Abstract
Clostridium perfringens enterotoxin (CPE), a causative agent of food poisoning, is a pore-forming toxin disrupting the selective permeability of the plasma membrane of target cells, resulting in cell death. We previously identified claudin as the cell surface receptor for CPE. Claudin, a component of tight junctions, is a tetratransmembrane protein and constitutes a large family of more than 20 members, not all of which serve as the receptor for CPE. The mechanism by which the toxin distinguishes the sensitive claudins is unknown. In this study, we localized the region of claudin responsible for interaction with CPE to the C-terminal part of the second extracellular loop and found that the isoelectric point of this region in sensitive claudins was higher than insensitive claudins. Amino acid substitutions to lower the pI resulted in reduced sensitivity to CPE among sensitive claudins, whereas substitutions to raise the pI endowed CPE-insensitive claudins with sensitivity. The steric structure of the claudin-binding domain of CPE reveals an acidic cleft surrounded by Tyr306, Tyr310, Tyr312, and Leu315, which were reported to be essential for interaction with the sensitive claudins. These results imply that an electrostatic attraction between the basic claudin region and the acidic CPE cleft is involved in their interaction.
Keywords: Cell/Adhesion, Membrane/Proteins, Protein/Protein-protein interactions, Protein/Domains, Protein/Structure, Receptors/Structure-Function, Toxins/Drugs/Xenobiotics/Bacterial
Introduction
Clostridium perfringens enterotoxin (CPE),2 a causative agent of food poisoning, damages intestinal epithelial cells by forming physical pores on the cell membrane. CPE consists of a single chain polypeptide of 319 amino acids. The toxin binds to a receptor on target cells via a C-terminal receptor-binding domain and reportedly organizes large molecular complexes with cellular components to make the pores, a process that is conducted by an N-terminal cytotoxic domain (1–6). Unlike other pore-forming toxins such as cholesterol-dependent cytolysins, CPE shows strict specificity for sensitive cells, implying the existence of a particular but not ubiquitous receptor for the toxin on CPE-sensitive cells (7–9). A tetratransmembrane protein was isolated as the CPE receptor in 1997 (10) and was later found to be a component of tight junctions and designated claudin (Cldn) (11). CPE kills only cells presenting Cldn on their surface. No CPE receptors other than Cldn have been identified to date. It was demonstrated that the noncytotoxic C-terminal part of CPE (C-CPE, residues 184–319 of CPE) disrupted the strand structure of the tight junction and increased paracellular permeability by sequestering Cldn (12), indicating that Cldn is essential to the function and structure of the tight junction. This raises the possibility that CPE or C-CPE could provide a useful tool for targeting Cldn-presenting cells or modulating the Cldn-dependent paracellular permeability. In fact, attempts have been made to use CPE in the treatment of several forms of cancer that highly express certain types of Cldns (13, 14). The use of C-CPE as a modulator of Cldns to enhance drug absorption through intestinal epithelial cells has also been attempted (15).
The Cldn family comprises more than 20 closely related transmembrane proteins. Not all of the members serve as the receptor for CPE; it was reported that Cldns 3, 4, 6, 7, 8, and 14, but not Cldns 1, 2, 5, and 10, were sensitive to CPE (16). Cldn is a tetratransmembrane protein and has two extracellular loops. CPE is considered to recognize the second extracellular loop of the sensitive Cldns. However, little is known about how CPE distinguishes the sensitive Cldns from the closely related insensitive Cldns. Therefore, we tried to narrow down the region of Cldn essential for CPE to recognize and look for common features in the CPE-sensitive Cldns. The results presented here imply that the C-terminal 12 amino acids of the second extracellular loop partly determined the sensitivity to CPE. Notably, the electric charge of this region was likely important for interaction with the toxin. These observations may provide important information for use of the toxin for cancer therapy and drug delivery through epithelial barriers.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-V5 monoclonal antibody and Alexa488-labeled anti-mouse monoclonal antibody were purchased from Invitrogen. Anti-HA monoclonal antibody (HA.11) was purchased from Covance (Berkeley, CA). Anti-FLAG M2 monoclonal antibody was from Sigma. Anti-β-actin polyclonal antibody was from Imgenex (San Diego, CA). Horseradish peroxidase-conjugated anti-rabbit polyclonal antibody was from Jackson (West Grove, PA), and horseradish peroxidase-conjugated anti-mouse polyclonal antibody was from ICN Pharmaceuticals (Solon, OH).
Plasmids
The plasmids and primers or oligonucleotides used in this study are listed in supplemental Table S1. All of the constructed recombinant genes were verified by sequencing before use.
pcDNA3-derived Plasmids for Expression in HEK293 Cells
cDNAs of human (hu) Cldn1, huCldn4, monkey (mk) Cldn4, Cldn4–1(A), and Cldn1–4-1(A), all of which were tagged with the FLAG peptide at their C termini and inserted in pMEpyori18Sf− (17), were kindly provided by Jun Katahira (Graduate School of Frontier Biosciences, Osaka University). These cDNAs were excised by digestion with XhoI and XbaI and inserted into the corresponding site of pcDNA3 (Invitrogen). pcDNA3·Cldn1–4-1(C) was constructed as follows. Upstream and downstream gene fragments of Cldn1–4-1(C) were amplified by PCR with pcDNA3·huCldn1 as a template and a combination of the primers T7 and 141C-R for the upstream fragment and of SP6 and 141C-F for the downstream fragment. The two fragments were further subjected to PCR with T7 and SP6. The DNA fragment was inserted into the XhoI-XbaI site of pcDNA3 by enzymatic digestion and ligation. pcDNA3·Cldn1–4-1(D) was prepared in the same way with the primers indicated. For pcDNA3·Cldn4HA, two fragments were amplified by PCR with pcDNA3·mkCldn4 as a template and a combination of 4aHS, flanked by HpaI and SacII sites, and T7, and of S-4b, containing a SacII site and SP6. After digestion with SacII, the two fragments were ligated and then inserted into the XhoI-XbaI site of pcDNA3. These procedures result in an HpaI-SacII site in the Cldn4 gene between codons corresponding to amino acid residues 72 and 73. Separately, synthetic oligonucleotides (HA-F and HA-R) were annealed to each other. The resultant fragment encoding the HA tag (VNYPYDVPDYAENLYFQGAA) flanked by adhesive ends for HpaI and SacII was inserted into the corresponding site of the Cldn4 gene. Other pcDNA3-derived plasmids were made by a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla CA) with the templates of pcDNA3·Cldn4–1(A) for pcDNA3·Cldn4–1(B) and pcDNA3·Cldn4–1(C) and pcDNA3·Cldn1–4-1(C) for pcDNA3·Cldn1–4-1(B).
pMEneo-derived Plasmids for Expression in L929 Cells
The cDNA of huCldn4 was amplified by PCR and inserted into pEF6/V5-His TOPO TA (Invitrogen). The Cldn4 gene was excised by digestion with SpeI and XbaI and inserted into the corresponding site of pMEneo (18). The EcoRV-PmeI fragment of pEF6/V5-His TOPO TA was inserted into the EcoRV site of the pMEneo-derived vector so that the gene for the V5 tag was inserted downstream of the Cldn4 gene. The DNA obtained was named pMEneo·huCldn4. cDNAs of huCldn5 and huCldn10 were obtained from the National Institutes of Health Mammalian Gene Collection (Image ID 5242567 and 4246806 for huCldn5 and huCldn10, respectively), and cDNA of mouse (ms) Cldn7 was obtained from the RIKEN Mouse Genome Encyclopedia DNABookTM (DNA ID 0610043B04, DNAFORM). DNA fragments amplified by PCR with the huCldn5, huCldn10, and msCldn7 cDNAs, and the indicated primers were cloned into pEF6/V5-His TOPO TA. The resultant plasmids were designated pEF6·huCldn5, pEF6·huCldn10, and pEF6·msCldn7. The DNA fragments encoding these Cldns were excised from each plasmid by digestion with SpeI and EcoRV and substituted for the Cldn4 gene of pMEneo·huCldn4 in the corresponding region. pMEneo·Cldn4-5-4 was constructed as follows. Two independent PCRs were carried out with pEF6·huCldn5 as the template and 454-R2 and T7 as the primers and with the template pEF6·huCldn4 and primers 454-F2 and BGHrv. The two DNA fragments obtained were used as templates for PCR with T7 and BGHrv, and a chimeric DNA fragment for huCldn5 (amino acids 1–160) and huCldn4 (amino acids 161–209) was obtained. This fragment was used as a template for PCR with the primers 454-F1 and BGHrv, and another PCR was carried out with pEF6· huCldn4 as a template and primers 454-R1 and T7. The two DNA fragments were subsequently subjected to PCR with primers T7 and BGHrv. The DNA fragment encoding Cldn4-5-4 was inserted into the SpeI-EcoRV site of pMEneo·huCldn4 as described above. pMEneo·Cldn5-4-5, pMEneo·Cldn7-5-7, and pMEneo·Cldn5-7-5 were prepared in the same way. Other pMEneo-derived plasmids were made by the QuikChange site-directed mutagenesis kit with pMEneo·huCldn4 as the template for pMEneo·Cldn4FYAA and pMEneo·Cldn4NRDY and pMEneo·huCldn5 as the template for pMEneo·Cldn5DYNR.
Cell Culture
HEK293 cells and L929 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37 °C, 5% CO2 in air. The cell lines stably expressing Cldns or Cldn variants were established as follows; HEK293 or L929 cells cultured in 24-well plates were transfected with the aid of Lipofectamine 2000 (Invitrogen) with 2 μg/well of the pMEneo-derived or pcDNA3-derived plasmid, respectively. The clone resistant to 1 mg/ml of G418 was selected as an actual transfectant and maintained in Dulbecco's modified Eagle's medium containing 500 μg/ml of G418. The expression of Cldns or Cldn variants by the selected clones was checked before use by Western blotting with the anti-FLAG antibody for Cldns expressed in HEK293 cells or anti-V5 antibody for Cldns expressed in L929 cells. L929 cells expressing msCldns except msCldn7 were provided by Shoichiro Tsukita (Kyoto University). The expression levels in these cells were checked by Western blotting with anti-FLAG antibody because these Cldns were tagged with the FLAG peptide at the C terminus.
CPE and C-CPE
pET16bCPE or pETH10PER (10), which includes a His10-tagged CPE gene or His-tagged C-CPE gene, respectively, was introduced into the Escherichia coli BL21-CodonPlus (DE3)-RIL strain (Stratagene). The proteins were produced by cultivation of the bacteria in LB broth supplemented with 1 mm isopropyl β-d-thiogalactopyranoside. The bacterial cells were disrupted by sonication and suspended in 50 mm phosphate buffer, pH 8.0, containing 0.3 m NaCl and 10 mm imidazole. The produced proteins were purified from the bacterial cell extract by elution with a 10–500 mm imidazole gradient in the same buffer from a His-Select nickel affinity gel (Sigma) column.
Cytotoxic Assay
A 100-μl aliquot of cell suspension at a concentration of 2.0 × 106 cells/ml was mixed with the same volume of CPE in Dulbecco's modified Eagle's medium at an appropriate concentration in each well of 96-well flat-bottomed microtiter plates. After incubation of the mixture at 37 °C for 2 h, 4 μl of tetrazolium salt solution (WST-8; Kishida Chemical, Osaka Japan) was added as an indicator of viability, and the mixture was incubated for another 1 h. The absorbance of the mixture in the well was measured at a wavelength of 450 nm for the indicator color and 620 nm for background. The net value was obtained by subtraction of the latter value from the former. The survival rate of cells was calculated according to the following equation, where ACPE is the net absorbance of the CPE-treated sample, ANO is the net absorbance of the untreated sample, and ABLANK is the net absorbance of a well without cells. The survival rate can be calculated as follows, ACPE − ABLANK/ANO − ABLANK. The EC50 value was computed with Prism 4 (GraphPad Software).
Other Methods
The radioiodination of CPE or C-CPE and binding assay for Cldn-expressing cells (105 cells/assay) were performed as described previously (10). The specific radioactivity of 125I-labeled CPE ranged from 42 to 144 MBq/mg protein. The amount of proteins described here were determined with a micro BCA protein assay reagent kit (Thermo Fisher Scientific) or by the method of Bradford with protein assay CBB solution (Nacalai, Japan). For Western blotting, the samples were electrically transferred onto polyvinylidene difluoride membranes (Bio-Rad) following SDS-PAGE. The membranes were then treated with 5% skim milk, and the transferred proteins were probed with appropriate antibodies and visualized with an enhanced chemiluminescence system (ECL plus; GE Healthcare). The pI values of peptides were calculated with the PEPSTATS program of the EMBOSS software package (19). The image of C-CPE was drawn from the structural data (Protein Data Bank code 2QUO) with PyMOL. The surface charge of C-CPE was evaluated by Adaptive Poisson-Boltxmann Solver.
RESULTS
A previous report revealed that CPE recognizes the second loop of the sensitive Cldns by swapping extracellular loops between the CPE-sensitive Cldn3 and the CPE-insensitive Cldn1 (16). The chimeric Cldn possessing the second extracellular loop of the CPE-sensitive Cldn3 but not the CPE-insensitive Cldn1 was sensitive to CPE. Other groups also concluded that CPE recognizes the second loop of Cldns (20, 21). It remains unclear whether the first extracellular loop of Cldn is involved in interaction with CPE in situ. Therefore, as a first step, we constructed a recombinant Cldn4 (Cldn4HA), which has an HA tag in the first extracellular loop (supplemental Fig. S1A). In this system, anti-HA antibody could be used as a specific ligand for the first loop of Cldn4HA. We established HEK293 cells stably expressing Cldn4HA and examined them for binding of anti-HA antibody and C-CPE, which carries the receptor-binding domain but not the cytotoxic domain. Flow cytometric analysis demonstrated a positive correlation indicating that biotinylated C-CPE and anti-HA antibody independently bound to Cldn4HA (supplemental Fig. S1B). The binding of the biotinylated C-CPE was inhibited by unlabeled C-CPE but not by anti-HA antibody (supplemental Fig. S1, C and D). These results imply that the first loop of claudin may not be involved in the interaction of CPE with Cldns, confirming previous observations.
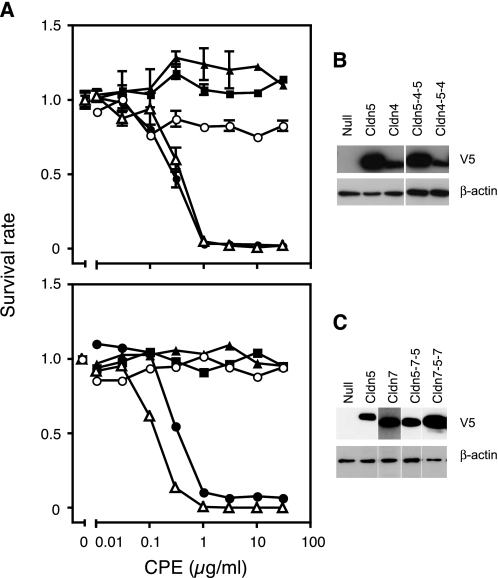
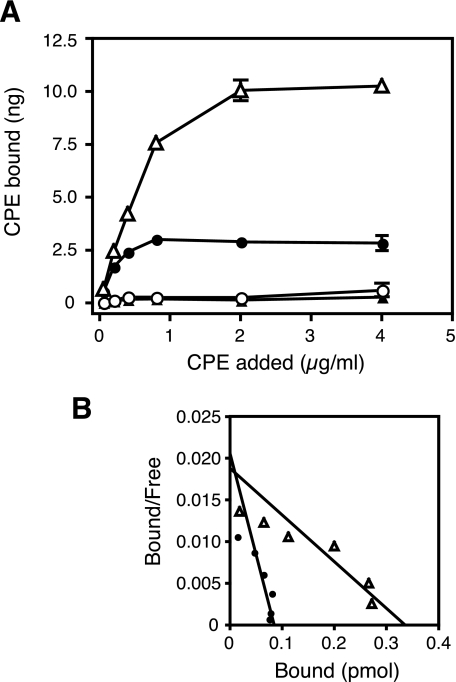
Because we could not find any evidence of the involvement of the first loop in the recognition of CPE, we then focused on the second loop. We prepared CPE-insensitive HEK293 cells expressing Cldns, which have various forms of the chimeric second loop (Table 1 and supplemental Fig. S2) and examined them for CPE sensitivity. It was confirmed by Western blotting of the cell fractions that the Cldns were presented on the cell membrane (supplemental Fig. S2). In the course of these experiments, we found that Cldn1, which had been considered to be a typical CPE-insensitive Cldn, actually responded to the toxin at higher concentrations. Therefore, we calculated the EC50 values of the cells expressing the chimeric Cldns against CPE from the results of the cytotoxicity assay and compared their sensitivities. As a result, the Cldns were grouped into a highly sensitive class (EC50 < 1 μg/ml), a slightly sensitive class (1 ≤ EC50 < 30 μg/ml), and an insensitive class (EC50 ≥ 30 μg/ml). According to these criteria, Cldn1 was classified as low sensitive, not insensitive. Among the chimeric Cldns, Cldn4–1(B) was highly sensitive to CPE, whereas Cldn4–1(A) was insensitive. Cldn1–4-1(B) was also highly sensitive, whereas Cldn1–4-1(C) and (D) were as slightly sensitive as Cldn1. These results imply that the region from Asn149 to Met160 of Cldn4 determines sensitivity to CPE. We named this region the CPE sensitivity-related region (CPE-SR). The reason why Cldn4–1(A), although expressed enough on the cell membrane (supplemental Fig. S2), became insensitive to CPE was unknown. This chimeric mutation may influence the structure around the second loop and downstream transmembrane region. Next, we tested whether the CPE-SR of Cldn4 changes another CPE-insensitive Cldn, Cldn5 (Fig. 1) (9, 10). We used L929 cells instead of HEK293 cells for the following experiments, because in the first experiments, HEK293 cells were found to occasionally gain sensitivity to CPE after serial passages for long periods. We established L929 cells stably expressing Cldn5-4-5, a chimera in which the CPE-SR of Cldn4 was inserted into Cldn5 in place of the corresponding region, and examined them for CPE sensitivity. The cells expressing Cldn5-4-5 were found to be as sensitive to CPE as the cells expressing Cldn4 (Fig. 1). In contrast, when the CPE-SR of Cldn4 was replaced with the corresponding region of Cldn5, Cldn4 became insensitive to CPE (Fig. 1, Cldn4-5-4). This was also the case with Cldn5 and CPE-sensitive Cldn7. The CPE-SR of Cldn7 made Cldn5 sensitive (Fig. 1, Cldn5-7-5), whereas replacing the CPE-SR of Cldn7 with Cldn5 resulted in insensitivity to CPE (Fig. 1, Cldn7-5-7). According to the calculated EC50 values, Cldn5-4-5 and Cldn5-7-5 could be classified as highly sensitive (Table 2). 125I-Labeled CPE bound to the cells expressing Cldn4 or Cldn5-4-5 but not to the cells expressing Cldn5 or Cldn4-5-4 (Fig. 2). These results indicate that the sensitivity to CPE of the cells expressing various Cldns reflected the binding of CPE to Cldns. The Kd values of Cldn4 and Cldn5-4-5 for CPE were 4.42 × 10−9 and 1.91 × 10−8 m, respectively. These values are consistent with those estimated for the CPE-sensitive Cldns (9, 10, 16, 22). The amount of the bound CPE was higher in Cldn5-4-5-expressing cells than in Cldn4-expressing cells. This is probably due to the former expressing more Cldn than the latter, as shown in Fig. 1B, the results of which were obtained with the same clones of Cldn-expressing cells.
TABLE 1.
Cldns with the chimeric second loop
The chimeric Cldns were expressed in HEK293 cells and examined for sensitivity to CPE, which was classified into three categories: highly sensitive (High), EC50 < 1 μg/ml; slightly sensitive (Low), 1 μg/ml ≤ EC50 < 30 μg/ml; and insensitive, 30 μg/ml ≤ EC50. The Cldn1 and Cldn4 genes used in these experiments were derived from human and monkey, respectively. The underlining in the sequences indicates the second extracellular loop predicted by Sosui. The CPE-SR is indicated by bold type. The EC50 values are the means ± S.D. (μg/ml) calculated from three independent experiments.
| Claudin | Amino acid sequence | EC50 | Sensitivity |
|---|---|---|---|
| Wild type | |||
| huCldn1 | 139WYGNRIVQEFYDPMTPVNARYEFGQALFTGW169 | 4.32 ± 2.94 | Low |
| mkCldn4 | 138WTAHNIIQDFYNPLVASGQKREMGASLYVGW168 | 0.18 ± 0.09 | High |
| Chimera | |||
| Cldn4-1(A) | WTAHNIIQDFYNPLVASGQKREFGQALFTGW | >30 | Insensitive |
| Cldn4-1(B) | WTAHNIIQDFYNPLVASGQKREMGQALFTGW | 0.50 ± 0.28 | High |
| Cldn4-1(C) | WTAHNIIQDFYNPLVASGQKREMGAALFTGW | 0.11 ± 0.01 | High |
| Cldn1-4-1(A) | WYGNRIVQEFYNPLVASGQKREMGASLYVGW | 0.65 ± 0.23 | High |
| Cldn1-4-1(B) | WYGNRIVQEFYNPLVASGQKREMGAALFTGW | 0.46 ± 0.29 | High |
| Cldn1-4-1(C) | WYGNRIVQEFYDPLVASGQKREMGAALFTGW | 3.42 ± 0.93 | Low |
| Cldn1-4-1(D) | WYGNRIVQEFYDPMVASGQKREMGAALFTGW | 2.08 ± 0.39 | Low |
FIGURE 1.
CPE sensitivity of L929 cells expressing various Cldn chimeras. A, upper panel, CPE sensitivity of the cells expressing no Cldn (filled squares), Cldn5 (filled triangles), Cldn5-4-5 (open triangles), Cldn4 (filled circles), or Cldn4-5-4 (open circles) was examined by the cytotoxicity assay with the WST-8 reagent. Lower panel, CPE sensitivity of the cells expressing no Cldn (filled squares), Cldn5 (filled triangles), Cldn5-7-5 (open triangles), Cldn7 (filled circles), or Cldn7-5-7 (open circles) was examined. The experiments were repeated three times, and representative results are shown. Each point represents the mean ± S.D. (upper panel, n = 3) or the intermediate value (lower panel, n = 2). B and C, the expression level of each Cldn was confirmed by Western blotting with anti-V5 antibody, to which 2 × 104 cells/lane were subjected. The β-actin of each sample was stained as a control.
TABLE 2.
CPE sensitivity of cells expressing various Cldns
Cultured cells expressing various Cldns were subjected to the cytotoxicity assay as described under “Experimental Procedures,” and the EC50 values were determined. The cells were classified into three categories as mentioned in the text. CPE-SRs are shown in bold type. The pI values are for CPE-SR. The asterisks indicate the consensus aromatic amino acids upstream of CPE-SR. mh, mouse-human chimera. mkCldn4 and huCldn1 were expressed in HEK293 cells. Other Cldns were expressed in L929 cells.
| Claudin | Amino acid sequence | pI | EC50 |
|---|---|---|---|
| ** | |||
| Highly sensitive | |||
| msCldn4 | NVIRDFYNPMVASGQKREMGAS | 9.70 | 0.21 |
| huCldn4 | NIIQDFYNPLVASGQKREMGAS | 9.70 | 0.083 |
| mkCldn4 | NIIQDFYNPLVASGQKREMGAS | 9.70 | 0.18 |
| msCldn3 | TIIRDFYNPLVPEAQKREMGAG | 6.53 | 0.20 |
| msCldn7 | QIVTDFYNPLTPMNVKYEFGPA | 6.40 | 0.29 |
| msCldn8 | SIIRDFYNPLVDVALKRELGEA | 6.49 | 0.69 |
| Slightly sensitive | |||
| msCldn14 | DVVQNFYNPLLPSGMKFEIGQA | 6.41 | 4.7 |
| msCldn2 | GILRDFYSPLVPDSMKFEIGEA | 4.18 | 4.4 |
| msCldn1 | GIVQEFYDPLTPINARYEFGQA | 4.18 | 12 |
| huCldn1 | RIVQEFYDPMTPVNARYEFGQA | 4.18 | 4.3 |
| Insensitive | |||
| msCldn5 | IVVREFYDPTVPVSQKYELGAA | 4.18 | >30 |
| huCldn5 | IVVREFYDPSVPVSQKYELGAA | 4.18 | >30 |
| huCldn10 | KITTEFFDP-LFVEQKYELGAA | 3.93 | >30 |
| Cldn mutants examined in this study | |||
| huCldn4-5-4 | NVIRDFYDPSVPVSQKYELGAS | 4.18 | >30 |
| huCldn5-4-5 | IVVREFYNPLVASGQKREMGAA | 9.70 | 0.40 |
| mhCldn7-5-7 | QIVTDFYDPSVPVSQKYELGPA | 4.18 | >30 |
| mhCldn5-7-5 | IVVREFYNPLTPMNVKYEFGAA | 6.40 | 0.13 |
| huCldn5DYNR | IVVREFYNPSVPVSQKRELGAA | 9.70 | 1.4 |
| huCldn4NRDY | NIIQDFYDPLVASGQKYEMGAS | 4.18 | 14 |
FIGURE 2.
Effects of CPE-SR on binding of 125I-CPE to L929 cells expressing Cldns. L929 cells were incubated with 125I-labeled CPE at various concentrations, and the amounts of bound CPE were determined as described under “Experimental Procedures.” The experiments were repeated at least three times, and representative results are shown. A, saturation curves of the binding of 125I-CPE to cells expressing Cldn4 (filled circles), Cldn5 (filled triangles), Cldn4-5-4 (open circles), or Cldn5-4-5 (open triangles). Each plot represents the mean ± S.D. (n = 3). B, Scatchard plots of the data shown in the A. The data of the cells expressing Cldn4 (filled circles) and Cldn5-4-5 (open triangles) are shown. Numbers of moles of His-tagged CPE were calculated with a molecular mass of 37,850 Da. The Scatchard lines were drawn based on the instructions of Prism 4 software.
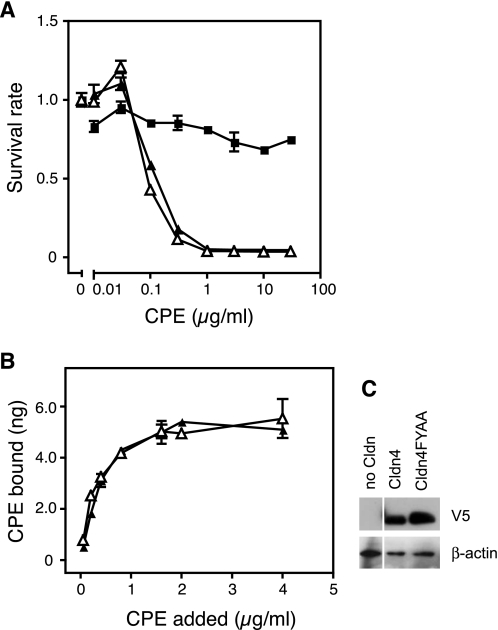
We have identified the CPE-SR as a key region for Cldns to serve as the receptor for CPE. To understand what element of the CPE-SR is recognized by CPE, we examined two possibilities: Cldns may have an intrinsic ability to bind to CPE, which the CPE-SR of the insensitive Cldns negates, or the CPE-SR of sensitive Cldns may have a common feature with which to interact with the toxin. Concerning the first possibility, we focused on a previous report suggesting that two consensus aromatic amino acids located in the second loop of Cldns directly interact with aromatic amino acids in the receptor-binding region of CPE (21). These aromatic amino acids, FY or FF, are located immediately upstream of the CPE-SRs of Cldns (Table 2) and are suggested to be involved in the transinteraction of Cldns between opposing cells to form tight junction strands (23), indicating that they are exposed outside the Cldn molecule. If this is the case, it is possible that FY or FF, which are common in members of the Cldn family, provide a contact site on Cldn for CPE, and the CPE-SR of CPE-insensitive Cldns might interfere with their interaction through the aromatic residues. To address this issue, we examined whether the FY residues of Cldn are actually involved in interaction with CPE. We established L929 cells expressing Cldn4FYAA, in which the consensus residues (Phe147 and Tyr148) were replaced with Ala and examined them for CPE sensitivity (Fig. 3). The cells with Cldn4FYAA were found to be as sensitive as those expressing Cldn4. In addition, the toxin bound to Cldn4FYAA-expressing cells as well as Cldn4-expressing cells. These results exclude the first possibility.
FIGURE 3.
Cldn4FYAA functions as a receptor for CPE. A and B, CPE sensitivity (A) and 125I-CPE-binding (B) of L929 cells expressing no Cldn (filled squares), Cldn4 (filled triangles), or Cldn4FYAA (open triangles) were examined by cytotoxicity assay with the WST-8 reagent. The experiments were repeated twice, and representative results are shown. Each point represents the mean ± S.D. (n = 3). C, the expression level of each Cldn was confirmed by Western blotting with anti-V5 antibody, to which 2 × 104 cells/lane were subjected. The β-actin of each sample was stained as a control.
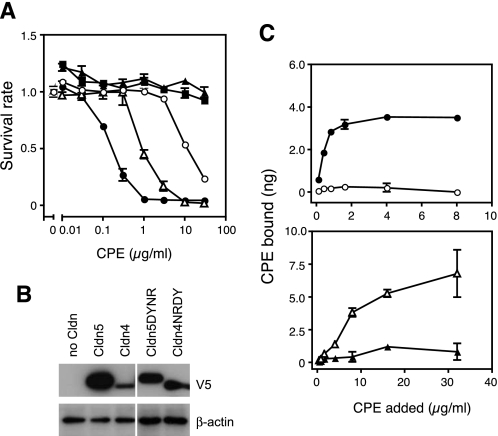
We next examined the alternative possibility that the CPE-SRs of the sensitive Cldns have a common feature with which to interact with CPE and tried to find in the CPE-SRs a consensus sequence or motif that likely determines CPE sensitivity. For this purpose, we first carried out cytotoxic assays with cells expressing various Cldns and classified them as highly sensitive, slightly sensitive, and insensitive and then compared the sequences of the CPE-SRs (Table 2). These results showed that all of the Cldns kept in the laboratory could be classified into three classes on the basis of sensitivity to CPE as described above, and no consensus sequences or motifs were evident in the CPE-SRs of each class. Instead, we found that CPE-SRs of highly sensitive Cldns had relatively high pI values, whereas those of slightly sensitive or insensitive Cldns showed lower pI values (Table 2). The difference in pI values of CPE-SR is derived from different amino acids at the first and tenth positions in CPE-SR, e.g. Asn and Arg for Cldn4 versus Asp and Tyr for Cldn5 (Table 2). To examine the relationship between pI values of CPE-SR and sensitivity, we constructed genes for Cldn4 and Cldn5 derivatives, whose first and tenth amino acids in the CPE-SR were exchanged so that the Cldn4 derivative, Cldn4NRDY, has a CPE-SR with a pI value equivalent to that of Cldn5, and the Cldn5 derivative, Cldn5DYNR, has a CPE-SR with a pI equivalent to Cldn4 (Table 2). These Cldns were expressed in L929 cells, and their sensitivity to CPE was examined (Fig. 4). As expected, the mutation to raise the pI value of CPE-SR made Cldn5 sensitive, whereas that to lower the pI value made Cldn4 ∼100 times less sensitive (Fig. 4). Cldn5DYNR was 10 times more sensitive than Cldn4NRDY, although they were both classified into the slightly sensitive group according to our criteria (Table 2). The binding assay also revealed that Cldn5DYNR but not Cldn5 served as a CPE receptor, and Cldn4NRDY reduced the ability to bind to the toxin, at least to levels below the limit of detection for the binding assay. Additionally, we carried out binding assays with Cldn4-expressing cells and C-CPE in the presence of various concentrations of NaCl to examine the effect of ionic strength on the Cldn-CPE interaction. Increasing the concentration of NaCl from 0.14 to 1.0 m in the reaction environment reduced the amount of C-CPE bound to the cells by ∼40% (data not shown).
FIGURE 4.
Electrostatic characteristics of the CPE-SR determines the sensitivity of Cldns to CPE. L929 cells expressing Cldns were subjected to the cytotoxicity assay (A) and 125I-CPE binding assay (C). Cells expressing Cldn4 (filled circles), Cldn5 (filled triangles), Cldn4NRDY (open circles), Cldn5DYNR (open triangles), or not expressing Cldn (filled squares) were examined. Each point represents the mean ± S.D. (n = 3). B, the expression level of each Cldn was confirmed by Western blotting with anti-V5 antibody on 2 × 104 cells. The β-actin of each sample was stained as a control.
DISCUSSION
In this study, we tried to understand the mechanism by which CPE identifies sensitive Cldns, which make up a large family of more than 20 members. For this purpose, we established cultured cells stably expressing a variety of Cldns including chimeric or amino acid-substituted mutants. Western blotting revealed that the expression levels of Cldns varied among the established cell lines. However, we found that they had little influence on the sensitivity to CPE, probably because the sensitivity of detection in Western blotting is much lower than that of the cytotoxicity assay. Occasionally, the sensitive Cldns in HEK293 or L929 cells induced sensitivity to the toxin, even if they were barely detected by Western blotting (see supplemental Fig. S3 as a typical example). We established several lines for each Cldn and chose one that expressed enough Cldn to be detected by Western blotting. Thus, we consider that the differences in sensitivity to CPE are hardly attributable to those in the expression level of Cldns in the experiments presented here.
The results of the cytotoxicity assay classified Cldns into three groups according to the sensitivity to CPE: highly sensitive (EC50 < 1 μg/ml), slightly sensitive (1 ≤ EC50 < 30 μg/ml), and insensitive (EC50 ≥ 30 μg/ml). The cytotoxicity assay with the chimera of Cldn1 and Cldn4 revealed that the CPE-SR (Asn149–Met160 for Cldn4) determines the sensitivity of Cldns to the toxin. Furthermore, the pI values of the CPE-SR were apparently related to the sensitivity, with the more sensitive Cldns having higher pI values. Cldn5, which is insensitive to the toxin, became sensitive with amino acid substitutions to raise the pI value of the CPE-SR. In contrast, Cldn4, a typical receptor for CPE, became less sensitive with substitutions to lower the pI value of the CPE-SR. The binding of 125I-labeled CPE to the cells exhibited a close relationship to the sensitivity of the expressed Cldns, indicating that the toxin actually binds to sensitive but not insensitive Cldns.
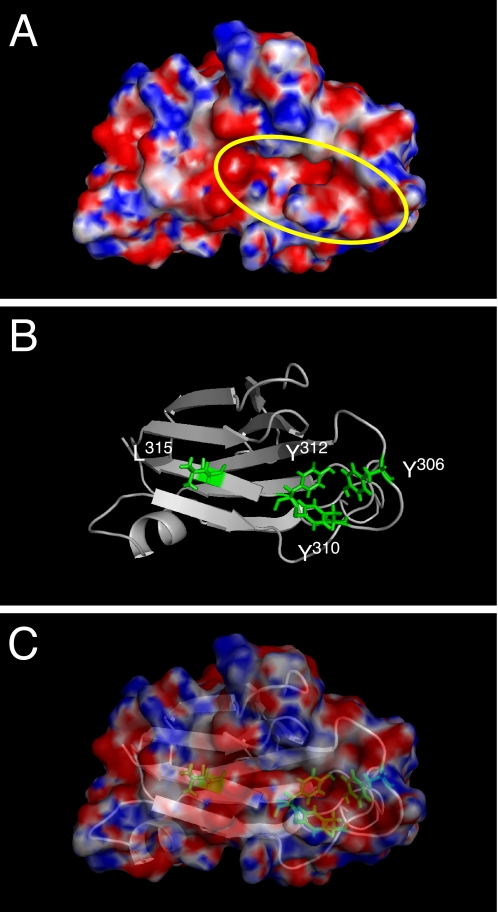
Analyses characterizing the receptor-binding domain of CPE have revealed that the 16–17 amino acids at the C-terminal end are important for binding to Cldn4 (15, 21, 24). Notably, Tyr306, Tyr310, Tyr312, and Leu315 in this region were shown to be involved in interaction with Cldn4 (25–27). Ling et al. (21), who recently identified a common motif for binding to Cldn using a phage display library, demonstrated the importance of Tyr and Leu residues. The steric structure of the C-terminal portion (residues 194–319) of CPE demonstrates that Tyr306, Tyr310, and Tyr312 reside on a large loop bulging out of the molecule (28). Additionally, the surface charge model reveals that these Tyr residues together with Leu315 form a cleft space, the bottom of which is negatively charged compared with the surrounding area (Fig. 5). Therefore, we consider the electrostatic attraction between the negatively charged cleft of CPE and the positively charged CPE-SR of the sensitive Cldns to partially play a role in the mutual interaction.
FIGURE 5.
Negatively charged cleft of the C-terminal CPE (residues 194–319) surrounded by Tyr306, Tyr310, Tyr312, and Leu315. A, surface electrostatic potential is depicted. The yellow oval indicates the cleft space, which is considered to be involved in the interaction with Cldn. Negatively and positively charged areas are indicated in red and blue, respectively. B, ribbon model of the structure with Tyr306, Tyr310, Tyr312, and Leu315 shown as green sticks. C, the merged image of A and B.
Cldns are major components of the tight junction, which organizes paracellular barriers to delimitate functional compartments of each tissue of the animal body. Specific Cldns are known to be overexpressed in various cancer cells (13, 14). CPE kills eukaryotic cells, and C-CPE opens the paracellular barrier by binding to sensitive Cldns. Therefore, they are considered probable candidates for a modulator of the paracellular barrier for drug delivery or for a Cldn-targeting agent for cancer therapy (13–15). In addition, some research groups have tried to understand the interaction between Cldns and CPE at the molecular level to ultimately modulate the selectivity or specificity of CPE against Cldns, which may make the toxin more useful. Fujita et al. (16) presented the first evidence that the second extracellular loop is responsible for binding to CPE, showing an in vitro interaction between the second loop of Cldn3 and CPE or C-CPE. However, the binding of C-CPE to peptide fragments comprising the second extracellular loop of Cldns could not be reproduced (21, 28). Ling et al. (21) succeeded in demonstrating the in vitro interaction using fragments consisting of the second loop and the subsequent transmembrane domain. These results imply that the steric structure of the second loop organized with the transmembrane domain is necessary for interaction with CPE or C-CPE. In this context, we tried to understand the interaction between Cldns and CPE by using the full-length toxin and mammalian cells expressing full-length Cldns, so as to examine the events that may occur in situ.
Recently, another group identified a motif, NPL(V/L)(P/A), as an essential sequence of the second extracellular loop of Cldns for interaction with CPE (20). This motif in Cldn3 corresponds to the N-terminal half of the CPE-SR. However, we observed that Cldns that do not possess the motif were sensitive to CPE (e.g. DPLTP for msCldn1, DPMTP for huCldn1, and SPLVP for msCldn2). Their conclusions were based on results obtained using an array with synthetic peptides corresponding to the second extracellular loop, which seem to be unreliable because the typical receptor Cldn4 was judged to be negative for binding to GST-CPE116–319. In addition, there were some contradictory results in that paper; Cldn5 was considered sensitive to CPE with low affinity, whereas in our experience, Cldn5 is definitely insensitive to CPE, and huCldn5 and msCldn5 did not confer CPE sensitivity to L929 cells. The binding of 125I-labeled CPE to the cells expressing huCldn5 was not detected. A further understanding of interactions between CPE and Cldns at a molecular level may be required to explain these discrepancies.
In this study, we conclude that electrostatic characteristics are important for Cldn and CPE to interact. This may provide information helpful to the use of CPE or C-CPE in a drug delivery system or cancer therapy. It remains to be elucidated how the electrostatic characteristics are involved in the interaction between Cldns and CPE. Although Cldn1, Cldn2, and Cldn5 possess CPE-SRs with pI values of 4.18 (Table 2), the former two are sensitive, whereas the latter is insensitive to CPE. Cldn4NRDY, also with a pI of 4.18 in the CPE-SR, still responded to the toxin. These results suggest that other factors are involved in the interaction between Cldn and CPE. To confirm or extend our conclusions, we are now attempting to determine the steric structure of a CPE-Cldn complex.
Supplementary Material
Acknowledgments
We greatly appreciate the gifts of cDNAs for huCldn1, huCdn4, mkCldn4, Cldn4–1(A), and Cldn1–4-1(A) from Dr. Jun Katahira (Graduate School of Frontier Biosciences, Osaka University) and L929 cells expressing msCldns from Dr. Shoichiro Tsukita (Kyoto University).
The work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S3.
- CPE
- C. perfringens enterotoxin
- Cldn
- claudin
- C-CPE
- C-terminal part of CPE
- hu
- human
- HA
- hemagglutinin
- ms
- mouse
- CPE-SR
- CPE sensitivity-related region.
REFERENCES
- 1.Hanna P. C., Mietzner T. A., Schoolnik G. K., McClane B. A. (1991) J. Biol. Chem. 266, 11037–11043 [PubMed] [Google Scholar]
- 2.Hanna P. C., Wieckowski E. U., Mietzner T. A., McClane B. A. (1992) Infect. Immun. 60, 2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horiguchi Y., Akai T., Sakaguchi G. (1987) Infect. Immun. 55, 2912–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kokai-Kun J. F., Benton K., Wieckowski E. U., McClane B. A. (1999) Infect. Immun. 67, 5634–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kokai-Kun J. F., McClane B. A. (1997) Infect. Immun. 65, 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClane B. A. (2001) Toxicon 39, 1781–1791 [DOI] [PubMed] [Google Scholar]
- 7.Alouf J. E. (2006) in The Comprehensive Sourcebook of Bacterial Protein Toxins (Alouf J. E., Popoff M. R. eds) 3rd Ed., pp. 507–515, Academic Press, Burlington, MA [Google Scholar]
- 8.Alouf J. E., Billington S. J., Jost B. H. (2006) in The Comprehensive Sourcebook of Bacterial Protein Toxins (Alouf J. E., Popoff M. R. eds) 3rd Ed., pp. 643–658, Academic Press, Burlington, MA [Google Scholar]
- 9.Horiguchi Y., Uemura T., Kozaki S., Sakaguchi G. (1985) FEMS Microbiol. Lett. 28, 131–135 [Google Scholar]
- 10.Katahira J., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N. (1997) J. Cell Biol. 136, 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S. (1998) J. Cell Biol. 141, 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda N., Furuse M., Sasaki H., Yonemura S., Katahira J., Horiguchi Y., Tsukita S. (1999) J. Cell Biol. 147, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kominsky S. L. (2006) Exp. Rev. Mol. Med. 8, 1–11 [DOI] [PubMed] [Google Scholar]
- 14.Morin P. J. (2005) Cancer Res. 65, 9603–9606 [DOI] [PubMed] [Google Scholar]
- 15.Kondoh M., Masuyama A., Takahashi A., Asano N., Mizuguchi H., Koizumi N., Fujii M., Hayakawa T., Horiguchi Y., Watanbe Y. (2005) Mol. Pharmacol. 67, 749–756 [DOI] [PubMed] [Google Scholar]
- 16.Fujita K., Katahira J., Horiguchi Y., Sonoda N., Furuse M., Tsukita S. (2000) FEBS Lett. 476, 258–261 [DOI] [PubMed] [Google Scholar]
- 17.Horiguchi Y., Inoue N., Masuda M., Kashimoto T., Katahira J., Sugimoto N., Matsuda M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11623–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe R., Kinoshita T., Masaki R., Yamamoto A., Takeda J., Inoue N. (1996) J. Biol. Chem. 271, 26868–26875 [DOI] [PubMed] [Google Scholar]
- 19.Rice P., Longden I., Bleasby A. (2000) Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 20.Winkler L., Gehring C., Wenzel A., Müller S. L., Piehl C., Krause G., Blasig I. E., Piontek J. (2009) J. Biol. Chem. 284, 18863–18872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling J., Liao H., Clark R., Wong M. S., Lo D. D. (2008) J. Biol. Chem. 283, 30585–30595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katahira J., Sugiyama H., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N. (1997) J. Biol. Chem. 272, 26652–26658 [DOI] [PubMed] [Google Scholar]
- 23.Piontek J., Winkler L., Wolburg H., Müller S. L., Zuleger N., Piehl C., Wiesner B., Krause G., Blasig I. E. (2008) FASEB J. 22, 146–158 [DOI] [PubMed] [Google Scholar]
- 24.Takahashi A., Kondoh M., Masuyama A., Fujii M., Mizuguchi H., Horiguchi Y., Watanabe Y. (2005) J. Control Release 108, 56–62 [DOI] [PubMed] [Google Scholar]
- 25.Takahashi A., Komiya E., Kakutani H., Yoshida T., Fujii M., Horiguchi Y., Mizuguchi H., Tsutsumi Y., Tsunoda S., Koizumi N., Isoda K., Yagi K., Watanabe Y., Kondoh M. (2008) Biochem. Pharmacol. 75, 1639–1648 [DOI] [PubMed] [Google Scholar]
- 26.Harada M., Kondoh M., Ebihara C., Takahashi A., Komiya E., Fujii M., Mizuguchi H., Tsunoda S., Horiguchi Y., Yagi K., Watanabe Y. (2007) Biochem. Pharmacol. 73, 206–214 [DOI] [PubMed] [Google Scholar]
- 27.Ebihara C., Kondoh M., Harada M., Fujii M., Mizuguchi H., Tsunoda S., Horiguchi Y., Yagi K., Watanabe Y. (2007) Biochem. Pharmacol. 73, 824–830 [DOI] [PubMed] [Google Scholar]
- 28.Van Itallie C. M., Betts L., Smedley J. G., 3rd, McClane B. A., Anderson J. M. (2008) J. Biol. Chem. 283, 268–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.