Abstract
PLAA (ortholog of yeast Doa1/Ufd3, also know as human PLAP or phospholipase A2-activating protein) has been implicated in a variety of disparate biological processes that involve the ubiquitin system. It is linked to the maintenance of ubiquitin levels, but the mechanism by which it accomplishes this is unclear. The C-terminal PUL (PLAP, Ufd3p, and Lub1p) domain of PLAA binds p97, an AAA ATPase, which among other functions helps transfer ubiquitinated proteins to the proteasome for degradation. In yeast, loss of Doa1 is suppressed by altering p97/Cdc48 function indicating that physical interaction between PLAA and p97 is functionally important. Although the overall regions of interaction between these proteins are known, the structural basis has been unavailable. We solved the high resolution crystal structure of the p97-PLAA complex showing that the PUL domain forms a 6-mer Armadillo-containing domain. Its N-terminal extension folds back onto the inner curvature forming a deep ridge that is positively charged with residues that are phylogenetically conserved. The C terminus of p97 binds in this ridge, where the side chain of p97-Tyr805, implicated in phosphorylation-dependent regulation, is buried. Expressed in doa1Δ null cells, point mutants of the yeast ortholog Doa1 that disrupt this interaction display slightly reduced ubiquitin levels, but unlike doa1Δ null cells, showed only some of the growth phenotypes. These data suggest that the p97-PLAA interaction is important for a subset of PLAA-dependent biological processes and provides a framework to better understand the role of these complex molecules in the ubiquitin system.
Introduction
Cdc48/p97 is a hexameric AAA ATPase involved in many ubiquitin (Ub)4-dependent functions (1). As with other AAA ATPases such as Vps4 and NSF, p97 disrupts protein complexes with its segregase activity (2). This activity is important for liberating denatured proteins undergoing endoplasmic reticulum-associated protein degradation from the ER membrane and conveying them to the proteasome. p97 fulfills this and other functions by associating with a variety of accessory proteins (3), some of which such as the E3 ligase Ufd2, the deubiquitinating enzyme Otu1, or the deglycosylating enzyme PNGase, can modify substrates. Other factors act as adaptors to recognize ubiquitinated substrates via distinct Ub binding domains. Exactly how p97 is properly programmed with the correct accessory proteins is not yet clear; however, different regions of p97 mediate binding to distinct sets of accessory proteins, some of them in mutually exclusive configurations. For example, the Ufd1-Npl4 complex, the Ubx family, and the Otu1 peptidase associate with the N-terminal domain of p97; Ufd2, Doa1/PLAA, and PNGase associate with the C terminus of p97. PNGase binds the C terminus of p97 via a PUB domain, which is found in a variety of other proteins that likely add functionality to p97 (4–6). In the case of Ufd2 and Doa1, each of these factors can compete for binding, which results in hexameric Cdc48-p97 complexes containing either Doa1 or Ufd2 (7). Recent studies demonstrate that the distal C terminus of p97 can be tyrosine-phosphorylated thus offering a potential way to regulate association with this subset of adaptors (6, 8). Interestingly, this is one of the most phosphorylated sites in proliferating cells (9).
The p97 accessory protein PLAA and its yeast homolog, Doa1/Ufd3, possess three conserved domains, including a seven-bladed WD40 β-propeller of unknown function, a central PFU domain that binds Ub, and a C-terminal PUL (PLAP, Ufd3p, and Lub1p) domain previously shown to mediate binding to Cdc48/p97 (Fig. 1A) (10–13). The PUL domain was originally defined as a conserved portion in orthologs PLAA, Doa1, and Lub1 (10). Loss of Doa1 results in a broad range of defects that can be largely traced to loss of Ub levels because overexpression of Ub suppresses the growth defects and some other phenotypes of doa1Δ null mutants (7, 11, 12, 14–19). How the levels of Ub are maintained by Doa1 is unclear, although genetic studies indicate that the function of Doa1 is antagonized by Ufd2 because loss of Ufd2 also suppresses some of the growth defects of Doa1 loss (7). Doa1 has been shown to have distinct functions that are independent of its effect on Ub levels. This includes a role in sorting ubiquitinated proteins into the vacuolar/lysosomal lumen via the MVB pathway (17). It is not clear whether any or all of Doa1 functions require association with Cdc48, although loss of the entire PUL domain that mediates association ablates all Doa1 function in yeast. Here, we show with structure/function studies that Doa1 has functions both dependent and independent of its association with Cdc48/p97. These data show that although Doa1 performs some of its functions with Cdc48, it also has a distinct set of functions that do not require association. Furthermore, our data suggest the existence of other factors besides Doa1 and Ufd2 that interact with the C terminus of Cdc48 that are important for Cdc48 function.
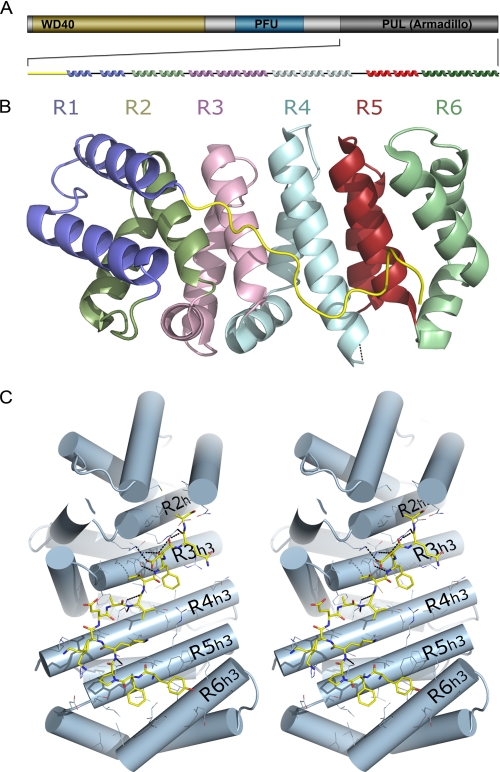
FIGURE 1.
Structure of the PUL domain. A, domain map of PLAA that defines N-terminal WD40 β-propeller (yellow), central PFU domain (blue), and C-terminal PUL domain (gray). Secondary structure elements are shown colored according to their ARM. B, overall structure of PUL domain containing six Armadillo repeats individually colored and labeled. All figures were generated with PyMol. C, stereoscopic view of the N-terminal extension in yellow stick format bound across the concave surface of the Armadillo fold, shown as light blue cylinders. The third helices of each Armadillo unit is labeled.
EXPERIMENTAL PROCEDURES
Expression and Purification of PUL Domain
The PUL domain of PLAA (residues 454–738) was cloned from a Mammalian Gene Collection cDNA template (AT56-D1) into the pET28-LIC vector (GenBankTM accession number EF456735). Competent BL21 (DE3) cells (C6000-03, Invitrogen) were transformed and grown using the LEX system (Harbinger BEC) at 37 °C in 2-liter bottles (89000-242, VWR) containing 1800 ml of TB (T0918, Sigma) supplemented with 150 mm glycerol, 100 μm kanamycin, and 600 μl of antifoam 204 (A-8311, Sigma). At A600 = 6.0, the temperature was reduced to 15 °C, and 1 h later the culture was induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside (IPT001, BioShop) and incubated overnight (16 h) at 15 °C. Cell pellets were resuspended in 30 ml/liter bacterial culture of Lysis Buffer (20 mm Tris, pH 8.0, 0.5 m NaCl, 5% glycerol, 1 mm β-mercaptoethanol, 0.1 μm phenylmethylsulfonyl fluoride), and cells were lysed by sonication (3000, Misonix) on ice for a 10-min total sonication time. After centrifugation at 40,000 × g for 30 min, the clarified cell lysate was applied on a 3-ml column packed with TALON metal affinity resin (Clontech). The column was washed with 10 column volumes of wash buffer (20 mm Tris, pH 8.0, 0.5 m NaCl, 5% glycerol, 10 mm imidazole). Protein was eluted with 2 column volumes of Elution Buffer (20 mm Tris, pH 8.0, 500 mm NaCl, 5% glycerol, 250 mm imidazole). The His tag was cut with thrombin (2 units/mg protein) overnight at 4 °C. The protein was further purified by gel filtration on a HighLoad 16/60 Superdex 200 column (GE Healthcare). Fractions containing protein were concentrated by ultrafiltration using an Amicon ultracentrifugal filter with 5-kDa cutoff.
Crystallization and Structure Determination
Crystals were grown at 20 °C using the sitting drop method by mixing equal volumes of protein (40 mg/ml) and Crystallization Buffer (30% PEG-4000, 0.2 m MgCl2, 0.1 m Tris buffer, pH 8.5). Suitable crystals were cryoprotected by immersion in well solution supplemented with 20% (volume/volume) glycerol prior to dunking and storage in liquid nitrogen. Single-wavelength anomalous diffraction data from a crystal of a selenium-methionine derivative of the PLAP PUL domain in complex with p97 peptide was collected at Beamline 23-ID-B at the Argonne Photon Source and processed using the HKL2000 program suite (20). Solve and Resolve were used to locate the selenium substructure and to build the initial model (21, 22). Iterative model building using the graphics program Coot (23) and maximum likelihood and TLS refinement with the program REFMAC5, part of the CCP4 suite (24), led to a model with an R factor of 18.8% (Rfree 24.8%) for data between 1.9 and 41.5 Å. Parameters for translation/liberation/screw refinement were generated using the TLSMD web server (25). The final model, including 8251 protein atoms, 477 solvent atoms, and a magnesium atom, exhibits excellent stereochemistry with no Ramachandran violations as judged by PROCHECK and the MolProbity Ramachandran plot (26). Statistics of data collection, processing, and refinement are provided in Table 1. The coordinates and structure factors were deposited into the RCSB PDB data base with ID code 3EBB.
TABLE 1.
Data collection, phasing, and refinement statistics
The highest resolution shell is 1.97 to 1.90 Åand is shown in parentheses. r.m.s.d. indicates root mean square deviation.
| PUL domain and p97 peptide | |
|---|---|
| Data set | |
| Space group | C1 2 1 |
| Unit cell a, b, c, β | a = 103.384, b = 68.651, c = 143.975, β = 103.45 |
| Beamline | APS 23-ID-B |
| Wavelength | 0.97945 Å |
| Resolution | 1.90-50.0 |
| Unique reflections | 76,756 |
| Data redundancy | 9.7 (3.1) |
| Completeness | 98.1% (99.6) |
| I/σI | 24.7 (3.21) |
| Rsym | 0.06 (0.51) |
| Rmeas | 0.22 (1.46) |
| Refinement | |
| Resolution | 1.90-41.49 |
| Reflections used | 72,714 |
| All atoms (solvent) | 8729 (477) |
| Rwork/Rfreea | 18.8/24.8 |
| r.m.s.d. bond length | 0.011 |
| r.m.s.d. bond angle | 1.25 |
| Mean B factor | 17.33 |
| Ramachandran plot | |
| Favored | 99.4 |
| Allowed | 100 |
| Disallowed | 0 |
a Rfree was calculated with 5% of the data.
Fluorescence Polarization Assay
Fluorescence polarization assays were performed in 384-well plates, using Synergy 2 microplate reader from BioTek. A peptide corresponding to the last 10 residues of the C terminus of p97 protein (TEDNDDDLYG) was synthesized, N-terminally labeled with fluorescein, and purified by Tufts University Core Services (Boston). Binding assays were performed in a 10-μl volume at a constant labeled peptide concentration (30 nm) by titrating the PLAA C-terminal PUL domain (residues 454–738) and the central PLAA PFU domain (residues 281–408) (at concentrations ranging from low to high) into 50 mm Hepes, pH 7.5, 50 mm KCl, and 0.01% Tween 20. A maximum fluorescence polarization of about 250 millipolarization units was detected upon binding of the PUL domain to the labeled peptide, whereas typically about 90–100 millipolarization units was detected for free labeled peptide. To determine dissociation constant values, the data were fit to a hyperbolic function using SigmaPlot software.
Functional Yeast Experiments
A URA3 pRS316-based plasmid expressing wild type DOA1 (pPL3498) was used for mutagenesis to make the I483A, D538R, and L571A doa1ΔCdc48 mutant (pPL4068). The GST-Cdc48 C-terminal fusion protein was made by subcloning a BamHI/EcoRI fragment encoding residues 208–835 of Saccharomyces cerevisiae Cdc48 into pGEX3X. Wild type and mutant Doa1 PUL domain was made by cloning a PCR fragment encoding residues 461–715 into pET151 downstream of a V5 epitope tag.
The doa1Δ::HIS3 (PLY3709) cells, ufd2Δ cells, ufd2Δ doa1Δ cells, and congenic wild type cells (BY4742) were used as described previously (17). Congenic wild type and cdc48ΔC-term cells were generated by integrating an HA-HIS5 fragment derived from pFa6a-3HA-HISMX6 (27) that was preceded by a stop codon just after the end of the Cdc48 open reading frame (wild type) or after codon 710 of Cdc48, respectively. Growth assays, immunoblots, GST binding assays, and analysis of Vph1-GFP-Ub sorting were performed as described previously (17). Levels of Ub were measured from immunoblots of two sets of transformants. The level of chemiluminescence was collected using a Fluorchem camera system (Alpha Innotech, San Leandro, CA) and quantified using ImageJ.
RESULTS
Fluorescence Polarization Assay
Previous studies have shown that the C-terminal PUL domain of yeast Doa1 mediates interaction with Cdc48 (7, 12). More specifically, the last 10 residues of p97, which are largely conserved in yeast Cdc48, were able to directly interact with a C-terminal fragment of PLAA containing the central PFU domain and C-terminal PUL domain (6). Binding studies were performed to confirm that the C-terminal PUL domain of human PLAA bound to the p97 C terminus. The supplemental Fig. S1 shows that the distal C-terminal p97 peptide (TEDNDDDLYG) bound recombinant PUL domain (PLAA residues Met454–Leu738) with a Kd of 5 μm consistent with previous measurements for p97 and yeast Doa1 PFU-PUL fragment (6). No peptide binding was observed to the central PFU domain of human PLAA (residues 338–453).
Structure of PLAA PUL Domain
The PLAA PUL domain was crystallized in a complex with a peptide corresponding to the last 10 residues of p97. Two complexes per asymmetric unit were found in the space group C2 (Table 1). The structure was determined by single-wavelength anomalous diffraction and was refined at 1.9 Åresolution to an R factor of 18.8% (Rfree 24.8%).
The PUL domain adopts an extended fold composed of six Armadillo repeats (ARM1 to ARM6) connected by short loops; each repeat is composed of three α-helices (Fig. 1, A and B). The six contiguous repeats form a superhelix that features a long, positively charged groove that is formed by the third helix of each repeat. There are three irregularities in the Armadillo repeat domain structure, including the absence of helix-1 in the first two ARM repeats and a disordered helix-2 in the fourth ARM. The first 17 residues of the domain (Lys531–Gln545) form an extended coiled loop that folds back and packs tightly against the inner curvature of the Armadillo domain. Tyr534 and Phe535 interact in a ridge between helix-3 of the fifth and sixth ARM repeats. The N-terminal segment Pro536–Ala546 extends across helix-3 of the remaining ARM repeats. Seven hydrogen bonded backbone amides, three buried aromatic side chains, and numerous aliphatic van der Waals interactions suggest that the N-terminal segment is fixed to the core, globular fold of the PUL domain, possibly stabilizing the Armadillo repeats (Fig. 1C).
Interaction with p97
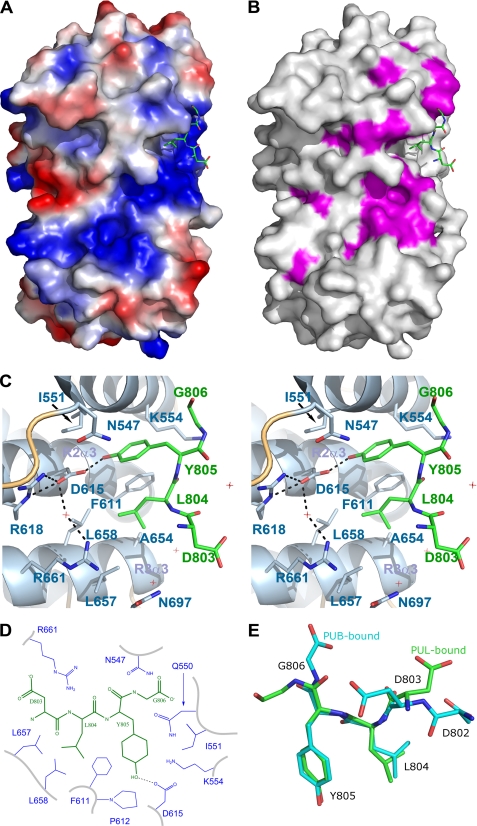
Our crystal structure clearly reveals the mode of binding for the p97 partner. Electron density is well defined for the last four residues of the peptide (DLYG806), which bind to the inside curvature of the PUL domain, adjacent to the extended N-terminal segment, in a pocket formed by the third helices of the first three ARM repeats (Fig. 2, A–C, and supplemental Fig. S2). The interaction is limited to only two residues of p97. The p97 side chains of Leu804 and Tyr805 point into this pocket, where Asp615 and Leu658 form the floor, and Asn547, Gln550, Ile551, Lys554, Phe611, Arg661, Leu657, Ala654, and Asn697 form the walls (Fig. 2, C and D). The hydroxyl group of Tyr805 forms a hydrogen bond with the carboxylate of Asp615, which is also bridged to the guanidinium of Arg618 and Arg661 via a water molecule (Fig. 2C). The burial of the tyrosine side chain by Phe611, Lys554, and Asp615, and the backbone of the first helix of the domain appears to be responsible for the bulk of the observed affinity. Residues that form van der Waals interactions with the peptide are schematized (Fig. 2D). The terminal Gly806 is variably ordered and is likely to be predominantly solvent-exposed; in one molecule of the asymmetric unit, it forms a long hydrogen bond with the amide side chain of Gln550. Asp803 is also variably ordered and solvent-exposed. There are no significant interactions between the peptide backbone and the PUL domain.
FIGURE 2.
Co-structure of the PUL domain with the C-terminal p97 peptide. A, electrostatic surface representation was generated with a gradient from −10 (red) to 10 (blue) kT/e of the PUL domain only. The bound p97 peptide is show in green stick format. B, surface representation of the PUL domain is shown in light gray with conserved residues in purple. C, stereoscopic view of the terminal p97 peptide residues LYG806 bound to the PUL domain shown with helices in schematic format and labeled. PUL residues within proximity are shown in stick format and labeled. The hydrogen bond is shown as a black dashed line; water molecules are shown as red cross-hairs. D, schematic representation of p97 (black) interactions with PLAA (blue). E, three-dimensional alignment of the PLAA (green) and PNGase (cyan)-bound p97 peptides.
The conformation of the p97 peptide bound to the PUL domain is strikingly similar to its conformation bound to the PNGase PUB domain, which does not have an Armadillo fold (6). The side chains of Leu804 and Tyr805, which have an ω angle of 120° in both PUB and PUL structures (Fig. 2E), similarly point into their pockets, burying 366 and 293 Å2, respectively. But although the terminal Gly806 is mostly disordered in PUL structures, it appears capped in PUB, being involved in an ionic bond with an arginine and hydrogen bond with an asparagine (6). These differences suggest that unlike the PUB domain, the PUL domain can also bind non-terminal, or internal, LYG motifs. The larger number of hydrogen bonds between the p97 peptide and the PUB domain (eight compared with only one in the PUL domain) is consistent with the estimated 1 order of magnitude greater affinity compared with the PUL domain (6).
Although the p97 pocket is hydrophobic, the neighboring ridge in the PUL domain is marked by a positively charged stretch that extends about 40 Åacross the concave inner surface, contributed by one basic residue from each of the ARM helix-3 helices. Interestingly, the channel formed by the inner groove is composed of the most conserved residues of the domain. Together, these observations strongly suggest that the PUL domain has evolved to bind peptide ligands that extended beyond the p97 peptide-binding pocket and that are significantly negatively charged. Although acidic, the majority of the added p97 peptide other than the terminal LYG segment was disordered. Crystal contacts are not within the vicinity of the positively charged ridge in the PUL domain. Together, these observations suggest that the PUL domain may have ligands other than p97. Moreover, the PUL domain unlikely functions in a role similar to importin-like proteins, which usually bind positively charged nuclear localization signal motifs.
Site-specific Mutations
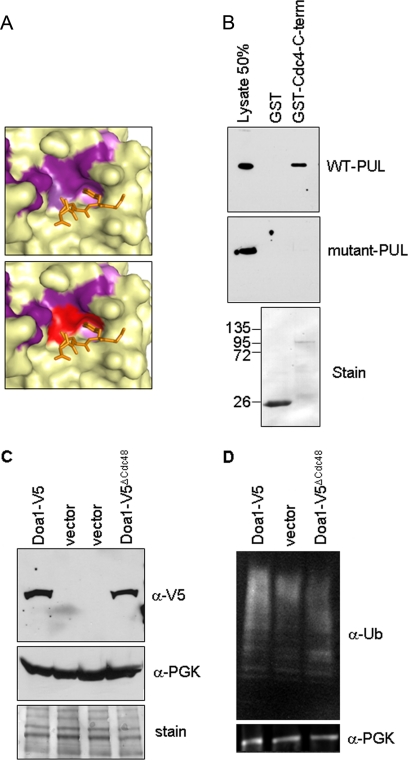
We sought to better understand the molecular functions and biological role of PLAA by testing the consequence of disrupting the interaction between the yeast orthologs of PLAA and p97, Doa1, and Cdc48, respectively. Conserved residues in PLAA PUL domain that contributed to binding the LYG tail of p97 are Ile551, Asp615, and Leu658 (Figs. 3 and 4A). The corresponding residues in yeast Doa1 were mutated as follows: I483A, D538R, and L571A to disrupt this interface. Wild type and mutant yeast PUL domains (defined by residues 461–715) were expressed as V5 epitope-tagged proteins in bacteria that were subjected to binding studies using GST alone or GST fused to the C-terminal 627 residues of Cdc48. Although the wild type PUL domain bound to Cdc48, the mutant PUL domain did not, thus confirming the predictions from the crystal structure of PLAA PUL/p97 (Fig. 4B). We refer to the Doa1 I483A,D538R,L571A triple mutant as Doa1ΔCdc48 to distinguish it as a mutant lacking its ability to bind Cdc48.
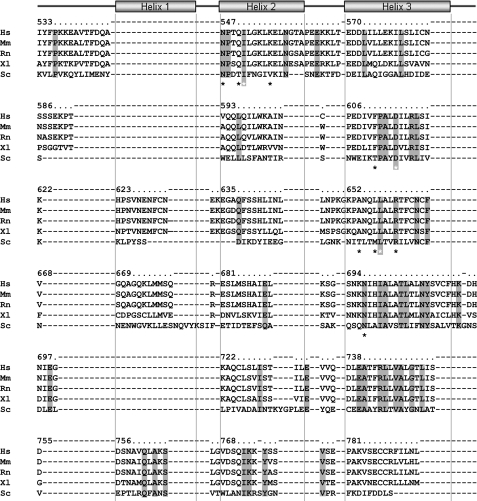
FIGURE 3.
Sequence alignment. Structure-based sequence alignment of PUL domain orthologs (Hs, Homo sapiens (Q9Y263); Ms, Mus musculus (P27612); Rn, Rattus norvegicus (P54319); Xl, Xenopus laevis (Q08B76) and Sc, Saccharomyces cerevisiae (P36037); Swiss-Prot accession number are in parentheses) are shown with fully conserved residues highlighted in gray. Asterisks indicate conserved residues of the p97 binding pocket; gray-highlighted, white asterisks indicate residues that were mutated in yeast Doa1. Numbering corresponds to human PLAA. Representation of the Armadillo helices are shown as rectangles.
FIGURE 4.
Effects of mutating the Cdc48-binding site in the PUL domain of Doa1. A, expanded view of the p97 binding pocket in the PUL domain of PLAA. Conserved residues are colored in purple (top panel) with the residues targeted for mutagenesis to make the doa1ΔCdc48 mutant in red (bottom panel). B, recombinant epitope-tagged PUL domains of Doa1 and Doa1ΔCdc48 were subjected to binding studies with GST alone (ø) or GST-Cdc48 C terminus. Bound fractions were immunoblotted with a 50% equivalent of input lysate. C, immunoblots from doa1Δ cells transformed with vector, wild type (WT) DOA1, or plasmid expressing Doa1ΔCdc48. Cell lysates were blotted for Doa1-V5 (anti-V5 antibody) or 3-phosphoglycerate. Stained membrane is shown as loading control. D, immunoblots from doa1Δ cells transformed with the indicated plasmids using anti-Ub antibodies or anti-3-phosphoglycerate antibodies.
Functional Studies in Yeasts
To test the functional consequence of loss of Cdc48 binding by Doa1, we transformed doa1Δ yeast with low copy plasmids expressing V5 epitope-tagged wild type Doa1 or the Doa1ΔCdc48 mutant. Both proteins were expressed under the control of the DOA1 promoter, and previous studies showed that the wild type V5 epitope-tagged Doa1 protein, which carries the tag at the C terminus), is fully functional (17). Immunoblotting cell lysates showed that the V5-Doa1ΔCdc48 was expressed at comparable levels to the wild type Doa1 (Fig. 4C). One of the major hallmarks of Doa1 loss is the depletion of ubiquitin levels. To assess the role of Cdc48 interaction in this function, we also immunoblotted for total Ub levels. The doa1ΔCdc48 cells showed only a moderate reduction in Ub compared with wild type cells (68 ± 8% S.D. that of wild type cells), and levels were elevated above those in doa1Δ null cells (38 ± 6% that of wild type cells) (Fig. 4D).
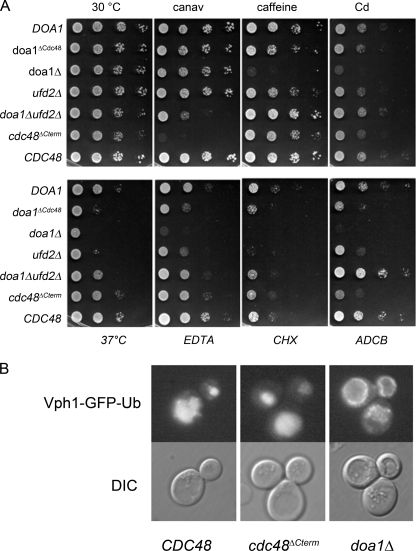
We next tested the doa1ΔCdc48 mutant in a number of growth assays under conditions previously found to dramatically affect doa1Δ cells (11, 12, 14–19, 28). As expected, doa1Δ null cells showed growth defects when subjected to elevated temperature (37 °C) EDTA, caffeine, cadmium, ADCB (a toxic proline analog), and low doses of cycloheximide (Fig. 5A). In contrast, wild type cells (doa1Δ cells transformed with the V5-DOA1 plasmid) grew well under these conditions. Surprisingly, doa1Δcdc48 cells only showed a major growth defect at 37 °C, and intermediate growth defects were found on EDTA, cycloheximide, and ADCB. Growth defects of doa1Δcdc48 cells on caffeine or cadmium were not observed. Thus, whereas the full function of Doa1 requires association with Cdc48, Doa1 can apparently execute some of its functions without the need to directly bind Cdc48.
FIGURE 5.
Functional uncoupling of the Cdc48-Doa1 interaction. A, yeast transformants with the indicated plasmids were serially diluted and plated on minimal plates containing the indicated drugs. Growth assays of wild type, doa1Δ cells, and cells where Cdc48 lacks its last five residues required for Doa1 interaction (cdc48ΔCterm) are shown. Drug plates contained 1 mm EDTA, 0.5 μg/.ml cycloheximide (CHX), 0.2% caffeine, 75 μg/ml canavanine (canav), or 75 μg/ml l-azetidine-2-carboxylic acid (ADCB). Cd, cadmium. B, sorting of Vph1-GFP-Ub, a marker of the MVB sorting pathway was assessed in doa1Δ cells, cdc48ΔC-term cells and matched CDC48::HIS cells. Intravacuolar delivery of Vph1-GFP-Ub was only blocked in the doa1Δ cells. DIC, differential interference contrast.
To confirm that Doa1 has a function independent of its association with Cdc48, we also conducted growth assays using cells in which the Doa1-binding site was deleted in Cdc48, cdc48ΔCterm. These cells were made by integrating a stop codon followed by the ADH1 3′-untranslated region and HISMX6 marker at the 3′ end of the Cdc48 open reading frame resulting in the deletion of the last five residues. As a control, we compared this strain to a similarly engineered strain, but where integration of the stop codon cassette left the Cdc48 open reading frame intact. Remarkably, doa1ΔCdc48 cells showed a set of phenotypes very distinct from doa1Δ null cells. In particular, growth on EDTA, caffeine, cadmium, and cycloheximide and growth at 37 °C were normal in cdc48ΔCterm cells confirming that the functions that Doa1 provides for growth under these conditions does not require association with Cdc48 (Fig. 5A). Because Cdc48 binds both Ufd2 and Doa1 competitively, we also compared the cdc48ΔC-term phenotype to ufd2Δ and doa1Δ ufd2Δ double mutant cells. Here again, the phenotype of the cdc48ΔC-term was distinct from the doa1Δ ufd2Δ double mutants indicating that the function of Doa1 and Ufd2 can be uncoupled from Cdc48. This was clearly seen with growth on ADCB and canavanine, where the cdc48ΔC-term cells showed defective growth, although the doa1Δ ufd2Δ cells showed robust growth.
Our previous experiments with Doa1 demonstrated a role in MVB sorting that was independent of the effect of Doa1 on ubiquitin levels (17). We speculated that Doa1 fulfilled this role by acting in concert with Cdc48. In particular, we found that the sorting of a large membrane protein complex, the vacuolar ATPase tagged with Ub using a Vph1-GFP-Ub fusion, was defective in delivery to the vacuole lumen. These studies demonstrated a unique and more direct role for Doa1 and provided the possibility that a Doa1-Cdc48 complex was required to break apart ubiquitinated membrane protein complexes prior to their sorting into the MVB pathway. Based on our structural understanding of the Doa1-Cdc48 interaction, we tested this idea by following the sorting of Vph1-GFP-Ub in cdc48ΔC-term cells that lack the Doa1-PFU Cdc48 interaction (Fig. 5B). Here, however, sorting of Vph1-GFP-Ub was indistinguishable from wild type cells. In contrast, Vph1-GFP-Ub sorting was still defective in doa1Δ cells as previously documented (17). Thus, whereas Doa1 provides an important function with regard to sorting proteins along the MVB pathway, its association with Cdc48 is not critical.
DISCUSSION
PLAA (Doa1) was discovered, along with Ufd1, -2, and -4, as an important component of the yeast Ub fusion-dependent degradation pathway (28). It was eventually found that these proteins interact with the hexameric p97 ATPase, which uses these and other adaptor proteins to dictate the fate of a variety of substrates, either facilitating proper folding or Ub-dependent proteasomal degradation. Doa1/Ufd3 is critical for proper function of a number of Ub-dependent processes; however, the precise biochemical activity Doa1/PLAA provides in vivo is not clear. One possibility is that it modulates or counteracts the ubiquitinating E3 ligase activity of Ufd2 associated with p97 (7). Genetic and biochemical data support this idea because deletion of UFD2 alleviates some of the growth defects caused by DOA1 loss. Furthermore, Ufd2 competes for the same site on p97 (the LYG-containing C terminus). Recently, it has been reported that the core region of Ufd2 consists of multiple Armadillo-like repeats, raising the possibility that this region interacts with the p97 C terminus in a manner similar to the PUL domain of Doa1/PLAA (29). The multivalent nature of assembled p97 would theoretically be able to support simultaneous binding of Ufd2, PLAA, and other C-terminally associated factors such as PNGase, Rnf1, and Ubxd1 (3, 4). Yet in vivo, Cdc48 complexes with Ufd2 have no simultaneous Doa1 bound (7). Other mechanisms such as weak homo-oligomeric associations of Ufd2 or the PLAA PUL domain that would favor occupancy of the p97 C terminus with only Ufd2 or PLAA may account for the final assembly of different p97 complexes. Evidence for such higher order oligomeric interfaces between PUL domains, however, is lacking, and further work will be required to determine how the p97 C terminus is exclusively bound to Ufd2 or PLAA or other factors.
Competition between Ufd2 and PLAA could be important for regulating p97-mediated ubiquitination signaling pathways. If Ufd2 is a vigorous E3 ligase, its unrestrained activity could lead to lowered global levels of free Ub. Although the simple model of competition for binding could potentially explain the antagonistic genetic interaction of Doa1 and Ufd2, this simple model is not supported by data showing that expression of Doa1 truncations containing the Cdc48-binding PUL domain are not functional. These data support the idea that displacement of Ufd2 is not the only important Doa1 function (12, 16).
In mammalian cells, the regulation of binding of PLAA, PNGase, and Ufd2 can be mediated by phosphorylation of the conserved Tyr805 residue of p97. Despite no structural homology between PLAA and PNGase Tyr805 of p97 plays an indispensable role in these interactions, forming ionic bonds, hydrogen bonds, and hydrophobic interaction. Phosphorylation of Tyr805 disrupts interaction with both PNGase and PLAA, and it will be interesting to determine whether Ufd2 association is similarly perturbed. Regulation of Tyr805 may provide a phosphorylation-driven switch that could prevent all C-terminal interactions or could swap one set of p97 interactors for another.
PLAA was proposed to regulate activation of phospholipase A2 (30). Using antibodies to the amphipathic α-helical melittin, a mouse cDNA variant that encoded a unique, melittin-like protein sequence downstream of the WD40 domain in lieu of the PFU or PUL domains was originally identified (GenBankTM accession number M57958). An alternative amphipathic helix within the PUL domain of PLAA has been proposed to work as a melittin-like phospholipase A2 activator (31). This region corresponds to the second α-helix in the PUL domain defined as residues Gly560–Gln595. As an isolated peptide, this region would likely also form an amphipathic α-helix that would activate phospholipase A2. However, in the context of the PUL domain, most of the hydrophobic side chains face inward toward the core. The tightly folded nature of the PUL domain makes it unlikely that such a region would be able to act independently as an amphipathic α-helix within the context of full-length PLAA. Thus, there is little biochemical rationale for PLAA serving as a true activator of phospholipase A2 activity.
Stacks of ARM repeats are usually rigid and form banana-shaped domains that generate good binding surfaces, particularly on the inside curvature (32). Three-dimensional structure comparisons of the PUL domain using the DALI server (33) identified 17 proteins with Z-scores greater than 10, the top three of which were importin-α, β-catenin, and Hsp70-binding protein, which all have classical Armadillo repeats. Despite structural homology, these proteins have distinct molecular functions and biological roles. Interestingly, a number of ligands are known for importin-α and β-catenin. Importin-α has the capacity to bind many different proteins with a loosely conserved NLS motif. β-Catenin is the central effector of the canonical Wnt signaling pathway, interacting via its Armadillo repeat region with numerous partners at the membrane, in the cytosol, and in the nucleus. There is no evidence suggesting that p97 is the only ligand of the PUL domain of PLAA, leaving the possibility that like other ARM repeat proteins additional binding partners may exist. Surprisingly, only two p97 residues appear to contribute to the bulk of the interactions with the PUL domain and are the basis of the observed affinity (5 μm). Our structure of the PUL-p97 complex supports the notion of additional PLAA ligands because there are conserved surface residues that lie outside the p97 peptide binding pocket. Indeed, while our manuscript was being prepared, a recently described crystal structure of a portion of the yeast Doa1 PUL domain alone was published along with the in vivo analysis of several point mutations (34). Although these point mutations did not lie within the binding pocket we define for the DLYG Cdc48 ligand, they mapped to conserved residues that lie in an adjacent groove and indeed caused a loss of Doa1 function in vivo, which could be due to the loss of binding factors in addition to Cdc48.
Mutations confined to the Cdc48 DLYG binding pocket defined in our crystal structure led to selective changes in the overall function of Doa1. The doa1ΔCdc48 cells displayed some of the phenotypes observed for doa1Δ cells, such as susceptibility to high temperature and EDTA. However, other Doa1 functions did not require Cdc48 interaction via the PUL domain such as resistance to caffeine and cadmium. The doa1ΔCdc48 cells also had partial resistance to ADCB, a toxic compound that is transported via the cell surface Gap1 permease and a crude measure of its Ub-dependent sorting. The converse mutation, Cdc48 lacking the last five residues that mediate binding to Doa1, also resulted in a diverse set of phenotypes that were different from doa1Δ and doa1Δ ufd2Δ cells. Together, our results indicate that Doa1 partially functions in a complex with Cdc48 but that Doa1 also has a functional life outside of this context. At least one of these functions includes its role in facilitating MVB sorting of ubiquitinated membrane proteins because Cdc48 mutants that are devoid of Doa1 binding are normal in this regard. Given the dramatic effect on Doa1 function when the entire PUL domain is deleted and the genetically separable effects of the cdc48ΔC-term mutation from doa1Δ and ufd2Δ mutations, we suspect that both the Doa1 PUL domain and the Cdc48 C terminus have potential interacting partners that have yet been identified.
Supplementary Material
Acknowledgments
We thank Yanjun Li, Nasrin Rastgoo, and Yulia Slessarev for assistance in cloning, mutagenesis, and purification, respectively. We thank Gino Cingolani for scientific discussions. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck, the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM58202 (to R. C. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2.
The atomic coordinates and structure factors (code 3EBB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Ub
- ubiquitin
- ARM
- Armadillo
- PFU
- PLAA family ubiquitin binding
- PNGase
- peptide:N-glycosidase
- MVB
- multivesicular body.
REFERENCES
- 1.Jentsch S., Rumpf S. (2007) Trends Biochem. Sci. 32, 6–11 [DOI] [PubMed] [Google Scholar]
- 2.Shcherbik N., Haines D. S. (2007) Mol. Cell 25, 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeung H. O., Kloppsteck P., Niwa H., Isaacson R. L., Matthews S., Zhang X., Freemont P. S. (2008) Biochem. Soc. Trans. 36, 62–67 [DOI] [PubMed] [Google Scholar]
- 4.Madsen L., Seeger M., Semple C. A., Hartmann-Petersen R. (2009) Int. J. Biochem. Cell Biol. 41, 2380–2388 [DOI] [PubMed] [Google Scholar]
- 5.Allen M. D., Buchberger A., Bycroft M. (2006) J. Biol. Chem. 281, 25502–25508 [DOI] [PubMed] [Google Scholar]
- 6.Zhao G., Zhou X., Wang L., Li G., Schindelin H., Lennarz W. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8785–8790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rumpf S., Jentsch S. (2006) Mol. Cell 21, 261–269 [DOI] [PubMed] [Google Scholar]
- 8.Li G., Zhao G., Schindelin H., Lennarz W. J. (2008) Biochem. Biophys. Res. Commun. 375, 247–251 [DOI] [PubMed] [Google Scholar]
- 9.Egerton M., Ashe O. R., Chen D., Druker B. J., Burgess W. H., Samelson L. E. (1992) EMBO J. 11, 3533–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer L. M., Koonin E. V., Aravind L. (2004) Cell Cycle 3, 1440–1450 [DOI] [PubMed] [Google Scholar]
- 11.Ghislain M., Dohmen R. J., Levy F., Varshavsky A. (1996) EMBO J. 15, 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- 12.Mullally J. E., Chernova T., Wilkinson K. D. (2006) Mol. Cell. Biol. 26, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell N. S., Wilkinson K. D. (2004) Biochemistry 43, 4844–4854 [DOI] [PubMed] [Google Scholar]
- 14.Kunze D., MacCallum D., Odds F. C., Hube B. (2007) Microbiology 153, 1026–1041 [DOI] [PubMed] [Google Scholar]
- 15.Lis E. T., Romesberg F. E. (2006) Mol. Cell. Biol. 26, 4122–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogiso Y., Sugiura R., Kamo T., Yanagiya S., Lu Y., Okazaki K., Shuntoh H., Kuno T. (2004) Mol. Cell. Biol. 24, 2324–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren J., Pashkova N., Winistorfer S., Piper R. C. (2008) J. Biol. Chem. 283, 21599–21611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfe D., Reiner T., Keeley J. L., Pizzini M., Keil R. L. (1999) Mol. Cell. Biol. 19, 8254–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandina I., Smirnov A., Kolesnikova O., Entelis N., Krasheninnikov I. A., Martin R. P., Tarassov I. (2007) FEBS Lett. 581, 4248–4254 [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 26.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 28.Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. (1995) J. Biol. Chem. 270, 17442–17456 [DOI] [PubMed] [Google Scholar]
- 29.Tu D., Li W., Ye Y., Brunger A. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15599–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark M. A., Ozgür L. E., Conway T. M., Dispoto J., Crooke S. T., Bomalaski J. S. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5418–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribardo D. A., Kuhl K. R., Peterson J. W., Chopra A. K. (2002) Toxicon 40, 519–526 [DOI] [PubMed] [Google Scholar]
- 32.Hatzfeld M. (1999) Int. Rev. Cytol. 186, 179–224 [DOI] [PubMed] [Google Scholar]
- 33.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao G., Li G., Schindelin H., Lennarz W. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16197–16202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.