Abstract
Hepatic clearance of triglyceride-rich lipoproteins depends on heparan sulfate and low density lipoprotein receptors expressed on the basal membrane of hepatocytes. Binding and uptake of the lipoproteins by way of heparan sulfate depends on the degree of sulfation of the chains based on accumulation of plasma triglycerides and delayed clearance of triglyceride-rich lipoproteins in mice bearing a hepatocyte-specific alteration of N-acetylglucosamine (GlcNAc) N-deacetylase-N-sulfotransferase 1 (Ndst1) (MacArthur, J. M., Bishop, J. R., Stanford, K. I., Wang, L., Bensadoun, A., Witztum, J. L., and Esko, J. D. (2007) J. Clin. Invest. 117, 153–164). Inactivation of Ndst1 led to decreased overall sulfation of heparan sulfate due to coupling of uronyl 2-O-sulfation and glucosaminyl 6-O-sulfation to initial N-deacetylation and N-sulfation of GlcNAc residues. To determine whether lipoprotein clearance depends on 2-O-and 6-O-sulfation, we evaluated plasma triglyceride levels in mice containing loxP-flanked conditional alleles of uronyl 2-O-sulfotransferase (Hs2stf/f) and glucosaminyl 6-O-sulfotransferase-1 (Hs6st1f/f) and the bacterial Cre recombinase expressed in hepatocytes from the rat albumin (Alb) promoter. We show that Hs2stf/fAlbCre+ mice accumulated plasma triglycerides and exhibited delayed clearance of intestinally derived chylomicrons and injected human very low density lipoproteins to the same extent as observed in Ndst1f/fAlbCre+ mice. In contrast, Hs6st1f/fAlbCre+ mice did not exhibit any changes in plasma triglycerides. Chemically modified heparins lacking N-sulfate and 2-O-sulfate groups did not block very low density lipoprotein binding and uptake in isolated hepatocytes, whereas heparin lacking 6-O-sulfate groups was as active as unaltered heparin. Our findings show that plasma lipoprotein clearance depends on specific subclasses of sulfate groups and not on overall charge of the chains.
Keywords: Extracellular Matrix/Heparan Sulfate, Lipid/Absorption, Lipid/Triacylglycerol, Lipoprotein/Metabolism, Organisms/Mouse, Proteoglycans Structure, Proteoglycans Synthesis
Introduction
Heparan sulfate proteoglycans act as receptors for various growth factors and chemokines, enzymes, and cell adhesion proteins (1, 2). They also act as endocytic receptors, facilitating the uptake of various ligands and metabolites (3–5). Although the mechanism of uptake has not been analyzed in detail in most cell types, available data suggest that ligands bound to the heparan sulfate chains most likely “piggy-back” into the cell during internalization of the proteoglycan. Binding and uptake via this mechanism have physiological relevance based on the discovery that triglyceride-rich lipoprotein uptake by hepatocytes via heparan sulfate proteoglycans represents a major clearance pathway in vivo for remnant lipoproteins derived from very low density lipoproteins (VLDLs)4 and chylomicrons (6).
In previous studies we showed that lipoprotein uptake in the liver was dependent on the degree of sulfation of the chains based on hypertriglyceridemia in mice defective in the expression of N-acetylglucosamine N-deacetylase-N-sulfotransferase-1 (Ndst1) in hepatocytes. Ndst1 belongs to a family of four enzymes and initiates the modification of heparan sulfate chains (7). An epimerase (HsC5Epi) converts adjacent d-glucuronic acid units to l-iduronic acid, and then a group of O-sulfotransferases add sulfate to C2 of uronic acids (Hs2st) and to C6 (Hs6st1–3) and C3 (Hs3st1–6) of glucosamine units. These modifications occur in contiguous blocks of sugars along the chain in an incomplete manner, resulting in domains of variable size and sulfation (8). Sets of modified sugars act as binding sites for different proteins, sometimes with great specificity and affinity (9). Because Ndsts act before these other enzymes, altering the level of N-sulfation leads to altered levels of sulfation at other positions along the chain. The changes that occur vary quantitatively depending on the specific cell type or tissue under study (10–14). In hepatocytes, inactivation of Ndst1 led to significantly decreased 2-O-sulfation of uronic acids with a lesser effect on 6-O-sulfation (6). Thus, altered lipoprotein uptake in Ndst1-deficient mice could result from altered sulfation at any of these positions. Understanding the requirement for specific groups of sulfate residues is important, as the relevant transferases might represent candidate genes for explaining hypertriglyceridemia of unknown etiology.
One way to analyze this problem further is to inactivate enzymes involved in O-sulfation. Because Ndst1 deficiency most dramatically reduced uronyl 2-O-sulfation, we decided to first examine the importance of Hs2st in lipoprotein clearance. Because systemic inactivation of the gene causes renal agenesis, over-mineralized skeletons, retardation of eye development, and neonatal death (15), we generated a conditional loxP-flanked allele of Hs2st and inactivated it selectively in hepatocytes. The loss of Hs2st activity led to hypertriglyceridemia, with characteristics identical to that observed in Ndst1-deficient animals. Competition studies using variously desulfated heparins showed that both N-sulfation and 2-O-sulfation played important roles in lipoprotein binding; 6-O-sulfation did not, consistent with studies of conditional mutants altered in Hs6st1.
EXPERIMENTAL PROCEDURES
Mice Derivation and Animal Husbandry
Mice bearing a conditional loxP-flanked allele of Hs2st were prepared in the thymidine kinase-neomycin phosphotransferase-based targeting vector described previously (16). Genomic DNA containing the first exon of Hs2st was obtained from a 129Sv/J BAC clone. At the time that this study was initiated the intron-exon boundaries of the gene were not fully established in 129Sv/J mice. Thus, the loxP recombination site was inadvertently inserted 167 nucleotides upstream from the ATG start codon in the short 5′-untranslated region segment (estimated by 5′ rapid amplification of cDNA ends to be 273 nucleotides). This exon also encodes 40 of 356 amino acids of the open reading frame of Hs2st including the signal peptide. The final targeting vector was linearized using SalI before transfection of E9.5 R1 129Sv/J embryonic stem cells by electroporation. Transfected cells were selected with G418, and homologous recombinants were identified by Southern blotting and PCR analysis. One cell clone with a normal karyotype was transfected with a Cre expression vector followed by ganciclovir selection to remove cells that had not undergone recombination. One recombinant ES cell line lacking the drug selection cassette and bearing two loxP sites was selected based on Southern blotting and PCR analysis, and cells were injected in C57Bl/6J blastocysts. Chimeric offspring were crossed with C57Bl/6 mice, and one line bearing the conditional allele was obtained (Hs2st1f/f). This strain has been backcrossed into a C57Bl/6 background for >10 generations.
Hs6st1f/f mice were generated by Izvolsky et al. (17) and provided by W. Cardoso (Boston University School of Medicine), to whom all requests should be directed. Hs6st1f/f and Hs2st1f/f mice were cross-bred to mice expressing the Cre recombinase expressed under the control of the rat albumin promoter (18).
All animals were housed under barrier conditions in Assessment and Accreditation of Laboratory Animal Care-approved vivaria in the School of Medicine, University of California San Diego following the standards and procedures approved by the local Institutional Animal Care and Use Committee. Mice were weaned at 3 weeks, maintained on a 12-h light-dark cycle, and fed water and standard rodent chow ad libitum. To measure fasting plasma lipids, chow was removed from the cages at ∼8 a.m. in the morning, and 4 h later samples of blood were drawn from the retroorbital plexus.
Genotyping
Mice were routinely genotyped by PCR. Primer sets were designed to distinguish wild type (Hs2st+), conditional (Hs2stf), and null alleles (Hs2st−) of Hs2st. The DNA sequences were Primer 1 (5′-gtgcggccgtggggtcc-3′), Primer 2 (5′-atggggctcctcaggattatgatgc-3′), and Primer 3 (5′-tgccctaggctcaggcatg-3′). The primer pair P1/P3 generated PCR bands at 1178, 876, and 360 bp, corresponding to Hs2stf, Hs2st+, and Hs2st− alleles, respectively. When the primer pair P1/P2 were used, the generated PCR bands at 747 and 609 bp correspond to Hs2stf and Hs2st+, respectively.
To estimate the extent of recombination in hepatocytes, Southern blots were performed on DNA isolated from purified hepatocytes using a DNeasy tissue kit (Qiagen). Genomic DNA (25 μg) was digested overnight with the EcoRI and separated on 1% agarose gels before transfer overnight to nitrocellulose. The membrane was UV-cross-linked and hybridized to an Hs2st-specific probe double-labeled with [α-32P]dCTP and [α-32P]dATP (EasyTides; PerkinElmer Life Sciences). The membrane was washed and exposed to x-ray film. Films were scanned, and bands were quantitated using Adobe Photoshop.
Heparan Sulfate Purification and Analysis by Glycan Reductive Isotope Labeling-Liquid Chromatography/Mass Spectrometry
Heparan sulfate was isolated from whole liver as previously described (6, 19). Briefly, livers were homogenized in phosphate-buffered saline and treated overnight with Pronase to degrade protein followed by purification of glycopeptides by anion exchange chromatography using DEAE-Sepharose (GE Healthcare ). The columns were washed with 0.25 m sodium chloride in 25 mm sodium acetate buffer, and the glycans were eluted with 1 m sodium chloride in buffer.
Disaccharide analysis was carried out using glycan reductive isotope labeling-liquid chromatography/mass spectrometry as described (20). Briefly, heparan sulfate chains were completely depolymerized with heparin lyases I, II, and III, and the resulting disaccharides were labeled with [12C]aniline. The labeled mixture of disaccharides was combined with 25 pmol each of disaccharide standards labeled with [13C]aniline, and the sample was subjected to reversed phase ion-pairing liquid chromatography followed by mass spectrometry to separate and identify individually labeled disaccharides. Ratiometric analysis of the 12C- and 13C-labeled disaccharides provided a quantitative way to determine the level of each disaccharide. The number of N-, 2-O-, and 6-O-sulfate groups was determined by summing each disaccharide containing the individual group and is expressed per 100 disaccharides.
Lipoprotein Analysis
Lipids were measured in blood samples drawn from overnight-fasted animals by retro-orbital sinus bleeds. Total cholesterol and triglyceride levels were determined enzymatically (Cobas Mira; Roche Diagnostics) and cholesterol high performance reagent (Roche Diagnostics) and triglyceride-SL (Diagnostic Chemicals Ltd.) kits.
Different lipoprotein subfractions were prepared from blood drawn by cardiac puncture and sequential preparative ultracentrifugation. Chylomicrons, VLDL, and their remnants were prepared from pooled plasma samples (n = 10 mice, volume = 5 ml) by centrifugation for 12 h at 45,000 × rpm in a Beckman 50.3Ti rotor (δ = 1.006 g/ml). Intermediate density lipoproteins were then collected from the infranatant fluid by adjusting the density to δ = 1.019 g/ml with sodium bromide. Low and high density lipoproteins were collected between δ = 1.019–1.063 and 1.063–1.21g/ml, respectively. The isolated lipoproteins were dialyzed against phosphate-buffered saline (21) and analyzed for lipid content.
VLDL Plasma Clearance
Human VLDL (δ < 1.006g/ml) was isolated from healthy, fasting volunteers by ultracentrifugation. Mice were fasted for 6 h and injected via the tail vein with 20 μg of human VLDL protein. Serial samples were taken by retro-orbital sinus bleeds at 2, 10, 30, 60, and 120 min after injection. The amount of human VLDL remaining in the plasma was determined by sandwich enzyme-linked immunosorbent assay, utilizing monoclonal antibody MB47 specific for human apoB-100 (22). MB47 does not bind to murine apoB-100. U-bottom 96-well plates were coated overnight with MB47 at 5 μg/ml in Tris-buffered saline followed by the addition of nonsaturating amounts of plasma (typically, 1:100 dilution of mouse plasma) to capture human VLDL. Bound VLDL was detected using biotinylated goat anti-human apoB-100 (BIODESIGN International) followed by alkaline phosphatase-labeled NeutrAvidin (Pierce). Plates were developed with Lumi-Phos 530 (Lumigen) and read in a DYNEX Technologies MLX Microtiter Plate Luminometer. Half-times were calculated using linear regression to extrapolate the clearance rates.
Vitamin A Fat Tolerance Testing
Vitamin A fat tolerance testing was done as described (23). Briefly, 27 μCi of [11,12-3H]retinol (44.4Ci/mmol; PerkinElmer Life Sciences) in ethanol was mixed with 1 ml of corn oil (Sigma). Each mouse received 200 μl of the mixture by oral gavage. Blood was sampled at the times indicated by retro-orbital sinus bleed, and radioactivity was measured in triplicate (10 μl serum) by scintillation counting.
Lipoprotein Uptake by Hepatocytes
Hepatocytes were isolated as described (24). Human VLDL (δ < 1.006 g/ml, 2.1 mg of protein) was iodinated using 3 mCi and Na125I (1.9 × 102 cpm/ng, PerkinElmer Life Sciences) using Iodogen (25). Under these conditions, ∼13% of the label was lipid-soluble. Isolated hepatocytes were treated with the indicated concentrations of 125I-VLDL in Dulbecco's modified Eagle's medium with 2.5 mg/ml lipoprotein-free serum (Biomedical Technologies, Inc., Boston, MA). Cells were incubated at 4 or 37 °C for 1 h, rinsed three times with phosphate-buffered saline, and then solubilized with 0.1 m NaOH. Protein concentration was determined by the Bradford assay (Bio-Rad), and radioactivity was measured by γ-counting. Degradation of 125I-VLDL was measured by precipitation of 0.25 ml of conditioned media with an equal volume of 50% trichloroacetic acid (26). After 30 min at 4 °C, the samples were centrifuged for 30 min. The supernatant was placed in a fresh glass tube, and 5 μl of 40% potassium iodide was added as a carrier. Hydrogen peroxide (30%) was added, and lipid-soluble material was extracted by the addition of 2 ml of chloroform. An aliquot of the upper aqueous phase containing iodotyrosine was counted as a measure of degradation.
In competition experiments, wild type hepatocytes were treated simultaneously with varying concentrations of heparin or chemically desulfated heparins and 125I-VLDL at 4 °C for 1 h. Porcine intestinal heparin (Mr = 12,000–15,000) was a kind gift from Dr. Patrick Shaklee (Scientific Protein Laboratories Inc., Milwaukee, WI). Desulfated heparins were from Neoparin (Alameda, CA). In some experiments wild type cells were incubated at 37 °C for 1.5 h with 15 milliunits of heparin lyases I, II, and III (a gift from Jian Liu, University of North Caroline). The media were then replaced with fresh media containing 125I-VLDL and heparin lyase I, II, and III. Binding and uptake were analyzed as described above.
Statistics
Statistical analyses were performed using PRISM (GraphPad Software). All data are expressed as the mean values ± S.D. unless otherwise indicated. Significance was determined using an unpaired Student's (two-tailed) t test unless indicated differently. Significance was taken as p < 0.05.
RESULTS
Conditional Targeting of Hs2st
Vertebrates contain one gene encoding Hs2st, which is ubiquitously expressed in all tissue examined to date. Mice with a gene-trap mutation of Hs2st (Hs2stgt/gt) die in the neonatal period, exhibiting bilateral renal agenesis and defects of the eye and the skeleton after midgestation (15). 2-O-Sulfated uronic acid residues were not detected in heparan sulfate preparations from various organs of Hs2stgt/gt mice, demonstrating a complete loss of function of Hs2st and confirming that only one Hs2st isozyme exists in the mouse genome.
To study the role of Hs2st in liver physiology, we produced a loxP-flanked allele of Hs2st (“Experimental Procedures”). Quantitative PCR of Hs2st transcripts in Hs2stf/f mice showed that the presence of the loxP sites had no effect on transcript levels. Furthermore, enzyme assays of extracts prepared from Hs2stf/f embryos had normal activity, and the composition of heparan sulfate was identical to that observed in wild type animals. Interbreeding of Hs2st1f/+ mice yielded pups in the expected Mendelian ratio (23% +/+, 54% f/+, and 23% f/f, n = 70), indicating that the presence of the loxP recombination sites has no deleterious effect on survival.
The conditional allele could be deleted by cross-breeding Hs2stf/+ and Zp3Cre mice, which led to heterozygosity at the locus in oocytes (see the supplemental data). Subsequently, inbreeding of heterozygotes showed that homozygous null animals (Hs2st−/−) suffered from kidney agenesis and died as neonates, exactly as described previously for the gene trap-targeted mice (15). Analysis of Hs2st enzyme activity and disaccharide analyses of Hs2st−/− neonates showed that homozygous null embryos were grossly defective in 2-O-sulfation compared with the wild type (supplemental Figs. S1 and S2).
Hepatocyte-specific Hs2st Gene Disruption Alters Heparan Sulfate in the Liver
To delete expression of Hs2st in hepatocytes, Hs2stf/f were cross-bred with transgenic mice expressing Cre recombinase under control of the rat albumin promoter (AlbCre) to drive selective inactivation of the gene in hepatocytes (18). Expression of the AlbCre transgene occurs after postnatal day 10. From these breedings we established a colony of hepatocyte-specific Hs2st knock-out mice (Hs2stf/fAlbCre+) with littermate controls of genotype Hs2stf/fAlbCre−. Hs2stf/fAlbCre+ animals had normal litters with the expected Mendelian ratio of genotypes, indicating loss of Hs2st did not lead to any gross deleterious effects. All genotypes were backcrossed more than 10 generations with C57Bl/6J mice prior to further studies.
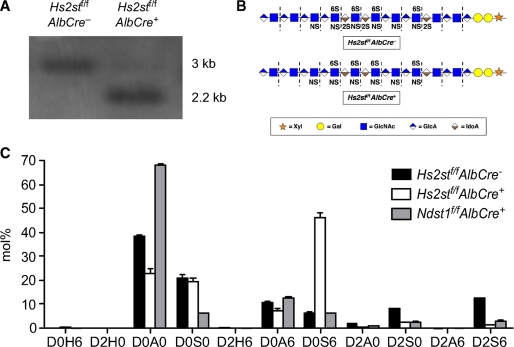
Hepatocytes were isolated from adult mice to measure the extent of Hs2st inactivation by Southern blotting. Cre-mediated recombination deleted exon 1, removing an EcoRI restriction site and yielding a fragment of 2.2 kb, whereas the wild type allele generated a 3.0 kb fragment. DNA from hepatocytes of 8-week old homozygous mutant mice (Hs2stf/f AlbCre+) showed that ∼90% of the Hs2stf/f alleles had undergone recombination (Fig. 1A).
FIGURE 1.
Hs2st conditional knock-out. A, hepatocyte genomic DNA was digested with EcoRI and analyzed by Southern blotting using a probe specific for Hs2st alleles (“Experimental Procedures”). The deleted allele gave a fragment of 2.2 kb, and the wild type allele gave a fragment of 3 kb. Quantification of the bands indicated 90–95% recombination had occurred in AlbCre+ hepatocytes. B, heparan sulfate chains from wild type and Hs2stf/fAlbCre+ mice are depicted using a standard symbol nomenclature for the individual sugar residues (71). The dashed lines indicate the sites where heparin lyases cleave the chains into individual disaccharides. C, heparan sulfate from livers obtained from Hs2stf/fAlbCre− (black bars), Hs2stf/fAlbCre+ (open bars), and Ndst1f/fAlbCre+ (shaded bars) mice were digested with heparin lyases I, II, and III, and the disaccharides were resolved by Glycan reductive isotope labeling liquid chromatography/mass spectrometry (“Experimental Procedures”) (n = 2 per genotype). D0H6, ΔUA-GlcNH2-6S; D2H0, ΔUA2S-GlcNH2; D0A0, ΔUA-GlcNAc; D0S0, ΔUA-GlcNS; D2H6, ΔUA2S-GlcNH26S; D0A6, ΔUA-GlcNAc6S; D0S6, ΔUA-GlcNS6S; D2A0, ΔUA2S-GlcNAc; D2S0, ΔUA2S-GlcNS; D2A6, ΔUA2S-GlcNAc6S; D2S6, ΔUA2S-GlcNS6S. ΔUA = 4,5-unsaturated uronic acid (20).
Inactivation of Hs2st induced significant changes in liver heparan sulfate composition. Heparan sulfate was isolated from whole livers of adult mice (n = 3 per genotype) and depolymerized into its constituent disaccharides by bacterial heparin lyases (Fig. 1B). The disaccharides were then quantitated using an aniline tagging method and liquid chromatography/mass spectrometry as described (20) (Fig. 1C). The levels of various sulfate esters (glucosamine N-sulfate, uronic acid 2-O-sulfate and glucosamine 6-O-sulfate) were then determined by summing the number of disaccharides bearing each type of modification (Table 1). The level of 2-O-sulfation of uronic acids (sum of D2H0, D2H6, D2A0, D2S0 and D2S6) decreased ∼5-fold, from 21 2-O-sulfate groups/100 disaccharides in wild type to 4 in the mutant. The residual sulfation arose presumably from incomplete inactivation of the gene and the presence of endothelial cells and other cell types in whole liver that did not undergo recombination (cf. Fig. 1A). Interestingly, the number of N-sulfate groups increased in Hs2stf/f AlbCre+ mice to 69/100 disaccharides from 48/100 disaccharides in the wild type and the extent of glucosamine 6-O-sulfation increased as well, from 19 sulfates/100 disaccharides in the wild type to 48/100 disaccharides in the mutant. These results are similar to those observed previously in mice carrying a systemic null allele of Hs2st (27) and Chinese hamster ovary cells lacking the uronyl 2-O-sulfotransferase (28). For comparison, we examined the composition of Ndst1f/f AlbCre+ mice using mass spectrometry (Fig. 1C and Table 1). Livers from Ndst1f/f AlbCre+ mice exhibited reduced levels of N-sulfation (18 sulfates/100 disaccharides, 2-O-sulfation (6/100 disaccharides) and 6-O-sulfation (9/100 disaccharides).
TABLE 1.
Distribution of sulfate groups in wild type and mutant hepatocytes
The amount of each class of sulfate group was determined from the disaccharide composition shown in Fig. 1C.
| Sulfate groups per 100 disaccharides |
|||
|---|---|---|---|
| Hs2stf/fAlbCre− | Hs2stf/fAlbCre+ | Ndst1f/fAlbCre+ | |
| N-Sulfate | 49 ± 0.8 | 69 ± 0.7 | 18 ± 0.7 |
| 2-O-Sulfate | 21 ± 0.2 | 4 ± 0.1 | 6 ± 0.2 |
| 6-O-Sulfate | 19 ± 0.7 | 48 ± 2.6 | 9 ± 0.4 |
Hs2stf/fAlbCre+ Mice Accumulate Triglyceride-rich Lipoproteins
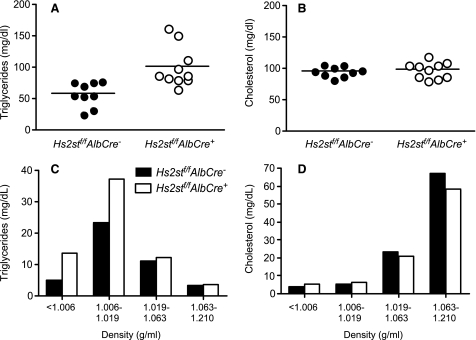
Analysis of plasma samples from fasted 8-week-old mice showed that total plasma triglyceride levels were ∼2-fold higher in Hs2st-deficient mice (58 ± 7 mg/dl in wild type Hs2stf/fAlbCre− mice versus 100 ± 10 mg/dl in the mutant; n = 10; p < 0.003) (Fig. 2A). Total plasma cholesterol was not affected (96 ± 3 mg/dl versus 98 ± 4; n = 10; p = 0.62) (Fig. 2B). Buoyant density ultracentrifugation of fasting plasma lipoproteins from Hs2stf/fAlbCre+ and Hs2stf/fAlbCre− mice showed that triglyceride-rich lipoproteins of δ < 1.006 and 1.006–1.019 g/ml accumulated in the mutant (Fig. 2C). There was no significant difference in cholesterol in these fractions and only a minor reduction in high density lipoprotein cholesterol (Fig. 2D). Analysis of lipoproteins of density δ < 1.006 g/ml by SDS-PAGE showed the presence of apoB-48, apoB-100, apoE, and apoCs characteristic of VLDL particles and chylomicron remnants (data not shown).
FIGURE 2.
Hypertriglyceridemia in Hs2stf/fAlbCre+ mice. Total triglycerides (A) and total cholesterol (B) were measured in plasma samples (n = 9 Hs2stf/fAlbCre−, n = 10 Hs2stf/fAlbCre+, mixed males and females). Horizontal bars indicate mean values. Triglyceride (C) and cholesterol (D) content of lipoproteins were fractionated according to density by preparative ultracentrifugation. Each value is from a set of pooled plasma derived from 10 mice.
Hs2st-deficient Mice Exhibit Slower Plasma Clearance of Triglycerides and VLDL
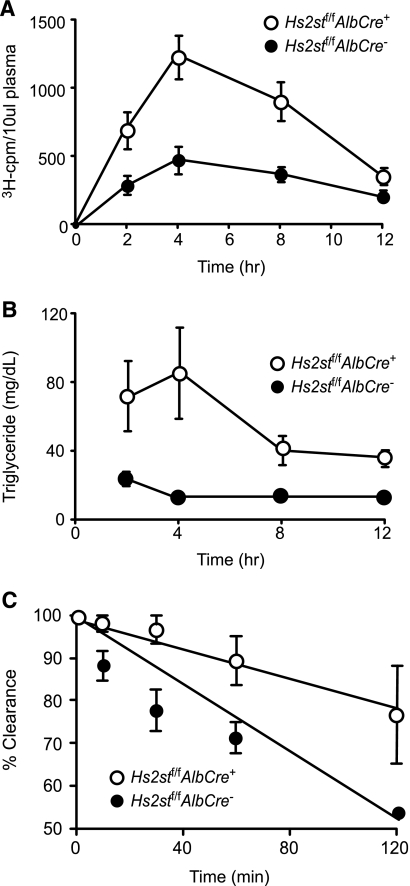
Clearance rates of intestinally derived triglyceride-rich lipoproteins were measured in mice lacking hepatocyte Hs2st (Fig. 3A). Overnight-fasted mice were given a bolus mixture of [3H]retinol and corn oil by gavage, and blood was sampled from the retroorbital sinus at various time points. The retinol is converted to retinyl esters in the intestinal mucosa and packaged with dietary lipids into chylomicron particles. After hydrolysis of the triglycerides in the periphery by lipoprotein lipase, the remnant particles containing [3H]retinyl esters are cleared in the liver. Thus, the disappearance of [3H]retinyl esters from the circulation is a measure of hepatic clearance. The retinyl ester excursion curves showed that Hs2stf/fAlbCre+ mice cleared intestinally derived lipoproteins at a significantly slower rate than Hs2stf/fAlbCre− mice (area under the curve = 4123 ± 1214 for the wild type versus 9554 ± 2074 for the mutant; p = 0.0033), but most of the particles were cleared by 12 h (Fig. 3A). Plasma triglycerides also cleared more slowly in the mutant (Fig. 3B).
FIGURE 3.
Hs2stf/fAlbCre+ mice exhibit delayed plasma clearance of dietary triglycerides and VLDL. A, retinyl ester excursions were measured at the times indicated in Hs2stf/fAlbCre+ mice (open circles, n = 3) and Hs2stf/fAlbCre− (closed circles, n = 3). Fasted animals were given 200 μl of corn oil containing [3H]retinol by oral gavage. Blood samples were taken at the indicated times, and radioactivity in 10 μl serum samples was determined by liquid scintillation counting. Clearance was significantly delayed in the Hs2stf/fAlbCre+ mice (p < 0.0001). B, plasma triglyceride levels were measured. C, plasma clearance of human VLDL was measured by enzyme-linked immunosorbent assay using human apoB-100-specific monoclonal antibody MB47 (see “Experimental Procedures”). Filled circles, (Hs2stf/fAlbCre− mice; n = 6 mice); open circles, (Hs2stf/fAlbCre+ mice; n = 6 mice). The difference in t½ between the genotypes was significant (p = 0.0007).
Clearance of exogenously provided VLDL was also affected based on experiments in which a bolus of human VLDL was injected intravenously. In these experiments the circulating level of human VLDL was determined by enzyme-linked immunosorbent assay using monoclonal antibody MB47, which is specific for human apoB100 (22) (Fig. 3C). The rate of turnover was reduced in mutant mice, yielding apparent half-lives of 125 ± 10 min in wild type mice and 259 ± 117 min in mutant mice (calculated by extrapolation from the curves, n = 6 per genotype; p < 0.0001). Although the absolute rates of clearance differ in these experiments from previous values obtained in Ndst1f/fAlbCre+ mice, the -fold difference between mutant and wild type is approximately the same (6). The source of variation is unknown but may be related to differences in VLDL preparation and nutritional status of the animals. Nevertheless, these experiments show that hepatocyte Hs2st plays a crucial role in the clearance of both intestinal- and liver-derived lipoprotein particles.
Binding and Uptake of 125I-VLDL Particles Is Reduced in Hs2st-deficient Hepatocytes
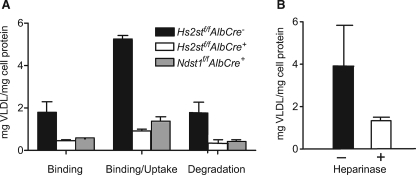
To determine how altering Hs2st might affect binding and uptake of VLDL in isolated hepatocytes, cells were isolated from Hs2stf/fAlbCre− and Hs2stf/fAlbCre+ mice and incubated with 125I-VLDL. Hs2stf/fAlbCre+ hepatocytes showed decreased binding at 4 °C compared with Hs2stf/fAlbCre− mice (1.8 ± 0.8 μg VLDL/mg of cell protein in the wild type versus 0.5 ± 0.1 in the mutant) and reduced binding/uptake of 125I-VLDL at 37° (5 ± 0.3 μg VLDL/mg of cell protein versus 0.9 ± 0.1 μg VLDL/mg cell protein, respectively) (Fig. 4A). Significant degradation of VLDL occurred during the 1-h incubation based on recovery of non-chloroform extractable, acid-soluble 125I counts (iodotyrosine) in the growth medium. Hs2stf/fAlbCre+ hepatocytes showed a dramatic decrease in degradation as well. Similar reduction in binding, uptake, and degradation were obtained when Ndst1f/fAlbCre+ was assayed (Fig. 4A), and the differences between the two types of mutants were not significant (p = 0.1345, p = 0.768, p = 0.928 for binding, binding/uptake, and degradation, respectively). Binding and uptake of 125I-VLDL particles was also measured in wild type hepatocytes after treatment with heparin lyases to remove cell surface heparan sulfate (Fig. 4B). A 3-fold decrease in uptake compared with untreated hepatocytes was observed (4 ± 2 μg VLDL/mg of cell protein versus 1.3 ± 0.2 μg VLDL/mg of cell protein), consistent with the studies of the mutants.
FIGURE 4.
Hs2st mediates binding, uptake, and degradation of VLDL. A, hepatocytes from Hs2stf/AlbCre− (wild type, filled bars), Hs2stf/fAlbCre+ (open bars), and Ndst1f/fAlbCre+ (gray bars) were incubated with 125I-VLDL at 4 °C, and the extent of binding was measured. Binding was reduced 3.7-fold in the mutants, and the difference between Hs2stf/fAlbCre+ and Ndst1f/fAlbCre+ hepatocytes was not significant (p = 0.1345). Binding and uptake (measured at 37 °C) were decreased ∼5.5-fold in Hs2stf/fAlbCre+ hepatocytes (p < 0.01), and again, no difference in uptake and degradation was noted when the two mutants were compared (p = 0.768). Degradation also was reduced in the mutants, and the difference between the mutants was not significant (p = 0.928). B, Hs2stf/fAlbCre− hepatocytes were incubated with heparin lyases I, II, and III, resulting in a 3-fold decrease in binding at 4 °C compared with untreated hepatocytes (p < 0.01).
Heparin Inhibits Binding and Uptake of 125I-VLDL
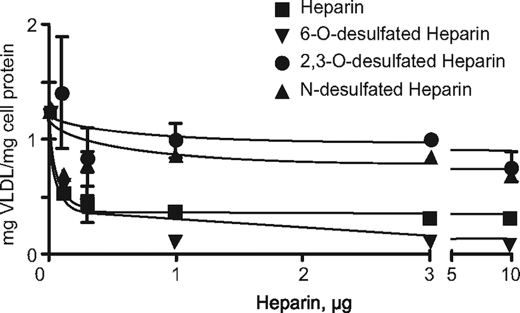
The altered binding of VLDL to heparan sulfate-deficient cells predicted that the addition of heparin, which is more highly sulfated than hepatocyte heparan sulfate, should block binding and uptake. Incubation of hepatocytes with increasing concentrations of heparin resulted in a decrease in binding (Fig. 5), with an extracted IC50 value of 0.1 μg/ml. Both N-desulfated and 2,3-O-desulfated heparin had only mild effects on VLDL binding, which supports the genetic studies showing that these groups were crucial for clearance in vivo and binding, uptake, and degradation in isolated cells. In contrast, 6-O-desulfated heparin had nearly the same potency as native heparin, with a calculated IC50 value of 0.1 μg/ml, suggesting that 6-O-sulfation is not required for binding of VLDL.
FIGURE 5.
Inhibition of binding by desulfated heparins. Hepatocytes from wild type animals were incubated with 125I-VLDL and various heparin preparations at 4 °C. The difference between 2,3-O-desulfated heparin and N-desulfated heparin or heparin and 6-O-desulfated heparin was not significant (p = 0.2 and p = 0.66, respectively). The difference between 2,3-O-desulfated heparin and heparin was significant (p = 0.016).
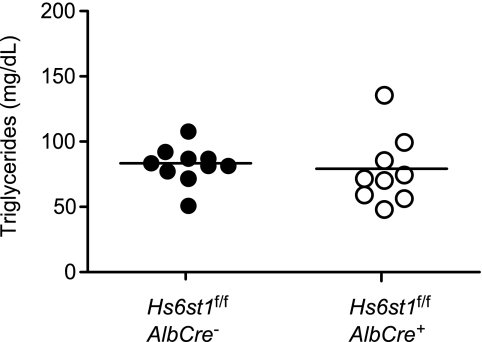
Sugaya et al. (29) recently showed that systemic deletion of Hs6st1 reduced the extent of 6-O-sulfation of hepatic heparan sulfate. Because the mutation results in embryonic lethality, litter runts, and multiple organ defects, we obtained Hs6st1f/f mice described by Izvolsky et al. (17) and cross-bred them to AlbCre mice to delete the gene specifically in the liver. Hs6st1f/fAlbCre+ mice were viable and did not accumulate plasma triglycerides (Fig. 6), in support of the heparin-inhibition studies (Fig. 5).
FIGURE 6.
Normal plasma triglycerides in Hs6st1-deficient mice. Fasting plasma samples were obtained from Hs6st1f/fAlbCre+ and Hs6st1f/fAlbCre− mice, and total triglycerides were measured (n = 10, Hs6st1f/fAlbCre−; n = 9 Hs6st1f/fAlbCre+). No significant difference in plasma triglycerides was noted.
DISCUSSION
Cell-surface heparan sulfate proteoglycans have been hypothesized to act as remnant receptors acting either independently or in concert with LDL receptor-related protein (30–39). In cell culture models, heparan sulfate proteoglycans can interact directly with lipoproteins bearing apoE, apoB, lipoprotein lipase, hepatic lipase, and endothelial lipase and facilitate their internalization by endocytosis (30, 36, 40). Infusion of mice with heparinase, heparin, suramin, or lactoferrin to neutralize heparan sulfate reduces plasma clearance rates and hepatic uptake of labeled VLDL (41–43), suggesting that the interaction of particles with heparan sulfate could be important in vivo. Recently, we provided genetic evidence that hepatocyte heparan sulfate fulfills this function in vivo by altering its biosynthesis through tissue-specific inactivation of Ndst1 (6). As this mutation affected the overall sulfation of the chain, the requirement for specific classes of sulfate groups along the chain was left unresolved.
In this report, we have advanced these studies by demonstrating that clearance depends on 2-O-sulfated uronic acids in the chain. This conclusion is based on accumulation of plasma triglycerides in mutant mice, reduced binding, uptake, and degradation of lipoproteins in hepatocytes isolated from the mutant and loss of inhibitory activity of heparin by removal of 2-O-sulfate groups. Clearance appears to depend on N-sulfation based on loss of inhibitory activity of N-desulfated, re-N-acetylated heparin. In contrast, the available data suggest that clearance occurs independently of 6-O-sulfate groups. The basis for this conclusion is 3-fold; (i) removal of 6-O-sulfate groups from heparin had no effect on its capacity to block binding, (ii) inactivation of Hs6st1 had no effect on plasma triglycerides, although this mutation does not result in complete loss of 6-O-sulfate groups in liver heparan sulfate (29), and (iii) inactivation of Hs2st results in increased levels of 6-O-sulfation (and N-sulfation), yet hypertriglyceridemia was manifest despite the higher charge density of the chains. To confirm these findings, additional studies are under way to accentuate the loss of 6-O-sulfate groups by crossing Hs6st1f/fAlbCre+ mice with Hs6st2−/− mice (29).
Several mechanisms have been suggested to explain how heparan sulfate interactions with apolipoproteins and/or enzymes affect lipoprotein processing in the liver. Mahley and Ji (44) have suggested that remnant lipoprotein particles entering the space of Disse bind to heparan sulfate proteoglycans via apoE, resulting in their sequestration before further processing and receptor-mediated endocytosis. Lipoprotein lipase and hepatic lipase also can act in this way, as both enzymes can bind to heparan sulfate and to the lipoprotein particles, thus acting as bridging molecules (3, 44). Triglyceride-rich lipoproteins also contain apoB and apoAV, which also bind to heparin. Apparently, one or more of these ligands contains a binding site that either involves direct contact with N-sulfoglucosamine units and 2-O-sulfated uronic acids or a conformation of the heparan sulfate chain dictated by the positioning of these groups. Many of these ligands can interact with heparan sulfate oligosaccharides of variable sulfation and most avidly to segments enriched in the trisulfated disaccharide, GlcNS6S-IdoA2S (Table 2). However, the available structural data does not allow us to determine which if any of these proteins serves as the relevant ligand for heparan sulfate-dependent clearance.
TABLE 2.
Apolipoproteins and lipases that bind heparin/heparan sulfate
| Ligand | Binding specificity | References |
|---|---|---|
| ApoE | Sequences rich in GlcNS6S-IdoA2S; heparin binding domains mapped | 55–58 |
| ApoB | Unknown, but interacts with heparin and heparin; heparin binding domains mapped | 59–61 |
| Hepatic lipase | Heparin oligosaccharides; heparin binding domains mapped | 62–64 |
| Lipoprotein lipase | Highest affinity for sequences rich in GlcNS6S-IdoA2S; modestly sulfated sequences | 65–67> |
| Endothelial lipase | Interacts with heparan sulfate | 68, 69 |
| ApoA-V | Interacts with heparin | 70 |
One of the interesting effects of altering 2-O-sulfation of heparan sulfate in hepatocytes is the dramatic increase in N-sulfation and 6-O-sulfation, which augments the overall charge of the chain. Despite “compensation,” triglycerides accumulate in the mutant, and VLDL uptake is reduced in isolated hepatocytes, suggesting that the ligand on the lipoprotein particles exhibits specificity for the arrangement of sulfate residues. Similar compensation occurs in Chinese hamster ovary cells containing a mutation in Hs2st (28), Drosophila melanogaster (45), Caenorhabditis elegans (46),5 and mice (27). The mechanism responsible for these coordinated changes in sulfation remains unknown. GlcNAc N-deacetylation and N-sulfation is generally thought to precede all of the other modification reactions and creates the preferred substrate for HsC5epi, which epimerizes glucuronic acids located to the reducing side of the N-sulfoglucosamine units to l-iduronic (7). The resulting IdoA then undergoes sulfation at C2 by Hs2st, although the enzyme can also work less efficiently on glucuronic acids (47–49). Although we have not measured GlcNAc N-deacetylase/N-sulfotransferase activity in mutant hepatocytes, in previous studies we showed that N-sulfotransferase activity in Hs2st-deficient CHO cells was unchanged, suggesting that the presence of 2-O-sulfated residues somehow decreases the rate or extent of the N-deacetylase/N-sulfotransferase reaction (28). This explanation suggests that the length of the N-sulfated segments (NS or S domains) might be greater in the absence of Hs2st, which is borne out by structural analysis of the heparan sulfate chains in the mouse gene trap mutant of Hs2st (27).
The selective elevation of plasma triglycerides without accumulation of plasma cholesterol in Hs2st-deficient mice is similar to that observed in patients with hypertriglyceridemia associated with elevated levels of VLDL particles (50). These patients persistently exhibit elevated plasma triglyceride levels, but plasma cholesterol and phospholipid levels usually remain close to normal levels. Mutations that affect triglyceride levels in humans include genes encoding Lpl, apoAV, apoCII, apoCIII, and GPIHBP1 (51–54). We can now add Hs2st to the growing list of genes involved in heparan sulfate proteoglycan formation that could affect triglyceride clearance. Patients predisposed to mild but clinically relevant hyperlipidemias should be screened for changes in 2-O-sulfation and polymorphisms in the relevant genes involved in 2-O-sulfation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL57345 and GM33063 (to J. D. E.). This work was also supported by American Heart Association Grant 0735038N (to J. R. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
H. Bülow, personal communication.
- VLDL
- very low density lipoprotein
- AlbCre
- transgene consisting of the rat albumin promoter fused to the Cre recombinase
- Ndst1
- N-acetylglucosamine N-deacetylase-N-sulfotransferase-1
- Hs2st
- heparan sulfate uronyl 2-O-sulfotransferase
- Hs6st1
- heparan sulfate glucosaminyl 6-O-sulfotransferase
- LDL
- low density lipoprotein.
REFERENCES
- 1.Conrad H. E. (1998) Heparin-binding Proteins, Academic Press, San Diego, CA [Google Scholar]
- 2.Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 3.Williams K. J., Fuki I. V. (1997) Curr. Opin. Lipidol. 8, 253–262 [DOI] [PubMed] [Google Scholar]
- 4.Belting M. (2003) Trends Biochem. Sci. 28, 145–151 [DOI] [PubMed] [Google Scholar]
- 5.Elson-Schwab L., Garner O. B., Schuksz M., Crawford B. E., Esko J. D., Tor Y. (2007) J. Biol. Chem. 282, 13585–13591 [DOI] [PubMed] [Google Scholar]
- 6.MacArthur J. M., Bishop J. R., Stanford K. I., Wang L., Bensadoun A., Witztum J. L., Esko J. D. (2007) J. Clin. Invest. 117, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 8.Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bame K. J., Lidholt K., Lindahl U., Esko J. D. (1991) J. Biol. Chem. 266, 10287–10293 [PubMed] [Google Scholar]
- 11.Ishihara M., Guo Y., Swiedler S. J. (1993) Glycobiology 3, 83–88 [DOI] [PubMed] [Google Scholar]
- 12.Toyoda H., Kinoshita-Toyoda A., Fox B., Selleck S. B. (2000) J. Biol. Chem. 275, 21856–21861 [DOI] [PubMed] [Google Scholar]
- 13.Ringvall M., Ledin J., Holmborn K., van Kuppevelt T., Ellin F., Eriksson I., Olofsson A. M., Kjellen L., Forsberg E. (2000) J. Biol. Chem. 275, 25926–25930 [DOI] [PubMed] [Google Scholar]
- 14.Ledin J., Staatz W., Li J. P., Götte M., Selleck S., Kjellén L., Spillmann D. (2004) J. Biol. Chem. 279, 42732–42741 [DOI] [PubMed] [Google Scholar]
- 15.Bullock S. L., Fletcher J. M., Beddington R. S., Wilson V. A. (1998) Genes Dev. 12, 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orban P. C., Chui D., Marth J. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6861–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izvolsky K. I., Lu J., Martin G., Albrecht K. H., Cardoso W. V. (2008) Genesis 46, 8–18 [DOI] [PubMed] [Google Scholar]
- 18.Postic C., Magnuson M. A. (2000) Genesis 26, 149–150 [DOI] [PubMed] [Google Scholar]
- 19.Grobe K., Inatani M., Pallerla S. R., Castagnola J., Yamaguchi Y., Esko J. D. (2005) Development 132, 3777–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence R., Lu H., Rosenberg R. D., Esko J. D., Zhang L. (2008) Nat. Methods 5, 291–292 [DOI] [PubMed] [Google Scholar]
- 21.Dulbecco R., Vogt M. (1954) J. Exp. Med. 99, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young S. G., Smith R. S., Hogle D. M., Curtiss L. K., Witztum J. L. (1986) Clin. Chem. 32, 1484–1490 [PubMed] [Google Scholar]
- 23.Ishibashi S., Perrey S., Chen Z., Osuga J., Shimada M., Ohashi K., Harada K., Yazaki Y., Yamada N. (1996) J. Biol. Chem. 271, 22422–22427 [DOI] [PubMed] [Google Scholar]
- 24.Horton J. D., Shimano H., Hamilton R. L., Brown M. S., Goldstein J. L. (1999) J. Clin. Invest. 103, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salacinski P. R., McLean C., Sykes J. E., Clement-Jones V. V., Lowry P. J. (1981) Anal. Biochem. 117, 136–146 [DOI] [PubMed] [Google Scholar]
- 26.Williams K. J. (2001) Methods Mol. Biol. 171, 457–477 [DOI] [PubMed] [Google Scholar]
- 27.Merry C. L., Bullock S. L., Swan D. C., Backen A. C., Lyon M., Beddington R. S., Wilson V. A., Gallagher J. T. (2001) J. Biol. Chem. 276, 35429–35434 [DOI] [PubMed] [Google Scholar]
- 28.Bai X., Esko J. D. (1996) J. Biol. Chem. 271, 17711–17717 [DOI] [PubMed] [Google Scholar]
- 29.Habuchi H., Nagai N., Sugaya N., Atsumi F., Stevens R. L., Kimata K. (2007) J. Biol. Chem. 282, 15578–15588 [DOI] [PubMed] [Google Scholar]
- 30.Ji Z. S., Brecht W. J., Miranda R. D., Hussain M. M., Innerarity T. L., Mahley R. W. (1993) J. Biol. Chem. 268, 10160–10167 [PubMed] [Google Scholar]
- 31.Ji Z. S., Fazio S., Mahley R. W. (1994) J. Biol. Chem. 269, 13421–13428 [PubMed] [Google Scholar]
- 32.Ji Z. S., Lauer S. J., Fazio S., Bensadoun A., Taylor J. M., Mahley R. W. (1994) J. Biol. Chem. 269, 13429–13436 [PubMed] [Google Scholar]
- 33.Ji Z. S., Mahley R. W. (1994) Arterioscler. Thromb. 14, 2025–2031 [DOI] [PubMed] [Google Scholar]
- 34.Al-Haideri M., Goldberg I. J., Galeano N. F., Gleeson A., Vogel T., Gorecki M., Sturley S. L., Deckelbaum R. J. (1997) Biochemistry 36, 12766–12772 [DOI] [PubMed] [Google Scholar]
- 35.Fuki I. V., Kuhn K. M., Lomazov I. R., Rothman V. L., Tuszynski G. P., Iozzo R. V., Swenson T. L., Fisher E. A., Williams K. J. (1997) J. Clin. Invest. 100, 1611–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Z. S., Dichek H. L., Miranda R. D., Mahley R. W. (1997) J. Biol. Chem. 272, 31285–31292 [DOI] [PubMed] [Google Scholar]
- 37.Zeng B. J., Mortimer B. C., Martins I. J., Seydel U., Redgrave T. G. (1998) J. Lipid Res. 39, 845–860 [PubMed] [Google Scholar]
- 38.Fuki I. V., Iozzo R. V., Williams K. J. (2000) J. Biol. Chem. 275, 25742–25750 [DOI] [PubMed] [Google Scholar]
- 39.Wilsie L. C., Orlando R. A. (2003) J. Biol. Chem. 278, 15758–15764 [DOI] [PubMed] [Google Scholar]
- 40.Williams K. J., Fless G. M., Petrie K. A., Snyder M. L., Brocia R. W., Swenson T. L. (1992) J. Biol. Chem. 267, 13284–13292 [PubMed] [Google Scholar]
- 41.Ji Z. S., Sanan D. A., Mahley R. W. (1995) J. Lipid Res. 36, 583–592 [PubMed] [Google Scholar]
- 42.Mortimer B. C., Beveridge D. J., Martins I. J., Redgrave T. G. (1995) J. Biol. Chem. 270, 28767–28776 [DOI] [PubMed] [Google Scholar]
- 43.Windler E., Greeve J., Robenek H., Rinninger F., Greten H., Jäckle S. (1996) Hepatology 24, 344–351 [DOI] [PubMed] [Google Scholar]
- 44.Mahley R. W., Ji Z. S. (1999) J. Lipid Res. 40, 1–16 [PubMed] [Google Scholar]
- 45.Kamimura K., Koyama T., Habuchi H., Ueda R., Masu M., Kimata K., Nakato H. (2006) J. Cell Biol. 174, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bülow H. E., Hobert O. (2004) Neuron 41, 723–736 [DOI] [PubMed] [Google Scholar]
- 47.Kusche M., Lindahl U. (1990) J. Biol. Chem. 265, 15403–15409 [PubMed] [Google Scholar]
- 48.Rong J., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. (2001) Biochemistry 40, 5548–5555 [DOI] [PubMed] [Google Scholar]
- 49.Bethea H. N., Xu D., Liu J., Pedersen L. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18724–18729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Havel R. J., Kane J. P. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Sly W. S., Childs B., Beaudet A. L., Valle D., Kinzler K. W., Vogelstein B. eds) 8th Ed., pp. 2705–2716, McGraw-Hill Inc., New York [Google Scholar]
- 51.Otarod J. K., Goldberg I. J. (2004) Curr. Atheroscler. Rep. 6, 335–342 [DOI] [PubMed] [Google Scholar]
- 52.van Dijk K. W., Rensen P. C., Voshol P. J., Havekes L. M. (2004) Curr. Opin. Lipidol. 15, 239–246 [DOI] [PubMed] [Google Scholar]
- 53.Jakel H., Nowak M., Helleboid-Chapman A., Fruchart-Najib J., Fruchart J. C. (2006) Ann. Med. 38, 2–10 [DOI] [PubMed] [Google Scholar]
- 54.Beigneux A. P., Franssen R., Bensadoun A., Gin P., Melford K., Peter J., Walzem R. L., Weinstein M. M., Davies B. S., Kuivenhoven J. A., Kastelein J. J., Fong L. G., Dallinga-Thie G. M., Young S. G. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Libeu C. P., Lund-Katz S., Phillips M. C., Wehrli S., Hernáiz M. J., Capila I., Linhardt R. J., Raffaï R. L., Newhouse Y. M., Zhou F., Weisgraber K. H. (2001) J. Biol. Chem. 276, 39138–39144 [DOI] [PubMed] [Google Scholar]
- 56.Dong J., Peters-Libeu C. A., Weisgraber K. H., Segelke B. W., Rupp B., Capila I., Hernáiz M. J., LeBrun L. A., Linhardt R. J. (2001) Biochemistry 40, 2826–2834 [DOI] [PubMed] [Google Scholar]
- 57.Bazin H. G., Marques M. A., Owens A. P., 3rd, Linhardt R. J., Crutcher K. A. (2002) Biochemistry 41, 8203–8211 [DOI] [PubMed] [Google Scholar]
- 58.Cardin A. D., Weintraub H. J. (1989) Arteriosclerosis. 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 59.Goldberg I. J., Wagner W. D., Pang L., Paka L., Curtiss L. K., DeLozier J. A., Shelness G. S., Young C. S., Pillarisetti S. (1998) J. Biol. Chem. 273, 35355–35361 [DOI] [PubMed] [Google Scholar]
- 60.Borén J., Olin K., Lee I., Chait A., Wight T. N., Innerarity T. L. (1998) J. Clin. Invest. 101, 2658–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flood C., Gustafsson M., Richardson P. E., Harvey S. C., Segrest J. P., Borén J. (2002) J. Biol. Chem. 277, 32228–32233 [DOI] [PubMed] [Google Scholar]
- 62.Merchant Z. M., Erbe E. E., Eddy W. P., Patel D., Linhardt R. J. (1986) Atherosclerosis 62, 151–158 [DOI] [PubMed] [Google Scholar]
- 63.Yu W., Hill J. S. (2006) Biochem. Biophys. Res. Commun. 343, 659–665 [DOI] [PubMed] [Google Scholar]
- 64.Sendak R. A., Berryman D. E., Gellman G., Melford K., Bensadoun A. (2000) J. Lipid Res. 41, 260–268 [PubMed] [Google Scholar]
- 65.Parthasarathy N., Goldberg I. J., Sivaram P., Mulloy B., Flory D. M., Wagner W. D. (1994) J. Biol. Chem. 269, 22391–22396 [PubMed] [Google Scholar]
- 66.Larnkjaer A., Nykjaer A., Olivecrona G., Th⊘gersen H., Ostergaard P. B. (1995) Biochem. J. 307, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spillmann D., Lookene A., Olivecrona G. (2006) J. Biol. Chem. 281, 23405–23413 [DOI] [PubMed] [Google Scholar]
- 68.Jaye M., Lynch K. J., Krawiec J., Marchadier D., Maugeais C., Doan K., South V., Amin D., Perrone M., Rader D. J. (1999) Nat. Genet. 21, 424–428 [DOI] [PubMed] [Google Scholar]
- 69.Fuki I. V., Blanchard N., Jin W., Marchadier D. H., Millar J. S., Glick J. M., Rader D. J. (2003) J. Biol. Chem. 278, 34331–34338 [DOI] [PubMed] [Google Scholar]
- 70.Lookene A., Beckstead J. A., Nilsson S., Olivecrona G., Ryan R. O. (2005) J. Biol. Chem. 280, 25383–25387 [DOI] [PubMed] [Google Scholar]
- 71.Varki A., Cummings R., Esko J. D., Freeze H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, 2nd ed., pp. 12–13, Cold Spring Harbor Laboratories Press, New York: [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.