic OMTs in the evolution of new biochemical functions.
Reportedly, the conventional conversion of a few adjacent residues around Glu-165 and Thr-133 of IEMT to those of COMT transferred IEMT activity back to the meta-methylation of caffeic acid (19, 24). However, those point mutations do not contribute to activity on monolignols (Fig. 3C). On the other hand, the variants created from saturation mutagenesis of Glu-165 and Thr-133 show no activity to caffeic acid (neither meta- or para-methylation) (Fig. 3D). Therefore, saturation mutation present here separates the two evolutionarily tightly linked biochemical properties of substrate specificity, and methylation regioselectivity of the parental OMTs, and entails novel function in the variants. Evidently, the mutant variants from our saturation mutagenesis represent a distinct evolutionary path from natural selection.
The novel enzyme variants we created greatly liberated the highly constrained substrate promiscuity of the parental IEMT. The triple mutants exhibited a broad spectrum of substrate preference on phenolics (Fig. 3D). Many examples of directed evolution have demonstrated that enzymes with promiscuous functions exhibit high plasticity; they might well be used as “generalists” to further tailor particular catalytic properties (33–35). One of our triple mutants, T133L/L139Q/E165F, catalytically prefers the guaiacyl lignin precursor, coniferyl alcohol, over sinapyl alcohol; its binding affinity for the former is about 2-fold higher than for the latter (see Table 1). Fine tuning this variant by additional rounds of mutagenesis, together with positive (for coniferyl alcohol) and negative selection (for sinapyl alcohol) might engender enzymes specific for methylating the guaiacyl lignin precursor, with potential use in specifically disrupting the biosynthesis of condensed lignin.
Para-methoxylation of monolignols abolishes oxidative radical coupling. In contrast to p-coniferyl alcohol, the 4-O-methoxy substituent did not produce any type of coupled dimer/oligomer (Fig. 4). These data directly demonstrate the importance of para-hydroxyl in the proposed one-electron oxidative dehydrogeneration; they also raise the possibility of disturbing lignin polymerization in vivo through efficiently substituting/modifying 4-O-hydroxyl of lignin precursors via our novel enzymes. It will be interesting to explore the outcome of expressing the mutant enzymes in planta.
Supplementary Material
Acknowledgments
We thank Dr. Eran Pichersky, University of Michigan, for sharing the C. breweri IEMT clone, and Dr. Scott R. Baerson, U. S. Department of Agriculture-the Agriculture Research Service, Natural Products Research Unit for the sorghum OMT clones. We also thank Drs. John Shanklin and William Studier, Brookhaven National Laboratory, for valuable discussions on this work.
This work was supported by the Office of Basic Energy Science, Department of Energy Grant DEAC0298CH10886 (to C. J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- COMT
- caffeic acid/5-hydroxyferulic acid 3-O-methyltransferase
- IEMT
- (iso)eugenol 4-O-methyltransferase
- OMT
- O-methyltransferase
- AdoMet
- S-adenosyl-l-methionine
- LC-MS
- liquid chromatography-mass spectrometry.
REFERENCES
- 1.Boerjan W., Ralph J., Baucher M. (2003) Annu. Rev. Plant Biol. 54, 519–546 [DOI] [PubMed] [Google Scholar]
- 2.Dixon R. A., Reddy M. S. S. (2003) Phytochem. Rev. 2, 289–306 [Google Scholar]
- 3.Weng J. K., Li X., Bonawitz N. D., Chapple C. (2008) Curr. Opin. Biotechnol. 19, 166–172 [DOI] [PubMed] [Google Scholar]
- 4.Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P. F., Marita J. M., Hatfield R. D., Ralph S. A., Christensen J. H., Boerjan W. (2004) Phytochem. Rev. 3, 29–60 [Google Scholar]
- 5.Freudenberg K. (1968) in Constitution and Biosynthesis of Lignin (Freudenberg K., Neish A. C. eds) pp. 45–122, Springer-Verlag, Berlin [Google Scholar]
- 6.Davin L. B., Lewis N. G. (2005) Curr. Opin. Biotechnol. 16, 398–406 [DOI] [PubMed] [Google Scholar]
- 7.Guo D., Chen F., Inoue K., Blount J. W., Dixon R. A. (2001) Plant Cell 13, 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gowri G., Bugos R. C., Campbell W. H., Maxwell C. A., Dixon R. A. (1991) Plant Physiol. 97, 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bugos R. C., Chiang V. L., Campbell W. H. (1991) Plant Mol. Biol. 17, 1203–1215 [DOI] [PubMed] [Google Scholar]
- 10.Edwards R., Dixon R. A. (1991) Arch. Biochem. Biophys. 287, 372–379 [DOI] [PubMed] [Google Scholar]
- 11.Pakusch A. E., Kneusel R. E., Matern U. (1989) Arch. Biochem. Biophys. 271, 488–494 [DOI] [PubMed] [Google Scholar]
- 12.Ye Z. H., Kneusel R. E., Matern U., Varner J. E. (1994) Plant Cell 6, 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue K., Parvathi K., Dixon R. A. (2000) Arch. Biochem. Biophys. 375, 175–182 [DOI] [PubMed] [Google Scholar]
- 14.Parvathi K., Chen F., Guo D., Blount J. W., Dixon R. A. (2001) Plant J. 25, 193–202 [DOI] [PubMed] [Google Scholar]
- 15.Osakabe K., Tsao C. C., Li L., Popko J. L., Umezawa T., Carraway D. T., Smeltzer R. H., Joshi C. P., Chiang V. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Popko J. L., Umezawa T., Chiang V. L. (2000) J. Biol. Chem. 275, 6537–6545 [DOI] [PubMed] [Google Scholar]
- 17.Zubieta C., Kota P., Ferrer J. L., Dixon R. A., Noel J. P. (2002) Plant Cell 14, 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim R. K., Bruneau A., Bantignies B. (1998) Plant Mol. Biol. 36, 1–10 [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Pichersky E. (1999) Arch. Biochem. Biophys. 368, 172–180 [DOI] [PubMed] [Google Scholar]
- 20.Baerson S. R., Dayan F. E., Rimando A. M., Nanayakkara N. P., Liu C. J., Schröder J., Fishbein M., Pan Z., Kagan I. A., Pratt L. H., Cordonnier-Pratt M. M., Duke S. O. (2008) J. Biol. Chem. 283, 3231–3247 [DOI] [PubMed] [Google Scholar]
- 21.Coiner H., Schröder G., Wehinger E., Liu C. J., Noel J. P., Schwab W., Schröder J. (2006) Plant J. 46, 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H. M., Rotter D., Hartman T. G., Pak F. E., Havkin-Frenkel D., Belanger F. C. (2006) Plant Mol. Biol. 61, 537–552 [DOI] [PubMed] [Google Scholar]
- 23.Gang D. R., Lavid N., Zubieta C., Chen F., Beuerle T., Lewinsohn E., Noel J. P., Pichersky E. (2002) Plant Cell 14, 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Pichersky E. (1998) Arch. Biochem. Biophys. 349, 153–160 [DOI] [PubMed] [Google Scholar]
- 25.Yuan L., Kurek I., English J., Keenan R. (2005) Microbiol. Mol. Biol. Rev. 69, 373–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tracewell C. A., Arnold F. H. (2009) Curr. Opin. Chem. Biol. 13, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reetz M. T., Carballeira J. D. (2007) Nat. Protoc. 2, 891–903 [DOI] [PubMed] [Google Scholar]
- 28.Bhuiya M. W., Liu C. J. (2009) Anal. Biochem. 384, 151–158 [DOI] [PubMed] [Google Scholar]
- 29.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 30.Sasaki S., Nishida T., Tsutsumi Y., Kondo R. (2004) FEBS Lett. 562, 197–201 [DOI] [PubMed] [Google Scholar]
- 31.Fournand D., Cathala B., Lapierre C. (2003) Phytochemistry 62, 139–146 [DOI] [PubMed] [Google Scholar]
- 32.Morreel K., Ralph J., Kim H., Lu F., Goeminne G., Ralph S., Messens E., Boerjan W. (2004) Plant Physiol. 136, 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khersonsky O., Roodveldt C., Tawfik D. S. (2006) Curr. Opin. Chem. Biol. 10, 498–508 [DOI] [PubMed] [Google Scholar]
- 34.Schmidt D. M., Mundorff E. C., Dojka M., Bermudez E., Ness J. E., Govindarajan S., Babbitt P. C., Minshull J., Gerlt J. A. (2003) Biochemistry 42, 8387–8393 [DOI] [PubMed] [Google Scholar]
- 35.Rothman S. C., Kirsch J. F. (2003) J. Mol. Biol. 327, 593–608 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.