Abstract
Mycocypins, clitocypins and macrocypins, are cysteine protease inhibitors isolated from the mushrooms Clitocybe nebularis and Macrolepiota procera. Lack of sequence homology to other families of protease inhibitors suggested that mycocypins inhibit their target cysteine protease by a unique mechanism and that a novel fold may be found. The crystal structures of the complex of clitocypin with the papain-like cysteine protease cathepsin V and of macrocypin and clitocypin alone have revealed yet another motif of binding to papain like-cysteine proteases, which in a yet unrevealed way occludes the catalytic residue. The binding is associated with a peptide-bond flip of glycine that occurs before or concurrently with the inhibitor docking. Mycocypins possess a β-trefoil fold, the hallmark of Kunitz-type inhibitors. It is a tree-like structure with two loops in the root region, a stem comprising a six-stranded β-barrel, and two layers of loops (6 + 3) in the crown region. The two loops that bind to cysteine cathepsins belong to the lower layer of the crown loops, whereas a single loop from the crown region can inhibit trypsin or asparaginyl endopeptidase, as demonstrated by site-directed mutagenesis. These loops present a versatile surface with the potential to bind to additional classes of proteases. When appropriately engineered, they could provide the basis for possible exploitation in crop protection.
Introduction
Inhibition of foreign protease activity is a widespread defense mechanism in plants against their pests, pathogens, and parasites (1). Protein inhibitors of proteases are present in a variety of plant tissues. They can be deployed alone or together with a variety of small molecules (2). It has been known for a long time that the expression of protease inhibitors is increased in injured plant leaves (3) and that their expression can be induced as a response to attack by insects or pathogens (2).
Given the negative environmental effects of chemical pesticides used in crop protection, it is important to explore alternative approaches, such as the incorporation of genes encoding protease inhibitors into plants. Transgenic plants expressing various protease inhibitors have shown enhanced levels of insect resistance; however, the adaptive capacity of insect digestive proteases limits the use of single protease inhibitors (4, 5). The use of hybrid protease inhibitors with multiple inhibitory activity could, however, affect the functional properties of the fused inhibitors (6). Incorporation of genes encoding a range of protease inhibitors is to run the risk of deleterious modification of plants, but the use of a single protease inhibitor with versatile functionality could be the way forward.
With these in mind, we have undertaken structural and mechanistic studies of the cysteine protease inhibitors clitocypin (Clt)3 from mushroom Basidiomycetes Clitocybe nebularis and macrocypins (Mcp) from Macrolepiota procera. Based on the lack of sequence similarity to other protease inhibitors, they form separate protease inhibitor families, I48 and I85, in the MEROPS classification (7) and were named mycocypins. They are ≈17-kDa proteins exhibiting high thermal and broad pH range stability, with completely reversible unfolding (8, 9). They all inhibit endopeptidases from the papain family, such as papain, cathepsins L, V, S, and K, in the low nanomolar range, and exopeptidases with higher inhibition constants. They exhibit different inhibitory specificities. The cysteine protease asparaginyl endopeptidase (AEP, legumain) is inhibited in the low nanomolar range by macrocypin 1 and macrocypin 3, whereas clitocypin inhibits AEP in the higher nm range. Macrocypin 4 does not inhibit AEP, but in contrast to the others, it inhibits serine protease trypsin in the micromolar range (8, 9).
In this respect mycocypins are similar to cystatins and thyropins. Cystatins inhibit cysteine papain-like proteases and AEP. The crystal structures have revealed that cystatins bind to papain-like proteases with a wedge composed of three regions, N-terminal trunk and two β-hairpin loops (10, 11), whereas their binding geometry to AEP is still unknown. Thyropins inhibit papain-like cysteine and aspartic proteases (12, 13). The crystal structure of the p41 fragment bound to cathepsin L (14) has revealed similarity of the three binding loops to those of cystatins. The three-loop mode of binding is shared also by chagasin, the parasite inhibitor of papain-like proteases from Trypanosoma cruzi (15). Common to them all is the fact that the binding loops bind into the non-primed and primed substrate binding regions and occlude the catalytic cysteine residue but do not interact directly with it. Bauhinia bauhinioides cruzipain inhibitor BbCI (16), a Kunitz-type inhibitor belonging to the I3 MEROPS family, inhibits papain-like endopeptidases cruzipain and cathepsin L and serine protease trypsin but not endopeptidase cathepsin V or exopeptidases cathepsins B and X. The structure of BbCI protease complexes is not known, although it has been suggested that the same reactive loop is involved in inhibition of cysteine and serine proteases (17).
To gain an insight into the inhibitory mechanism of mycocypins, we have determined the crystal structures of the complex of clitocypin with the papain-like cysteine protease cathepsin V and of macrocypin 1 and clitocypin alone. The study has revealed yet another motif for binding to papain-like cysteine proteases that occludes the catalytic residue. The fold of mycocypins is based on a six-stranded β-barrel that composes the core of the β-trefoil fold, providing a versatile surface capable of binding to various protease types. We have identified their binding regions to AEP and trypsin by site-directed mutagenesis and shown that the loops binding to papain-like proteases are different from those binding to AEP and trypsin.
EXPERIMENTAL PROCEDURES
Protein Expression and Isolation
Natural clitocypin was purified from the fruiting bodies of Basidiomycetes C. nebularis (9). Macrocypin, clitocypin, methionine-containing clitocypin mutant (Clt-L82M, Clt-I89M), and their mutants (Mcp-1/4 (Mcp1 with β5-β6 loop of Mcp4), Mcp-4/1 (Mcp4 with β5-β6 loop of Mcp1), Mcp-ΔG25, Mcp-G25A, Mcp4-N74R, and Clt-ΔG24, Clt-G24A, Clt-N69K) were expressed in Escherichia coli (8, 18). Mutants were produced by PCR site-directed mutagenesis using the appropriate pET vectors as templates followed by digestion with DpnI (Fermentas) and recovery of the vectors containing mutated inserts (19). The selenomethionine mutant was produced using minimum autoinduction media with the addition of selenomethionine in E. coli BL384 cells (20). Cathepsin V was expressed in Pichia pastoris (21).
Crystallization
Recently we have reported crystallization conditions and phasing attempts using clitocypin purified from natural source (22). 1-ml drops of 15 mg/ml solution of clitocypin in 15 mm MES buffer, pH 6.0, gave crystals when mixed with 1 ml of crystallization buffer (50 mm monopotassium dihydrogen phosphate, 20% (w/v) polyethylene glycol 8000, pH 3.76) using the vapor diffusion method. A number of data sets with a variety of resolutions were collected. The structure could not be solved because of the high heterogeneity of the natural clitocypin and unsuccessful derivatization of the crystals, whereas the absence of significant sequence similarity (23) to other proteins with known structures discouraged molecular replacement attempts.
The initial crystallization screening for clitocypin alone and in complex with the target proteases was performed with the recombinant double methionine (L82M,I89M) mutant of clitocypin, its selenomethionine mutants, and cathepsins L and V. In contrast to the natural clitocypin, the recombinant clitocypin alone gave no crystals. Of the complexes, only cathepsin V and the methionine mutant produced diffracting crystals. Cathepsin V and clitocypin were mixed in molar ratio 1:1.1 and concentrated to 50 mg/ml in 10 mm acetate, 100 mm NaCl, pH 5.5. Crystals of dimensions of 0.2 × 0.4 × 0.1 mm3 were obtained in 0.4 m Li2SO4, 12% polyethylene glycol 800, 20% glycerol after 4 months. The selenomethionine mutant of clitocypin in complex with cathepsin V gave better diffracting crystals in a much shorter time. The crystals were frozen in liquid nitrogen before data collection. Diffraction data were collected at Synchrotrone Elletra, Trieste, from a single crystal using wavelength 1.0 Å.
Macrocypin 1 was concentrated to 30 mg/ml in 10 mm acetate buffer, 200 mm NaCl, pH 5.5, and crystallized by the sitting drop method at 20 °C using commercial screens from Qiagen. Crystals grew overnight in various conditions (Bistris propane buffer, pH 6.5–7.5, 100–500 mm different sodium salts, 20% polyethylene glycol 3350 or 0.8–1.6 m sodium/potassium phosphate, pH 7.0–8.0). The best diffracting crystals were grown in Bistris propane buffer, pH 7.0, 200 mm NaI, 20% polyethylene glycol 3350. They were soaked in a saturated solution of NaI in the same buffer before flash-freezing. Diffraction data were collected on an in-house copper rotating anode Rigaku (RU 200). Another high resolution data set was collected at Synchrotrone Elletra, Trieste, Italy, from a crystal grown in Bistris propane buffer, pH 7.0, 200 mm sodium citrate, 20% polyethylene glycol 3350 and soaked in the saturated solution of sodium citrate in the same buffer. All crystals were larger than 0.5 mm in all three dimensions.
Structure Solution and Refinement
All data were processed using the HKL2000 package (24). The macrocypin structure was solved with single wavelength anomalous diffraction phasing from the data collected from the crystal soaked in the saturated solution of sodium iodide. The data set was collected to 2.2 Å on the in-house Rigaku rotating anode (RU 200) using Xenox mirrors. 615 images were collected from a single crystal with the linear R-merge of 13% and redundancy of 30. Single wavelength anomalous diffraction phasing was based on 15 iodine positions with occupancy ranging from 0.8 to 0.15 using automated SOLVE/RESOLVE scripts incorporated in the AutoSol module of the PHENIX suite (25). Automated model building and docking to the macrocypin sequence gave a solution with ∼120 of 159 amino acids built (data not shown). Despite the good data quality, we were unable to refine the structure, presumably due to the multiple conformations of several loop regions induced by the binding of iodine ions, quite a few of them with low occupancy positions inside the protein core. Therefore, another data set was collected with the crystals, grown in sodium citrate. We phased this data set with the partial structure of macrocypin from the iodine-soaked crystal using molecular replacement program Amore (26). Cycles of manual and automated building with ARP/warp (27) and refinement with Refmac (28) and MAIN (29) were performed until all residues were built in the electron density. The final structure was refined using MAIN against 1.64 Å resolution data (29).
The crystal structure of cathepsin V-clitocypin complex was determined by molecular replacement with Amore using cathepsin V (PDB code 1fh0) (30) as the search model. Four molecules of cathepsin V were positioned into the asymmetric unit. The 4-fold electron density averaging in MAIN (29) produced maps that enabled us to build manually substantial parts of the clitocypin structure. Fragments of the clitocypin model enabled manual superimposition of the macrocypin structure using the similarity between the two models, accelerating the model building. The positions of the two selenomethionine residues in the clitocypin sequence were helpful in the initial sequence assignment. The structure was refined using MAIN against 2.24 Å resolution data. Geometric parameters for S-CH3 bound to the active site cysteine residue were obtained from PURY server (31). Data collection and refinement statistics are summarized in Table 1.”
TABLE 1.
Data collection and refinement statistics
Numbers in parentheses are for the highest resolution shell. Data sets from only one crystal were used for determination of each structure.
| Macrocypin | Clitocypin-Cathepsin V | Clitocypin | |
|---|---|---|---|
| Data collection | |||
| PDB ID | 3H6Q | 3H6S | 3H6R |
| Space group | P3121 | P21212 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 77.1, 77.1, 60.9 | 98.2, 177.8, 60.9 | 46.5, 58.0, 58.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 111.2, 90 |
| Resolution (Å) | 50-1.64 | 27.5-2.22 | 30-1.94 |
| Rmerge (%) | 5.0 (14.2) | 3.7 (19.4) | 2.6 (9.8) |
| I/σI | 69.0 (13.1) | 43.9 (6.1) | 63.1 (16.8) |
| Completeness (%) | 98.6 (86.5) | 98.8 (76.8) | 97.9 (85.0) |
| Redundancy | 10.3 (8.3) | 3.9 (1.8) | 7.2 (6.9) |
| Refinement | |||
| Resolution | 27.7-1.64 | 27.5-2.22 | 30-1.948 |
| No. of reflections (work/free) | 24.490/1.282 | 76.800/4.054 | 20.034/1.040 |
| Rwork/Rfree | 16.2/19.3 | 18.3/23.4 | 18.6/24.1 |
| B factors | |||
| Protein | 19.0 | 26.5 | 28.7 |
| Water | 35.4 | 37.3 | 40.1 |
| No. of atoms | |||
| Protein | 1424 | 11500 | 2388 |
| Water | 321 | 856 | 278 |
| r.m.s. deviation | |||
| Bond lengths (Å) | 0.019 | 0.018 | 0.022 |
| Bond angles (°) | 2.00 | 1.70 | 2.05 |
Kinetic Measurements
Kinetic and equilibrium constants for the inhibition of cathepsin V were determined under pseudo-first order conditions in continuous kinetic assays at 25 °C and calculated by nonlinear regression analysis according to Morrison (32) or Henderson (33) as described previously (8, 9).
RESULTS
Structures of Macrocypin and Clitocypin
Macrocypin 1 crystallized in P3121 space group with one molecule in the asymmetric unit. The macrocypin crystals contain the complete sequence, numbered from Gly-1 to Glu-168. Positioning of nearly all the residues is clearly revealed by the electron density maps. The exceptions are the side chains of His-114, Tyr-140, and Lys-21, which are only partially defined, and the stretch of residues Ser-20, Lys-21, Ile-22, which is only loosely defined. Nine residues (Gly-1, His-17, Arg-55, Ile-75, Gln-78, Ser-80, Glu-100, Gln-110, Ile-158) were modeled in alternative conformations.
Clitocypin crystallized in the P21 space group with two molecules in the asymmetric unit. The positioning of nearly all the residues is clearly revealed by the electron density maps. The exceptions are the loop Gln-67—Tyr-75 in molecule 1, Gly-68—Asn-70 and the side chain of Gln-115 in molecule 2, and the first two N-terminal residues in both molecules. Because of the genetic heterogeneity, clitocypin isolated from the natural source contains a large number of isoforms in unknown ratios (23). As default we have, therefore, used the sequence of the clitocypin isoform used for the complex formation. However, when the electron density unambiguously showed disagreement with that sequence, we built an appropriate amino acid residue from an alternative sequence based on amino acid sequences deduced from clitocypin genetic data (H17S, Y62S, L82M, P84Q, I88M, A105T, T139N in both molecules, S46F and Q48R in molecule 1, and Q37K, N42S, and A57S in molecule 2).
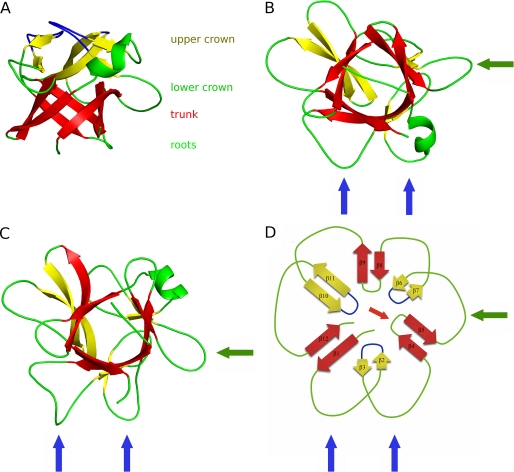
The macrocypin and clitocypin structures have the same fold. In the projection used in Fig. 1A the fold is reminiscent of a tree with a short, thick trunk, and a crown with branches expanding far from the center. The trunk part is an up-and-down β-barrel composed of six antiparallel β-strands (β1, β4, β5, β8, β9, β12). The strands are laid at an angle of less than 45 degrees to the axis of the barrel. The N and C termini are at the bottom. They form the roots of the tree together with the loops connecting strands β4-β5 and β8-β9. On the top three long regions between the strands β1-β4, β5-β8, and β9-β12 constitute the tree crown. Each contains a pair of antiparallel β-strands. In this manner two additional loops are formed between the strands from the crown and the trunk, adding another layer of loops that spread away from the trunk. Hence, the 3-fold arrangement of loops and strands is preserved in four layers of the structure: in the roots, the trunk, and the lower and upper layers of crown (Fig. 1, B–D). The loop region before strand β8 in the lower crown layer of macrocypin is folded into a short three-turn α-helix, whereas in clitocypin the loop preceding the strand β4 contains a short helical region. Although macrocypin and clitocypin have the same fold, the r.m.s. deviation between 116 equipositioned Cα atoms is 1.75 Å. The β-barrels are more similar, yielding r.m.s. deviations of 0.67 Å between 31 equipositioned Cα atoms. Macrocypin and clitocypin have a pseudo-3-fold symmetry (Fig. 1B–D), with the 3-fold rotational axis running through the six-stranded barrel.
FIGURE 1.
Structures of clitocypin (A and B), macrocypin (C), and schematic representation of trefoil fold (D). The trunk strands are always shown in red, and the crown strands are in yellow. A, the view from the side is shown. The structure resembles a tree structure, with two loops in the root region, a stem built of a six-stranded β-barrel, and two layers of loops (6 + 3) in the crown region. The first layer of the crown loops is shown in green, and the second is in blue. B and C, shown is the view along the barrel. The binding loops for cathepsins are marked with blue arrows, and the AEP loop is marked with green arrows. The two loops that bind to cysteine cathepsins belong to the lower layer of crown loops, whereas a single loop from the root region can inhibit trypsin or AEP.
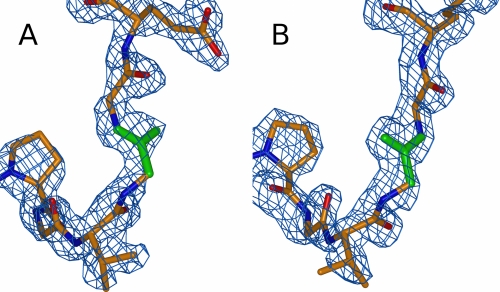
The structure of both clitocypin molecules in the asymmetric unit is basically the same (r.m.s. value of 0.28 Å), with one important exception. The peptide bond between Gly-24 and Gly-25 residues appears in two different orientations that are clearly seen in the electron density (Fig. 2, A and B), both in glycine-preferred regions of the Ramachandran plot. This suggests that this peptide bond is flexible and can appear in either orientation.
FIGURE 2.
Orientation of the Gly-24-Gly-25 peptide bond in the two clitocypin molecules. The 2Fobs − Fcalc electron density map is contoured at 1 σ. The bonds of glycine 24 are shown in green, whereas the rest of the chain is shown in red for oxygen, blue for nitrogen, and orange for carbon. The Gly-24–Gly-25 peptide bond is flexible and can exist in either orientation.
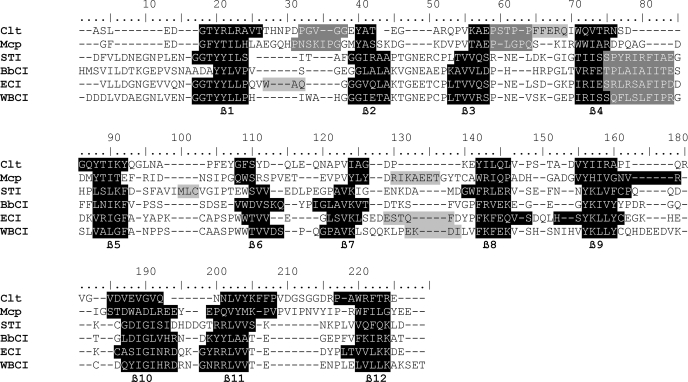
When the macrocypin structure was submitted to the protein structure comparison service SSM (34) at the European Bioinformatics Institute, the fold was identified as the β-trefoil fold present in proteins such as Kunitz-type soybean trypsin inhibitor (STI) (35), inhibitor isolated from Erythrina caffra (36), interleukins-1α and -1β (37), and fibroblast growth factors (38). The sequence alignment shows the low similarity of these proteins, in contrast to the structural alignment (Table 2), which shows that the secondary structure patterns are quite similar. The number of β-strands in macrocypin (12 strands) and clitocypin is the same as in STI, whereas their lengths differ significantly. STI has only two short strands (β6 and β7), whereas macrocypin and clitocypin have four (β2, β3, β6, β7). The highest structural as well as sequence similarity is in the regions composing the β-barrel. The fact that these are rather short stretches of sequence explains why homology searches based on sequence alignment have failed.
TABLE 2.
Structural alignment of sequences of clitocypin, macrocypin, and selected β-trefoil serine protease inhibitors
Sequences were aligned by the Protein structure comparison service SSM at the European Bioinformatics Institute (33). The parts of the sequence belonging to β-strands are printed in white on a black background, the binding loops are in white on a gray background, and the helical parts are in black on a gray background.
Structures of the Cathepsin V-Clitocypin Complex
The complete protein sequences are seen in the crystal structure of the complex of clitocypin with cathepsin V. The catalytic site of cathepsin V was blocked with a methyl methanethiosulfonate, leaving the S-CH3 group on the active site cysteine residue. This form of cathepsin V is much more stable and resulted in better diffracting crystals. The crystals have the P21212 space group and contain four pairs of molecules per asymmetric unit. The four structures of cathepsin V and clitocypin are closely related. The r.m.s. deviations over equivalent Cα atoms range from 0.18 to 0.25 for cathepsin V and 0.33 to 0.50 for clitocypin. Although the cathepsin V structures, apart from the ends of a few side chains, are unambiguously resolved from the electron density maps, three N-terminal residues and two loop regions (Gln-67—Asn-70 in molecules 1, 3, and 4 and Asp-138—Gly-141 in molecules 2, 3, and 4) of clitocypin lack adequate electron density or are only loosely defined.
Clitocypin binds into the active site of the target protease in the orientation of a fallen tree, with trunk and roots pointing sideways and up (Fig. 3, A and B). The wedge-shaped structure fills the active site cleft along its whole length, resulting in a buried area of 825 Å2. The interaction surface of clitocypin comprises basically two broad loop regions positioned at the lower edge of the crown. The loop structure and binding geometry are stabilized by numerous hydrogen bonds. Both loops originate from the first third of the clitocypin sequence; the first loop connects strands β1 and β2, and the second loop strands β3 and β4. The first loop is a broad, lasso-like structure cross-connected in the middle with a hydrogen bond between the Arg-12 side chain and the Gly-22 carbonyl. The second loop region is narrower and contains a short helix. The first loop binds into the non-primed substrate, and the second loop binds into the primed substrate binding site (Fig. 4). They occlude the catalytic cysteine in the middle and thereby prevent the approach of substrate molecules. Because the reactive site Cys-25 in the structure is modified, we cannot exclude the possibility that the interaction of clitocypin with the naked cysteine cathepsin may deviate slightly from the observed one.
FIGURE 3.
The cathepsin V-clitocypin complex. A, shown is the view along the active site cleft. B, shown is the view perpendicular to the active site cleft. The folds of cathepsin V and clitocypin are shown in gray and red. The catalytic cysteine is shown in yellow. Clitocypin binds into the active site of cathepsin V in the orientation of a fallen tree with the trunk and roots pointing sideways and up. The wedge shaped structure fills the active site cleft along its whole length.
FIGURE 4.
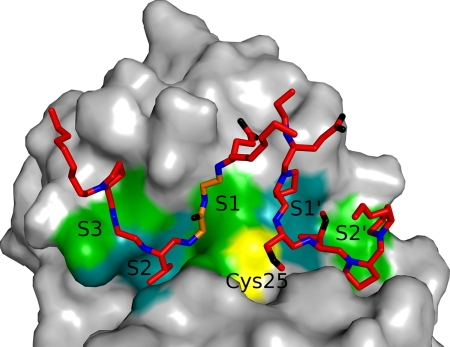
Binding loops of clitocypin in its complex with cathepsin V. Clitocypin loops are shown as sticks. Only main chain atoms, without the carbonyl oxygen atom and side chains, are shown. Nitrogen atoms are shown in blue, oxygen is in black, and carbon is in red, with the exception of the Gly-24-Gly-25 part, shown in orange. The surface of cathepsin V is shown in gray, apart from the catalytic cysteine shown in yellow and the S3, S2, S1, S1′ and S2′ binding site, shown in green and cyan. The chain of the first binding loop comes down the S3 binding area of cathepsin V, occupies the S2 binding site, and continues upward through the S1 binding site. The second binding loop of clitocypin approaches the S1′ and S2′ binding sites of cathepsin V from the top.
The chain from the first loop region comes down the S3 binding area (Pro-21, Gly-22) of cathepsin V, occupies the S2 binding site with Val-23, and continues through the S1 binding site, upwards and away from the cathepsin V surface. A hydrogen bond from the side chain amide of Asn-18 attaches clitocypin to the Gln-63 side chain of cathepsin V. A water molecule mediates additional contacts in the S3 binding area. Arg-12 plays a dual role; it stabilizes the lasso as well as attaching it to the cathepsin V surface by the Asn-66 main chain carbonyl, filling the interaction surface between the S3 binding area and the S1 binding site. Positioning of Val-23 builds antiparallel hydrogen bond arrangements with the Gly-68 of cathepsin V in a substrate-like manner. The peptide Val-23–Gly-24 bond is additionally fixed by the interaction between its hydrogen and the Asp-163 carbonyl from cathepsin V. In the S1 binding site, Gly-23 of cathepsin V is involved in hydrogen bonds between its carbonyl and the Gly-25 amide and between its amide hydrogen and the carboxylic oxygen atom of Glu-26. The peptide bond between Gly-24 and Gly-25 is flipped when compared with the structure of molecule 2 of clitocypin and macrocypin, presumably to form a hydrogen bond with Gly-23 of cathepsin V (Fig. 3). The other carboxylic oxygen atom of Glu-26 interacts with the amide of the cathepsin V Asn-66 side chain. However, the possibility that Glu-26 is partially neutral and interacts with the carbonyl oxygen atom of Gln-21 cannot be excluded. The binding of the first loop is further stabilized by the side chain amide group of Asn-18, which forms a hydrogen bond with the peptide bond carbonyl atom of Gln-21 of cathepsin V. It is notable that, of the hydrogen bond interactions between the enzyme and inhibitor, most are contributed by main chain atoms, at least on one side.
The second binding loop of clitocypin approaches the S1′ and S2′ binding sites of cathepsin V from the top. A single hydrogen bond between the carbonyl of Ser-42 and the side chain amide of Gln-145 of cathepsin V fastens the loop to the cathepsin V surface. In the first complex, an additional hydrogen bond is formed between the carboxylic group of Glu-48 and cathepsin V Ser-142. A layer of solvent molecules mediates the contacts between the N-terminal bottom of the short helix and cathepsin V. When we modeled the complex between macrocypin and cathepsin V by superimposing macrocypin on the clitocypin structure in the complex, it became evident that the binding loops do not fit into the active site. To find out whether the binding loops are the same in clitocypin and macrocypin, we expressed four mutants in which Gly-24 in the S3 binding area of clitocypin and Gly-25 in macrocypin were either replaced by alanine or deleted. We have also used these mutants to assess the relevance of the Gly-24-Gly-25 peptide flip. We assumed that the mutation to alanine or its deletion will reduce the flexibility of the main chain. The resulting clitocypin mutants yielded Ki values to cathepsin V that were 20 times higher than that of the native variant. It is notable that the major source of this difference is the slower association, whereas dissociation was not significantly affected (Table 3). This suggests a mechanism in which the peptide bond flip occurs before or concurrently with the inhibitor docking. The macrocypin mutants exhibit equivalent effects on their Ki constants, indicating that the loops that bind into the active site of cysteine cathepsin are equivalent in clitocypin and macrocypin. This implies that the binding loops of macrocypin exhibit substantial conformational flexibility during binding into the active site of their target enzymes.
TABLE 3.
Inhibition constants of cathepsin V by macrocypin, clitocypin, and their mutants
Kinetic data for interaction of macrocypin 1 and cathepsin V were reported previously (9).
| 10−6kon | 104koff | Ki | |
|---|---|---|---|
| m−1s−1 | s−1 | nm | |
| Clt | 1.26 ± 0.07 | 1.61 ± 0.65 | 0.08 ± 0.03 |
| Clt ΔG24 | 0.10 ± 0.06 | 1.8 ± 0.9 | 1.9 ± 0.9 |
| Clt G24A | 0.08 ± 0.01 | 1.2 ± 0.6 | 1.6 ± 0.7 |
| Mcp1 | 1.48 ± 0.01 | 10.3 ± 0.7 | 0.69 ± 0.06 |
| Mcp1 G25A | 0.13 ± 0.02 | 16.0 ± 5.0 | 12.5 ± 5.2 |
| Mcp1 ΔG25 | 0.15 ± 0.02 | 12.4 ± 2.4 | 8.5 ± 2.1 |
Inhibition of AEP
When mammalian asparaginyl endopeptidase was characterized, it was named according to its distinctive specificity (39). It suggests that AEP must have an S1 substrate binding site that is highly specific for asparagine. AEP is inhibited in the low nanomolar range (3–20 nm) by natural and recombinant clitocypin, natural macrocypin, and some isoforms of expressed macrocypin (macrocypins 1 and 3), whereas macrocypin 4 does not inhibit AEP at all. The availability of the mycocypins three-dimensional structure enabled the search for potential interacting areas to be narrowed down. Inspection of the aligned sequences of these isoforms in their surface regions focused attention on the β5-β6 loop, positioned in the lower crown region (residues 71–76 containing the sequence Ile-Asp-Asn-Ser-Ile). This part of the sequence is similar to the consensus sequence (S/T)N(D/S)(M/I) found in three inhibitory cystatins C, E, and F (Table 4) that bind to AEP in the nanomolar range (40). Interestingly, in macrocypin 4, the residue at position 72 is Lys, in contrast to the equipositioned Asn in macrocypins 1 and 3. To verify the role of the residues in these regions, like Alvarez-Fernandez et al. (40) in the case of cystatin C, we introduced mutations in the inhibitory sequence. The residues that differ between macrocypins 1 and 4 in the β5-β6 loop were exchanged. In addition, the corresponding clitocypin mutant (Clt-N69K) was also expressed. As expected, the mutant Mcp-1/4 (Mcp1 with β5-β6 loop of Mcp4) exhibited no inhibition, whereas the mutant Mcp-4/1 (Mcp4 with β5-β6 loop of Mcp1) increased inhibition against AEP (Table 5). Equivalently, the Clt-N69K mutant did not inhibit AEP. Thus, Asn-72 in macrocypins and Asn-69 in clitocypins are confirmed to be the residues responsible for the inhibition of AEP. Mycocypins are, in this respect, similar to cystatin C, which has two different binding sites, one for papain-like proteases and another for AEP (40).
TABLE 4.
Alignment of loops of macrocypin isoforms, clitocypin, cystatins V, E, and F, STI, and BbCI involved in AEP and trypsin inhibition
Residues that bind to the P1 pocket are marked with an asterisk. Mutated residues are shown bold. Cst, cystatin.
TABLE 5.
Inhibition constants of AEP, cathepsin V and trypsin by macrocypin, clitocypin and their mutants
NI, no inhibition.
| Ki AEP | Ki Cathepsin V | Ki Trypsin | |
|---|---|---|---|
| nm | nm | μm | |
| Mcp 1a | 3.38 ± 1.44 | 0.69 ± 0.06 | NI |
| Mcp 1/4 | NI | 3.43 ± 0.31 | 0.18 ± 0.02 |
| Mcp 4a | >1000 | 1.44 ± 0.11 | 0.16 ± 0.01 |
| Mcp 4/1 | 2.86 ± 0.38 | 10.2 ± 0.6 | NI |
| Mcp 4 K72R | NI | 6.9 ± 1.1 | 0.13 ± 0.02 |
| Clta | 21.5 ± 2.81 | 0.084 ± 0.03 | NI |
| Clt N70K | NI | 0.26 ± 0.09 | NI |
a Data reported previously (9).
Trypsin Inhibition
The binding geometry of several families of protein inhibitors of serine proteases, including the soybean Kunitz-type inhibitor (35), are known to adopt a substrate-like conformation known as the “canonical” binding mode (41). All Kunitz-type serine protease inhibitors inhibit trypsin with a highly homologous loop from the root region that mimics the substrate and is positioned between strands β4 and β5. This loop contains either lysine or arginine, which binds into the S1 pocket of trypsin. From the sequence and structure alignments it is evident that the classical β4-β5 loop is missing in macrocypin and clitocypin, and these proteins do not inhibit trypsin. Surprisingly, macrocypin 4 was found to inhibit trypsin with a Ki value in the micromolar range. The Ki values of the exchange mutants produced for AEP binding site identification (Table 5) show that the Lys-74 residue of macrocypin 4 is mandatory for inhibition of trypsin. The Mcp4 mutant with Lys-74 replaced by arginine (Mcp4-K74R) was similarly inhibitory (Table 5), thereby confirming the involvement of the loop β5-β6 positioned within the lower crown layer in binding to the trypsin active site. These mutants have no significant effect on the inhibition of cathepsin V (Table 5). The binding loop of macrocypins and clitocypins is, thus, positioned differently from the serine protease binding loop of known Kunitz-type inhibitors such as STI.
DISCUSSION
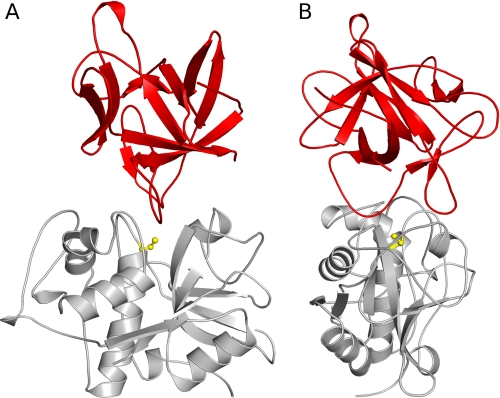
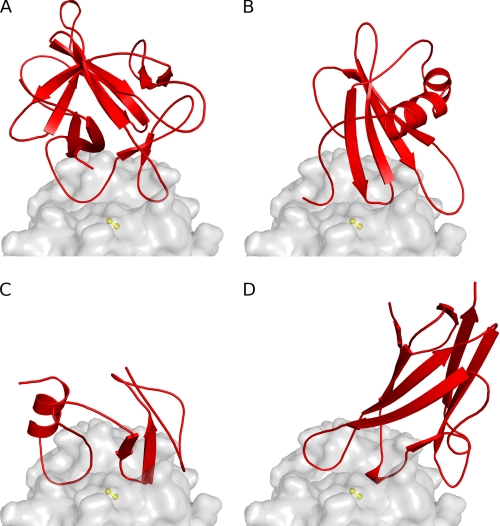
Like cystatins (10, 11), the p41 fragment (14) and chagasin (15), clitocypins, and macrocypins bind to cysteine proteases along the whole active site cleft (Fig. 5). These molecules have different folds, yet for docking to papain-like cysteine proteases, they utilize a similar architecture by which the activity of the target proteases is inhibited. Their constructs occlude the reactive site cysteine. On the non-primed substrate binding site they utilize a single chain. The first binding region in clitocypin is the loop Asp-19—Glu-25. Its position is similar to those of the N-terminal region in stefin A and the first loops of the p41 fragment and chagasin. This region contains a residue that, in a substrate-like manner, fills the S2 binding pocket. In contrast, the loops covering the primed binding areas are much less similar. The second binding region in clitocypins is a single, broad loop (Glu-39—Ile-50), whereas cystatins, the p41 fragment, and chagasin use two loop constructs. The two broad loops of mycocypins are stabilized by multiple hydrogen bonds and are much more rigid than the N-terminal trunk and two loops in cystatins. This explains why cystatins are capable of competing for binding with the additional features of exopeptidase such as occluding loop and mini chain, whereas mycocypins exhibit lower affinity or no binding at all.
FIGURE 5.
Comparison of the binding modes of inhibitors. The fold of inhibitors is shown in red (A, clitocypin; B, stefin A; C, p41 fragment; D, chagasin), whereas the endopeptidase cathepsin V is presented as a gray surface. The surface of the catalytic cysteine is shown in yellow. All inhibitors have different folds, yet for docking to the active site of papain-like cysteine proteases they utilize loops that fill the active site along its whole length and occlude the reactive site cysteine residue by two loops.
The mode of the binding of mycocypins to cysteine cathepsins differs markedly from the binding of the Kunitz type of β-trefoil folded inhibitors to serine proteases. The two binding loops from the crown region bind into the non-primed and primed substrate binding regions of cysteine proteases and occlude the catalytic cysteine residue. In contrast, only one binding loop from the root region of Kunitz inhibitors docks into the active site of a serine protease, in a substrate-like manner. A possible explanation for the differences in the modes of canonical inhibition of cysteine cathepsins and trypsin-like serine proteases may lie in the features of the active site cleft. Whereas in the trypsin-like proteases the S1 binding site is a pocket in the protein structure, in cysteine cathepsins the S1 binding site is positioned on the surface on one side of the active site cleft, shaped so that the P1 residue side chain points away from the protein core (42). Furthermore, analysis of the structural data has revealed that papain-like cysteine proteases have only three clearly defined substrate binding sites (S2, S1, S1′) and one conditional site (S2′), whereas the binding into regions beyond position 2 can only be considered substrate binding areas spread over the surface of the widening active site cleft (43). Cysteine cathepsins, thus, appear to lack the binding surface to which the P1 and neighboring residues could be tightly anchored in a substrate-like manner and, therefore, can probably not be inhibited by the single loop construct.
The flexible peptide bond, which can flip on docking to protease, is a unique feature among the cysteine protease inhibitors. Peptide flipping has already been observed in the mitogen-activated protein kinase p38α MAPK, where the flip of the Met-109 and Gly-110 peptide bond facilitates the higher specificity of certain inhibitors (44).
The trefoil fold supports 11 loops coming out of the six-stranded β-barrel. Nine are in the crown region (six are positioned at the lower level of the crown, and three enclose the top of the crown), and two are in the roots. Therefore, it is easier to comprehend that the six loops from the lower crown region can act in pairs, whereas the two loops from the root region lack that capability and must bind alone. In this respect the report of the binding site of B. bauhinioides cruzipain inhibitor (BbCI) to cysteine cathepsins is intriguing, as the authors (17) suggest that the same alanine residue positioned within the root region, which is responsible for binding to neutrophil elastase, is also crucial for the binding to cathepsin L and cruzipain (the partial cleavage of the serine protease interacting loop after incubation of BbCI with cruzipain was the key evidence supporting the hypothesis of the common interaction site). The binding of BbCI to trypsin is consistent with current structural knowledge, as the loop in which Ala-63 resides folds very similarly to the loop of STI, including the position and orientation of the Ala-63 residue. However, the single inhibitory loop is not consistent with the canonical inhibition mechanism of cysteine cathepsins evidenced here. Superimposition of the structure of BbCI on clitocypin in complex with cathepsin V showed that two broad loops in the BbCI structure are equivalent to the clitocypin binding loops and that the BbCI sequence contains two consecutive glycine residues, Gly-28—Gly-29, homologous to the peptide bond flip residues Gly-24 and Gly-25. Hence, these two loops are probably responsible for cathepsin L inhibition and not the loop containing the trypsin cleavage site (it should be noted, however, that as in the case of macrocypin, the tips of the loops require a slight adjustment to fit into the active site of a cysteine cathepsin). The absence of inhibition of cathepsin V by BbCI, given the similarity of cathepsins L and V, cannot, however, be explained.
Clitocypins and macrocypins exhibit no sequence similarity to other known proteins, which was the basis for establishing the I48 and I85 families supported by the sequence alignment score with an E value less than 0.001 (FASTA search with default BLOSUM50 matrix used in MEROPS). However, their structure has revealed that the basic element of their fold is the six-stranded β-barrel, the hallmark of the β-trefoil fold shared by the members of the I3 MEROPS family that includes serine protease inhibitors of the Kunitz type. The sequence similarity, based on superimposition of the structures, is low even within the 6-stranded β-barrel part (Fig. 4), thus, questioning the common origin of these two groups of proteins. This confirms that mycocypins (families I48 and I85) are indeed distinct from members of I3 family, whereas the structural similarity between these families provides support that they belong to the same clan IC.
The β-trefoil fold is armed with potent interacting loops that differ in shape and composition and are able to inhibit several classes of proteases including cathepsins, AEP, cruzipain, trypsin, chymotrypsin, elastase, subtilisin, and amylases. Several loops are involved in inhibition, whereas the same inhibitory loop can target different proteases. For example, the crown region loops β1-β2 and β3-β4 are used for inhibiting the papain-like cysteine proteases by mycocypins and, most probably, cruzipain and cathepsin L by BbCI (17). The root region loop β4-β5 is involved in inhibiting chymotrypsin (by the winged bean chymotrypsin inhibitor) (45), trypsin (by STI) (35), and porcine pancreatic elastase and human neutrophil elastase (by BbCI) (17), whereas the crown region loop β5-β6 is involved in inhibiting AEP and trypsin (by mycocypins) and the subtilisin savinase (by barley α-amylase/subtilisin inhibitor) (46). The numerous crown region loops, β1-β2, β3-β4, β6-β7, β9-β10 and β11-β12, are responsible for the interaction of barley α-amylase/subtilisin inhibitor with barley α-amylase (47). This makes β-trefoil inhibitors, in particular mycocypins, promising candidates for transgenic trials for the purposes of crop protection, where inhibitors with selectivity against only one class of proteases have failed because of the compensation of proteolytic activity by induced expression of other proteases insensitive to the transgenic inhibitor (48–50).
Acknowledgments
We are very grateful to Dr. Roger Pain for critical reading of the manuscript and Dr. Tatjana Popovič for help with kinetics. Vida Puizdar is acknowledged for providing asparaginyl endopeptidase and Dr. Katja Galeša for cloning and test expression of double methionine clitocypin mutant.
This work was supported by Slovenian Research Agency Grants P1-0048 (to D. T.) and P4-0127 (to J. K.), by a Young Researcher fellowship (to M. R.), and by the European Commission Framework VI Program (CAMP project, LSHG-2006-018830).
The atomic coordinates and structure factors (codes 3H6R, 3H6Q, and 3H6S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Clt
- clitocypin
- Mcp
- macrocypin
- AEP
- asparaginyl endopeptidase
- BbCI
- B. bauhinioides cruzipain inhibitor
- STI
- soybean trypsin inhibitor
- WBCI
- winged bean chymotrypsin inhibitor
- ECI
- inhibitor isolated from E. caffra
- Cst
- cystatin
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- MES
- 4-morpholineethanesulfonic acid
- r.m.s.
- root mean square.
REFERENCES
- 1.Ryan C. A. (1990) Annu. Rev. Phytopathol. 28, 425–449 [Google Scholar]
- 2.Frost C. J., Mescher M. C., Carlson J. E., De Moraes C. M. (2008) Plant Physiol. 146, 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green T. R., Ryan C. A. (1972) Science 175, 776–777 [DOI] [PubMed] [Google Scholar]
- 4.Ussuf K. K., Laxmi N. H., Mitra R. (2001) Curr. Sci. 80, 847–854 [Google Scholar]
- 5.Christou P., Capell T., Kohli A., Gatehouse J. A., Gatehouse A. M. (2006) Trends Plant Sci. 11, 302–308 [DOI] [PubMed] [Google Scholar]
- 6.Benchabane M., Goulet M. C., Dallaire C., Côté P. L., Michaud D. (2008) Plant Physiol. Biochem. 46, 701–708 [DOI] [PubMed] [Google Scholar]
- 7.Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) Nucleic Acids Res. 36, D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabotic J., Popovic T., Puizdar V., Brzin J. (2009) FEBS J. 276, 4334–4345 [DOI] [PubMed] [Google Scholar]
- 9.Brzin J., Rogelj B., Popovic T., Strukelj B., Ritonja A. (2000) J. Biol. Chem. 275, 20104–20109 [DOI] [PubMed] [Google Scholar]
- 10.Stubbs M. T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. (1990) EMBO J. 9, 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenko S., Dolenc I., Guncar G., Dobersek A., Podobnik M., Turk D. (2003) J. Mol. Biol. 326, 875–885 [DOI] [PubMed] [Google Scholar]
- 12.Lenarcic B., Turk V. (1999) J. Biol. Chem. 274, 563–566 [DOI] [PubMed] [Google Scholar]
- 13.Mihelic M., Turk D. (2007) Biol. Chem. 388, 1123–1130 [DOI] [PubMed] [Google Scholar]
- 14.Guncar G., Pungercic G., Klemencic I., Turk V., Turk D. (1999) EMBO J. 18, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ljunggren A., Redzynia I., Alvarez-Fernandez M., Abrahamson M., Mort J. S., Krupa J. C., Jaskolski M., Bujacz G. (2007) J. Mol. Biol. 371, 137–153 [DOI] [PubMed] [Google Scholar]
- 16.Hansen D., Macedo-Ribeiro S., Veríssimo P., Yoo Im S., Sampaio M. U., Oliva M. L. (2007) Biochem. Biophys. Res. Commun. 360, 735–740 [DOI] [PubMed] [Google Scholar]
- 17.Araújo A. P., Hansen D., Vieira D. F., Oliveira C., Santana L. A., Beltramini L. M., Sampaio C. A., Sampaio M. U., Oliva M. L. (2005) Biol. Chem. 386, 561–568 [DOI] [PubMed] [Google Scholar]
- 18.Sabotic J., Galesa K., Popovic T., Leonardi A., Brzin J. (2007) Protein Expr. Purif. 53, 104–111 [DOI] [PubMed] [Google Scholar]
- 19.Weiner M. P., Costa G. L. (1994) PCR Methods Appl. 4, S131–S136 [DOI] [PubMed] [Google Scholar]
- 20.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 21.Mihelic M., Teuscher C., Turk V., Turk D. (2006) FEBS Lett. 580, 4195–4199 [DOI] [PubMed] [Google Scholar]
- 22.Galesa K., Brzin J., Sabotic J., Turk D. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 10–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabotic J., Gaser D., Rogelj B., Gruden K., Strukelj B., Brzin J. (2006) Biol. Chem. 387, 1559–1566 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 26.Navaza J. (1994) Acta Cryst. 50, 157–163 [Google Scholar]
- 27.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.Turk D. (1992) Weiterentwicklung eines Programms fuer Molekülgraphik und Elektrondichte-Manipulation und Seine Anwendung auf Verschiedene Protein-Strukturaufklërungen. Ph.D. thesis, Technische Universitaet Muenchen, Germany [Google Scholar]
- 30.Somoza J. R., Zhan H., Bowman K. K., Yu L., Mortara K. D., Palmer J. T., Clark J. M., McGrath M. E. (2000) Biochemistry 39, 12543–12551 [DOI] [PubMed] [Google Scholar]
- 31.Andrejasic M., Praaenikar J., Turk D. (2008) Acta Crystallogr. D. Biol. Crystallogr. 64, 1093–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison J. F. (1982) Trends Biochem. Sci. 7, 102–105 [Google Scholar]
- 33.Henderson P. J. F. (1972) Biochem. J. 127, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 35.Song H. K., Suh S. W. (1998) J. Mol. Biol. 275, 347–363 [DOI] [PubMed] [Google Scholar]
- 36.Onesti S., Brick P., Blow D. M. (1991) J. Mol. Biol. 217, 153–176 [DOI] [PubMed] [Google Scholar]
- 37.Graves B. J., Hatada M. H., Hendrickson W. A., Miller J. K., Madison V. S., Satow Y. (1990) Biochemistry 29, 2679–2684 [DOI] [PubMed] [Google Scholar]
- 38.Ago H., Kitagawa Y., Fujishima A., Matsuura Y., Katsube Y. (1991) J. Biochem. 110, 360–363 [DOI] [PubMed] [Google Scholar]
- 39.Chen J. M., Dando P. M., Rawlings N. D., Brown M. A., Young N. E., Stevens R. A., Hewitt E., Watts C., Barrett A. J. (1997) J. Biol. Chem. 272, 8090–8098 [DOI] [PubMed] [Google Scholar]
- 40.Alvarez-Fernandez M., Barrett A. J., Gerhartz B., Dando P. M., Ni J., Abrahamson M. (1999) J. Biol. Chem. 274, 19195–19203 [DOI] [PubMed] [Google Scholar]
- 41.Bode W., Huber R. (1992) Eur. J. Biochem. 204, 433–451 [DOI] [PubMed] [Google Scholar]
- 42.Turk B., Turk V., Turk D. (1997) Biol. Chem. 378, 141–150 [PubMed] [Google Scholar]
- 43.Turk D., Guncar G., Podobnik M., Turk B. (1998) Biol. Chem. 379, 137–147 [DOI] [PubMed] [Google Scholar]
- 44.Fitzgerald C. E., Patel S. B., Becker J. W., Cameron P. M., Zaller D., Pikounis V. B., O'Keefe S. J., Scapin G. (2003) Nat. Struct. Biol. 10, 764–769 [DOI] [PubMed] [Google Scholar]
- 45.Ravichandran S., Sen U., Chakrabarti C., Dattagupta J. K. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1814–1821 [DOI] [PubMed] [Google Scholar]
- 46.Micheelsen P. O., Vévodová J., De Maria L., Ostergaard P. R., Friis E. P., Wilson K., Skjøt M. (2008) J. Mol. Biol. 380, 681–690 [DOI] [PubMed] [Google Scholar]
- 47.Vallée F., Kadziola A., Bourne Y., Juy M., Rodenburg K. W., Svensson B., Haser R. (1998) Structure 6, 649–659 [DOI] [PubMed] [Google Scholar]
- 48.Jongsma M. A., Bakker P. L., Peters J., Bosch D., Stiekema W. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8041–8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Leo F., Bonade-Bottino M. A., Ceci L. R., Gallerani R., Jouanin L. (1998) Plant Physiol. 118, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cloutier C., Jean C., Fournier M., Yelle S., Michaud D. (2000) Arch. Insect Biochem. Physiol. 44, 69–81 [DOI] [PubMed] [Google Scholar]