Abstract
In the recently identified cholesterol catabolic pathway of Mycobacterium tuberculosis, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (HsaD) is proposed to catalyze the hydrolysis of a carbon-carbon bond in 4,5–9,10-diseco-3-hydroxy-5,9,17-tri-oxoandrosta-1(10),2-diene-4-oic acid (DSHA), the cholesterol meta-cleavage product (MCP) and has been implicated in the intracellular survival of the pathogen. Herein, purified HsaD demonstrated 4–33 times higher specificity for DSHA (kcat/Km = 3.3 ± 0.3 × 104 m−1 s−1) than for the biphenyl MCP 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA) and the synthetic analogue 8-(2-chlorophenyl)-2-hydroxy-5-methyl-6-oxoocta-2,4-dienoic acid (HOPODA), respectively. The S114A variant of HsaD, in which the active site serine was substituted with alanine, was catalytically impaired and bound DSHA with a Kd of 51 ± 2 μm. The S114A·DSHA species absorbed maximally at 456 nm, 60 nm red-shifted versus the DSHA enolate. Crystal structures of the variant in complex with HOPDA, HOPODA, or DSHA to 1.8–1.9 Åindicate that this shift is due to the enzyme-induced strain of the enolate. These data indicate that the catalytic serine catalyzes tautomerization. A second role for this residue is suggested by a solvent molecule whose position in all structures is consistent with its activation by the serine for the nucleophilic attack of the substrate. Finally, the α-helical lid covering the active site displayed a ligand-dependent conformational change involving differences in side chain carbon positions of up to 6.7 Å, supporting a two-conformation enzymatic mechanism. Overall, these results provide novel insights into the determinants of specificity in a mycobacterial cholesterol-degrading enzyme as well as into the mechanism of MCP hydrolases.
Introduction
Mycobacterium tuberculosis is the leading cause of bacterial mortality, causing an estimated 2 million deaths/year (1). The mechanisms underlying the remarkable ability of this pathogen to survive for long periods of time within the host are poorly understood (2). Although it was well known that saprophytic mycobacteria could metabolize cholesterol (3), it was only recently demonstrated that pathogenic strains can also utilize this nutrient as a growth substrate (4, 5). Interestingly, cholesterol has been found in high concentrations within caseating granulomas in both humans and mice (6, 7), and bacteria have been observed congregating around cholesterol foci (7). Highlighting the importance of cholesterol in bacterial pathogenesis, the deletion of genes involved in cholesterol metabolism reduces the virulence of M. tuberculosis (5, 8). Therefore, further knowledge of cholesterol metabolism in M. tuberculosis is crucial to our understanding of bacterial virulence.
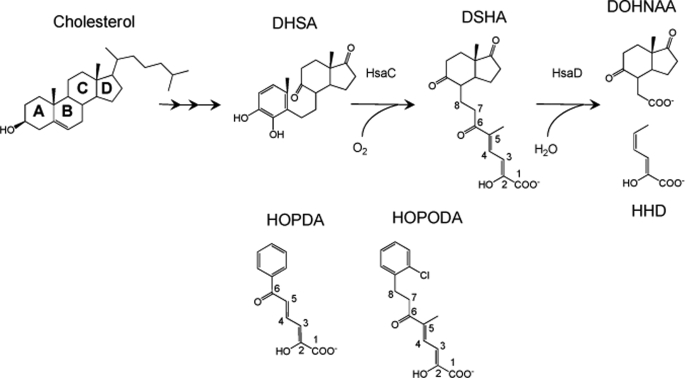
M. tuberculosis catabolizes cholesterol using a metabolic pathway similar to that identified in Rhodococcus jostii RHA1 (4, 9). In this pathway, the aerobic degradation of the four-ringed steroid nucleus occurs through the opening of ring B, aromatization of ring A, and hydroxylation of the phenolic metabolite. The resulting catechol undergoes oxygenolytic cleavage, yielding 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid (DSHA),4 (see Fig. 1 for IUPAC numbering) a meta-cleavage product (MCP; Fig. 1). The latter is then hydrolyzed by the MCP hydrolase, HsaD, to yield 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid (DOHNAA) and 2-hydroxy-hexa-2,4-dienoic acid (HHD). HHD is then metabolized to tricarboxylic acid cycle intermediates, whereas the metabolic fate of DOHNAA is unknown. In this pathway, HsaD is of particular interest, because transposon mutagenesis has indicated that it is critical for the intracellular survival of M. tuberculosis (10).
FIGURE 1.
The successive reactions catalyzed by HsaC and HsaD in the catabolism of cholesterol by M. tuberculosis and alternate substrates of HsaD. DSHA is referred to as 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid in the steroid literature. However, the carbon atoms are numbered here according to its IUPAC name, 2-hydroxy-5-methyl-8-(7a-methyl-1,5-dioxooctahydro-1H-inden-4-yl)-6-oxoocta-2,4-dienoic acid, to be consistent with the numbering in HOPDA and HOPODA. In this study, DSHA, HOPDA, and HOPODA were generated by ring cleavage of DHSA, 2,3-dihydroxybiphenyl, and DHDS, respectively.
MCP hydrolases are involved in the aerobic degradation of aromatic compounds, hydrolyzing vinylogous 1,5-diketones arising from the extradiol ring cleavage of catechols (11). Crystal structures of MCP hydrolases, including those of BphD from Burkholderia xenovorans LB400, MhpC from Escherichia coli, and a ht-HsaD (12–14), reveal that these enzymes belong to the α/β-hydrolase superfamily and contain a conserved Ser-His-Asp catalytic triad as well as an “oxyanion” hole, similar to a number of other hydrolytic enzymes (15). The substrate-binding pocket comprises two subsites: a hydrophilic polar subsite that binds the dienoate moiety of the MCP and a hydrophobic nonpolar subsite that accommodates the remainder of the MCP. In HsaD, the nonpolar subsite is at least twice as a large as that of other MCP hydrolases, possibly to fit the larger cholesterol metabolite DSHA (14).
Despite the similar catalytic machinery of MCP hydrolases and serine proteinases, the mechanism of the former appears to differ from the nucleophilic mechanism of the latter in two respects. First, the enolate substrate of MCP hydrolases undergoes tautomerization to a keto intermediate prior to hydrolysis (16). Second, the ketonized substrate (17) is believed to be hydrolyzed through a general base mechanism via a gem-diol intermediate (18, 19). Initial evidence for this intermediate was obtained from the incorporation of 18O from H218O into the MCP of MhpC as well as the exchange of 18O into a nonhydrolyzable substrate analogue (19). Subsequently, low amounts of this gem-diol intermediate were detected using 13C NMR spectroscopy (20) in reactions catalyzed by MhpC and BphD. A general base mechanism is more consistent with that of other α/β-hydrolases that activate small molecules (21).
Nevertheless, features of the MCP hydrolase mechanism remain to be resolved. First, the nature of a transient intermediate observed in the transformation of HOPDA by BphD, E:Sred, is unclear (13). E:Sred possesses a red-shifted absorption spectrum (λmax = 492 nm) with respect to that of the free HOPDA enolate (λmax = 434 nm). X-ray diffraction studies (13) of a similar intermediate (λmax = 506 nm) trapped using the S112A variant of BphD revealed torsion angles that were most consistent with a ketonized tautomer of HOPDA (E:Sk). Nevertheless, the refinements did not exclude the 2-enol tautomer, and a strained enolate (E:Sse) more satisfactorily accounts for the red-shifted absorption spectrum because steric hindrance to planarity about a double bond raises the ground state energy level but not the excited state (22). Second, the roles of catalytic residues remain unclear. The active site serine has been variously proposed to deprotonate the nucleophilic water (19) and to stabilize the oxyanion (23). Similarly, the active site histidine has been proposed to catalyze tautomerization (23) and to deprotonate the catalytic water (23). Finally, despite evidence supporting a general base mechanism, in the structures of BphD from B. xenovorans LB400 and MhpC from E. coli, no solvent species have been detected near the active site of the enzyme.
In this study, we determined the specificity of HsaD for various MCPs including its proposed physiological substrate, DSHA. To elucidate the interactions between enzyme and substrates, a S114A variant of HsaD was generated to slow substrate turnover, and the crystal structures of the variant·substrate complexes were analyzed. These results provide insights into the catalytic mechanism of MCP hydrolases.
EXPERIMENTAL PROCEDURES
Chemicals
HOPDA used for crystallization was synthesized as previously described (24). 2,3-Dihydroxybiphenyl and 2,3-dihydroxy-6-methyl-7,8-dihydrostilbene (DHDS) were synthesized by a combined directed ortho metalation cross-coupling strategy as described elsewhere (8, 25). 3,4-Dihydroxy-9,10-seco-nandrost-1,3,5(10)-triene-9,17-dione (DHSA) was obtained by incubating a culture of the ΔhsaC mutant of R. jostii RHA1 with cholesterol as described previously (8). All other chemicals were of analytical grade.
Preparation and Characterization of MCPs
MCPs were produced by dissolving the corresponding catechol in a small volume of ethanol and diluting to the desired volume with 100 mm potassium phosphate, pH 7.5 (ethanol constituted less than 0.1% of the solution). To this solution a sufficient amount of HsaC was added to completely transform DHDS and DHSA to HOPODA and DSHA, respectively. Upon the addition of HsaC to a diluted sample, complete cleavage of DHDS and DHSA was verified by ensuring that there was no further increase in absorbance of the ring-cleaved products at 396 nm (ϵ = 6.8 and 3.8 mm−1 cm−1 for HOPODA and DSHA, respectively). MCPs used for steady-state kinetic studies were prepared enzymatically using 100 mm potassium phosphate, pH 7.5, and used within 2 h.
To determine pKa values, HOPODA and DSHA were prepared as described above in unbuffered water. HsaC was removed by ultrafiltration using a stirred cell equipped with an YM10 membrane (Amicon). Solutions of ∼0.5 mm MCP were acidified to pH 3 using 12 n HCl. This solution was titrated with aliquots of 50 mm NaOH, and the pH was determined after the addition of each aliquot. The pKa values were determined from plots of pH versus the amount of base added.
The nonenzymatic transformation of MCPs in solution (100 mm potassium phosphate, pH 7.5, at 25 °C) was determined by monitoring the absorption spectra. The experiments were performed using two concentrations of each MCP (0.03, 0.3 mm). The half-lives were determined by fitting equations to the decay at 396 nm using Excel.
Protein Production and Purification
Ser114 of HsaD was substituted with alanine to generate the S114A variant using the QuikChange II-E site-directed mutagenesis kit (Stratagene) as per the manufacturer's instructions. In this reaction, the template was pT7HD1, a previously constructed pT7−7 derivative carrying hsaD under control of the T7 promoter (4). The primers were 5′-CTGGTGGGCAACGCGTTGGGCGGGG-3′ and 5′-CCCCGCCCAACGCGTTGCCCACCAG-3′. The nucleotide sequence of the mutated gene was confirmed using an ABI 373 Stretch DNA sequencer (Applied Biosystems, Foster City, CA) and a Big-Dye Terminator v3.1 kit. The mutated hsaD was subcloned into pEMBL18 downstream of the Plac promoter as an XbaI fragment, yielding pEMHSA.
HsaD was produced and purified from E. coli strain GJ1158 containing pT7HD1. The cells were grown at 30 °C in salt-depleted Luria-Bertani broth (10 g of tryptone extract, 5 g of yeast extract/liter) supplemented with 100 μg/ml ampicillin. The culture was grown to an optical density at 600 nm of ∼0.5, at which point NaCl was added to a final concentration of 300 mm. The cultures were incubated for an additional 20 h, and then the cells were harvested by centrifugation. The S114A variant was produced using E. coli strain DH5α containing pEMHSA. The cells were grown at 30 °C in LB broth supplemented with 100 μg/ml ampicillin. The culture was grown to an optical density at 600 nm of ∼0.5, and isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm. The culture was incubated for an additional 20 h before the cells were harvested.
The cell pellet obtained from 4 liters of culture was resuspended in 40 ml of 20 mm HEPES, pH 7.5, and disrupted using three passages through a Emulsiflex C-5 cell disrupter (ATA Scientific, Sutherland, Australia) operating at a pressure of 15,000 p.s.i. The cell debris was removed by ultracentrifugation at 120,000 × g for 60 min. The supernatant was removed and filtered through a 0.45-μm cellulose filter to yield ∼45 ml of raw extract. The raw extract was loaded onto 28 ml of Source 15Q anion exchange resin (GE Healthcare) packed in an AP-2 column (Waters Corp., Milford, MA) equilibrated with 20 mm HEPES, pH 7.5. The proteins were eluted using a linear gradient of 70–220 mm NaCl in 280 ml. HsaD eluted at ∼140 mm NaCl. Fractions of 10 ml containing activity (in the case of the wild-type enzyme) or the protein of the expected size (in the case of the variant) were pooled and concentrated to 10 ml using an Amicon stirred cell concentrator equipped with an YM10 ultrafiltration membrane (Millipore, Billerica, MA). The HsaD-containing solution was brought to 1 m (NH4)2SO4, and the sample was briefly centrifuged to harvest the pellet. The cloudy white precipitate was washed twice with 1 m (NH4)2SO4 solution; exchanged into 20 mm HEPES, pH 8.5, to resolubilize the enzyme; concentrated to >20 mg/ml; flash frozen as beads in liquid nitrogen; and stored at −80 °C. Coomassie Blue-stained denaturing gels revealed that preparations of the wild-type and variant proteins were >97% HsaD.
Kinetic Measurements
The enzyme-catalyzed hydrolysis of the MCP was monitored using a Varian Cary 5000 spectrophotometer equipped with a thermostatted cuvette holder (Varian Canada, Mississauga, Canada) maintained at 25.0 ± 0.5 °C. The amount of enzyme used in each assay was adjusted so that the progress curve was linear for at least 2 min. Initial velocities were determined from a least squares analysis of the progress curves using the kinetics module of the Cary WinUV software.
Specificity experiments were performed in a total volume of 1.0 ml of 100 mm ionic strength potassium phosphate, pH 7.5, at 25.0 ± 0.1 °C. The reaction was initiated by adding 5 μl of an appropriately diluted enzyme preparation to the reaction cuvette. The reactions were monitored at the wavelengths corresponding to the enolate. Initial velocities were determined over the following ranges of substrate concentration: 0.5–30 μm HOPDA, 10–300 μm HOPODA, and 2–60 μm DSHA. Steady-state kinetic parameters were evaluated by fitting the appropriate equations to the data using the least squares and dynamic weighting options of LEONORA (26).
Kd Determination
The dissociation constant of the S114A·DSHA complex was determined by titrating a 200-μl solution of 100 mm ionic strength potassium phosphate, pH 7.5, at 25.0 ± 0.1 °C containing 5 μm DSHA with S114A and monitoring the change in absorbance at 456 nm. The spectra were recorded using a Cary 5000 spectrophotometer. The dissociation constant was evaluated by fitting the binding equation (27) to the data using the curve fitting program and the nonlinear statistical analyses of R.
The half-life of S114A·DSHA was determined by monitoring the decay of the complex at 456 nm. The experiment was carried out in a total volume of 150 μl of 100 mm ionic strength potassium phosphate, pH 7.5, at 25.0 ± 0.1 °C containing 14 μm S114A and 6 μm DSHA. A single-exponential equation was fit to the data.
Crystallization of S114A and Preparation of Substrate Complex
Protein crystals were grown by sitting drop vapor diffusion with equal volumes (1 μl) of S114A (8 mg/ml) and precipitant (200 mm KSCN, 24% polyethylene glycol 3.35 K, and 100 mm bis-tris propane, pH 7.0). Disphenoid S114A crystals typically grew in 3–5 days at 19 °C. Complexes between S114A and HOPDA, HOPODA, and DSHA were obtained by soaking S114A crystals in 10 μl of precipitant supplemented with 15 mm of each ligand for 30–60 min.
Diffraction Data Measurement and Processing
The crystals were briefly transferred to a cryoprotectant solution (1:4 (v/v) mixture of glycerol and precipitant) and then flash-frozen in liquid nitrogen. For enzyme-ligand complexes, the cryoprotectant solution was supplemented with the ligand (15 mm).
S114A crystal x-ray data were collected in-house using a Bruker MicroStar x-ray generator with a Smart6000 CCD detector and integrated with the commercial software SAINT (Bruker-AXS). X-ray data from S114A·HOPDA crystals were collected at the Swiss Light Source on Beamline X06SA with a Pilatus 6m detector and integrated with MOSFLM (28). X-ray data from HOPODA and S114A·DSHA crystals were collected at the Diamond Light Source on Beamline I03 with a Quantum ADSC CCD detector and integrated with MOSFLM (28).
Structure Determination and Refinement
The initial phases of S114A were determined by molecular replacement using ht-HsaD (Protein Data Bank code 2vf2), refined to 2.3 Å, as a search template (14) with the program MOLREP (29). For the enzyme-ligand complexes the phases were determined as described above, using the refined S114A structure as a search template. Simulated annealing refinement was followed with iterative cycles of manual density fitting with COOT (30) and refinement with PHENIX (31). PHENIX eLBOW (31) was used to calculate the geometric restraints for HOPDA, HOPODA, and DSHA. The stereochemical properties and quality of the final model were assessed with the program MOLPROBITY (32). All of the structural figures, alignments, and graphical renderings were done using PYMOL (33).
RESULTS
Physical Properties of HOPODA and DSHA
The absorption spectrum of MCPs are strongly pH-dependent because of the C-2-OH group. Moreover, these compounds undergo a nonenzymatic transformation in aqueous buffer (20). The half-lives of HOPODA and DSHA were less than 30 h, significantly less than that of HOPDA. These data are summarized in Table 1 together with the pKa values of the enolic hydroxyl groups as determined by titration and the extinction coefficients of the respective enolate anions.
TABLE 1.
Properties of MCPs
| Substrate | λmax | ϵ | Half-lifea | pKab |
|---|---|---|---|---|
| nm | mm−1cm−1 | h | ||
| HOPDA | 434 | 25.7 | 58c | 7.3c |
| HOPODA | 396 | 6.8 | 25 | 7.5 |
| DSHA | 396 | 3.8 | 15.4 | 6.8 |
a Half-lives were determined in potassium phosphate buffer (I = 0.1 m), pH 7.5, at 25 °C.
b The pKa values of the respective enolates were determined in unbuffered water.
c Data shown are from Ref. 17.
Substrate Specificity of HsaD
Steady-state kinetic studies were conducted to assess the substrate specificity of HsaD. Over the substrate concentration ranges studied, HsaD displayed Michaelis-Menten behavior with all three MCPs. Consistent with the role of the enzyme in cholesterol degradation, HsaD had the highest specificity (kcat/Km) for DSHA, which was ∼33 times higher than for HOPODA but only ∼4 times higher than for HOPDA (Table 2). Although HsaD had a higher specificity for DSHA over HOPODA, the enzyme turned over these substrates with similar kcat values.
TABLE 2.
Steady-state kinetic parameters of HsaD with HOPDA, HOPODA, and DSHA
The experiments were performed using potassium phosphate buffer (I = 0.1 m), pH 7.5, at 25 °C. The values in parentheses indicate standard deviations.
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | mm−1s−1 | |
| HOPDA | 8 (1) | 0.066 (0.005) | 9 (1) |
| HOPODA | 310 (70) | 0.33 (0.06) | 1.06 (0.06) |
| DSHA | 17 (2) | 0.55 (0.02) | 33 (3) |
Characteristics of S114A in Solution
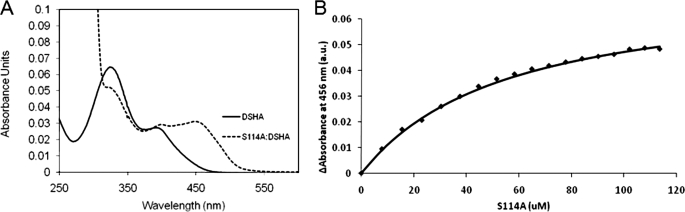
The catalytic serine of HsaD was substituted with alanine to generate a catalytically impaired variant. The addition of a 60-fold excess of S114A to a 5 μm solution of DSHA produced a species with an absorbance maximum of 456 nm (Fig. 2), 60 nm red-shifted with respect to that of the DSHA enolate tautomer. This species had a half-life of 3 h (supplemental Fig. S1), similar to that of the BphD S112A·HOPDA complex (34). Based on this, the upper limit of the kcat value of S114A for DSHA was 8.5 (± 0.9) × 10−5 s−1, 6500-fold lower than that of the wild-type enzyme. This slow turnover facilitated further study of the S114A·DSHA complex.
FIGURE 2.
Characterization of the S114A·DSHA complex by UV-visible absorption spectroscopy. A, spectra of S114A·DSHA and DSHA in solution are shown by dotted and solid lines, respectively. The complex absorbs maximally at 456 nm. B, the curve represents the fit of the binding equation to ΔA456 observed upon titrating DSHA with S114A (Kd = 51 ± 2 μm). The experiments were performed using potassium phosphate (I = 0.1 m, pH 7.5) at 25 °C.
Titration of the DSHA with S114A yielded a dissociation constant (Kd) of 51 ± 2 μm (Fig. 2, inset). In comparison, titration of a 2 μm solution of HOPDA with the variant enzyme resulted in modest spectral shifts; in the presence of a 10-fold excess of S114A, most of the HOPDA (∼90%) was present as the free enolate.
Crystallography
To characterize the interactions of HsaD with its substrate, crystallization studies were undertaken using the S114A variant. The variant failed to crystallize using conditions previously developed for ht-HsaD (14). After screening and optimization, diffracting disphenoid crystals of S114A could be reproducibly grown. Using these conditions, the structures of S114A were determined alone or in complex with HOPDA, HOPODA, or DSHA at resolutions between 1.8 and 2.1 Å. The crystals have space group F222 (a = 112Å, b = 118Å, c = 181Å) with two protomers/asymmetric unit. There was clear electron density in all structures for at least residues 7–288. For all of the models, >98% of the residues were in the favored region, and none were in the disallowed region of Ramachandran plots as defined by MolProbity (32). The statistics of the data collection and refinement for these structures are summarized in Table 3.
TABLE 3.
Summary of data collection and refinement statistics
The values in parentheses are for the highest resolution shell.
| S114 | S114A·HOPDA | S114A·HOPODA | S114A·DSHA | |
|---|---|---|---|---|
| X-ray source | Cu-Kα | Swiss light source (X06SA) | Diamond (I03) | Diamond (I03) |
| Space group | F 222 | F 222 | F 222 | F 222 |
| Unit cell parameters | a = 112.3, b = 118.8, c = 182.6, α = β = γ = 90 | a = 111.9, b = 118.0, c = 180.9, α = β = γ = 90 | a = 111.9, b = 118.0, c = 180.9, α = β = γ = 90 | a = 111.9, b = 118.0, c = 180.9, α = β = γ = 90 |
| Data collection statistics | ||||
| Resolution (Å) | 59.8-2.1 (2.2-2.1) | 29.49-1.80 (1.80-1.90) | 40.66-1.80 (1.80-1.90) | 40.59-1.90 (2.00-1.90) |
| No. of unique reflections | 35,484 (4560) | 55,165 (7928) | 55,043 (8086) | 46,107 (6736) |
| Rσ | 0.0753 (0.2759) | 0.080 (0.390) | 0.069 (0.413) | 0.094 (0.419) |
| I/σ (I) | 11.7 (3.8) | 16.8 (3.3) | 16.0 (3.1) | 9.9 (2.0) |
| Completeness (%) | 99.5 (99.2) | 98.6 (97.8) | 99.1 (100) | 98.4 (99.3) |
| Multiplicity | 4.2 (3.8) | 4.2 (4.2) | 5.0 (5.0) | 3.5 (3.5) |
| Refinement and model statistics | ||||
| Resolution (Å) | 49.7789-2.1 (2.11-2.10) | 29.49-1.80 (1.90-1.80) | 36.34-1.80 (1.90-1.80) | 40.59-1.90 (2.00-1.90) |
| No. reflections used (work + test) | 35,439 | 54,360 | 53,203 | 42,140 |
| Rworka | 0.192 | 0.167 | 0.194 | 0.209 |
| Rfreea | 0.239 | 0.192 | 0.210 | 0.231 |
| No. of residues (chain A/chain B) | 281/281 | 282/282 | 281/282 | 281/281 |
| No. of water molecules | 275 | 280 | 334 | 183 |
| Additional molecules | 2 thiocynate | 1 glycerol, 1 thiocynate | 2 thiocynate | 2 glycerol, 1thiocynate |
| Total No. of atoms | 4990 | 4871 | 4775 | 4725 |
| Root mean square deviation bond lengths (Å) | 0.009 | 0.008 | 0.008 | 0.013 |
| Root mean square deviation bond angles (°) | 0.746 | 0.783 | 0.716 | 0.933 |
| Wilson B-factor (Å2) | 16.6 | 16.5 | 21.1 | 28.5 |
| Mean B-factor (Å2) | 21.0 | 21.6 | 27.3 | 34.4 |
| Ramachandran statistics (%) | ||||
| Core region | 98.04 | 98.40 | 98.06 | 97.84 |
| Additional allowed region | 1.96 | 1.60 | 1.94 | 2.16 |
| Disallowed | 0.00 | 0.00 | 0.00 | 0.00 |
a Rwork and Rfree = Σh||Fo(h)| − |Fc(h)||/Σh|| for the working set and test set (5%) of reflections, where the Fo(h) and Fc(h) are the observed and calculated structure factor amplitudes for reflection h.
Structure of S114A
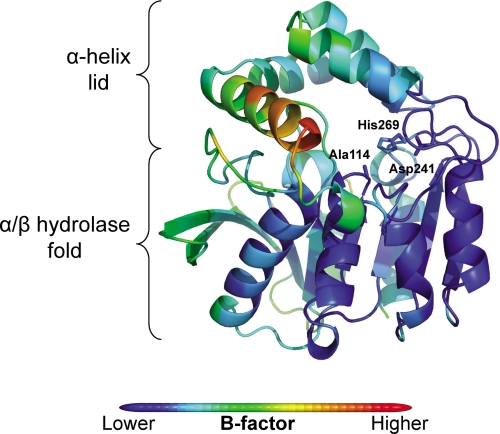
The structure of S114A is essentially identical to that of ht-HsaD (14) (root mean square deviation, 0.26 Å, Cα) except for the substituted active site residue. Briefly, this structure comprises a core α/β hydrolase domain with a four-helix lid domain, inserted between residues 137 and 233 in HsaD, which “caps” the active site (supplemental Fig. S2). As with other MCP hydrolases (12, 13), the residues of the lid domain in substrate-free S114A had an elevated temperature factor (26.2 ± 0.5 Å2 S.E.) compared with the rest of the structure (19.7 ± 0.2 Å2 S.E.), with the highest B-factor found in helix αL4 (residues 199–212; 36.2 ± 1.2 Å2 S.E.) (Fig. 3). The substituted active site residue is found within a tightly turning “nucleophilic elbow” (Gly112, Asp113, Ala114, Leu115, and Gly116) similar to other α/β hydrolases. This strand-turn-helix motif introduces unfavorable main chain torsional angles but contributes to the “oxyanion hole” formed by the backbone nitrogen atoms from Leu115 and Gly45. His269 of the catalytic triad forms a hydrogen bond via Nδ1 with Oδ2 of Asp241 (2.7 Å) and likely also interacts with Oδ1 of this residue (3.2 Å).
FIGURE 3.
Secondary structure and B-factor of S114A. The secondary structure of S114A was color-coded according to Cα B-factor from blue (lowest B-factor: 6.0 Å2) to red (highest B-factor: 67.4 Å2).
Structures of S114A·MCP Complexes
In the structure of S114A crystals soaked with HOPDA, there was clear electron density in the active site, supporting the presence of HOPDA in both protomers A and B (Fig. 4A). However, in crystals soaked with either DSHA or HOPODA, there was well defined electron density only in the active site of protomer B (Fig. 4, B and C). In protomer A of DSHA-soaked crystals, there was density supporting the presence of a ligand, but it was not sufficiently well resolved for accurate placement. Similar to previously published work, attempts to fit the 3E,5Z,2E,4E and 2Z,4Z 2-enol or the monoanionic (Z) 2-keto tautomers of the ligand were unsuccessful (34). The ligands were therefore initially refined as both the 2Z,4E enol and monoanionic (E) 2-keto tautomers. Protomer B of S114A·HOPDA was refined first, because this structure had the clearest ligand electron density. Although both tautomers fit the electron density of HOPDA equally well, the refinement of the ligand introduced considerable deviations of the torsion angles for the enol tautomer. Specifically, the refinement of the enol tautomer introduced a 67° deviation from planarity of the torsion angle about the C-4–C-5 bond (Table 4). By contrast, the keto tautomer refined without significant deviations in the expected torsional angle of the C-3=C-4 double bond. A similar trend was seen in protomer A of the S114A·HOPDA, protomer B of S114A·HOPODA, and S114A·DSHA structures, in which the C-4–C-5 bond refined with 51, 77, and 67° deviations from planarity, respectively (Table 4). Therefore, all of the ligands were refined as the monoanionic (E) 2-keto tautomers.
FIGURE 4.
Section of electron density map of S114A in complex with MCP. Stereo view showing 2Fo − Fc (blue, contour level = 1 σ) and Fo − Fc (green, contour level = 2.5σ). The electron density maps are shown for the ligand HOPDA (A), DSHA (B), and HOPODA (C). The protein structures are shown as white, whereas the ligands are shown as pink (HOPDA), purple (DSHA), or cyan (HOPODA). All of the structures are shown in ball and stick representation, with the nitrogen, oxygen, and sulfur atoms colored blue, red, and yellow, respectively.
TABLE 4.
Torsional angles of the refined 2-enol and 2-keto tautomers of HOPDA, HOPODA, and DSHA in S114A
| 2-Enol |
2-Keto |
|||||
|---|---|---|---|---|---|---|
| C2-C3 | C3-C4 | C4-C5 | C2-C3 | C3-C4 | C4-C5 | |
| Bonding | Double | Single | Double | Single | Double | Single |
| HOPDAchainB | 174 | 175 | 113 | 177 | 177 | 116 |
| HOPDAchainA | 164 | 168 | 129 | 149 | 173 | 139 |
| HOPODA | 173 | 165 | 103 | 176 | 165 | 98 |
| DSHA | 159 | 167 | 113 | 158 | 175 | 97 |
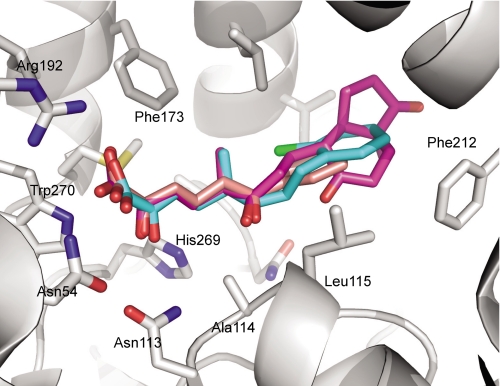
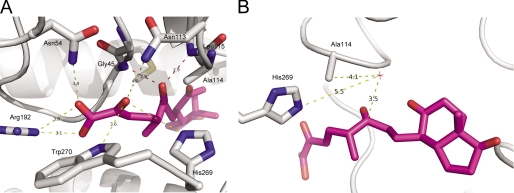
In each of the three S114A·MCP structures, the ligands orientated similarly in the active sites; the dienoate was bound to the polar subsite, and the rings (bicycloalkanone in the case of DSHA and phenyl in the case of HOPODA and HOPDA) were found in the nonpolar subsite (Fig. 5). More specifically, the C-6-oxo group of each ligand forms hydrogen bonds with the backbone amides of Gly45 (2.7–2.9 Å) and Leu115 (2.8–3.1 Å) (Fig. 6A). This interaction keeps the ligands in close proximity with active site residue 114. The C-5-methyl group of both DSHA and HOPODA was located within a hydrophobic pocket lined by the side chains of Phe173, Met177, and Leu158. When in the presence of a ligand, Arg192 was found to adopt two conformations: one similar to that observed in the absence of ligand and a second conformation involving a rotation of ∼90° about χ3 such that the guanidium group is in closer contact with ligand. This movement allowed the formation of a salt bridge (2.7–3.4 Å) between Nη1 of Arg192 and the carboxylic acid group of the substrate. In addition, the substrate carboxylic acid interacts with Asn54 (3.1–3.4 Å). Although likely too distant to form hydrogen bonds with the C-2-oxo group of HOPDA, HOPODA, and DSHA, the NH2 of Trp270 (3.6–4.1 Å), Nδ2 of Asn113 (3.7–4.0 Å), and Nϵ2 of His269 (3.5–4.0 Å) may contribute to a polar environment important for substrate binding and positioning (Fig. 6A). In all structures, a solvent species was present 3.2–3.4 Åfrom the C-6-carbonyl (Fig. 6B) and in two of them, S114A·DSHA and S114A·HOPODA, formed an angle (water O–C-6–O-6) that was within 10–20° of the Bürgi-Dunitz angle of 107°. Stabilized by hydrogen bonds with Nδ2 of Asn244 (3.0–3.1 Å), this water molecule has a relatively low B-factor (30.8 ± 12.3 Å2), suggesting essentially 100% occupancy. A similarly positioned solvent species was also present in the ligand-free S114A structure. The bicycloalkanone and phenylchloro moieties of DSHA and HOPODA, respectively, were located within the nonpolar subsite in a hydrophobic region formed by Phe212, Met208, Leu115, and Val155. However, the ring moieties of all MCPs were found to have elevated B-factors compared with the remainder of the ligand, suggesting that these groups are mobile. As such, there is weak electron density for these groups (Fig. 4, B and C).
FIGURE 5.
Overlay of the MCP substrates HOPDA, HOPODA, and DSHA in complex with S114A. The protein (white) is shown in with the side chains involved in ligand binding shown as sticks. The ligands are represented as sticks and are colored pink (HOPDA), cyan (HOPODA), and purple (DSHA). In all structures the nitrogen, oxygen, and sulfur atoms colored blue, red, and yellow, respectively.
FIGURE 6.
Interaction of DSHA with S114A. A, the structure of S114A (white) in complex with DSHA (purple) is shown in cartoon representations with the amino acid side chains involved in binding and DSHA displayed in stick representation. The interactions between S114A and the C-2 oxo and C-1 carboxylic acid are shown by yellow dashes, whereas the interactions between C-6 oxo and S114A are shown by red dashes. B, the structure of S114A (white) in complex with DSHA (purple). The water molecule (HOH89) is shown by a red star. All of the distances are in angstroms.
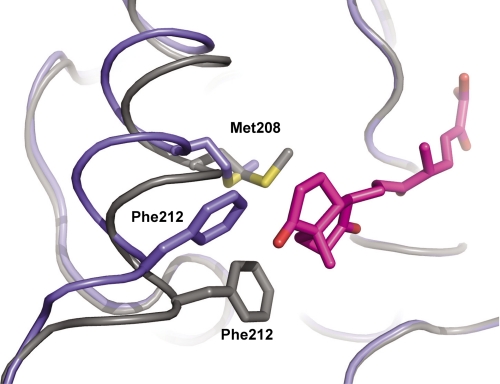
Comparison of the five ligand-containing protomers with the three ligand-free protomers of S114A revealed two conformations of the αL4 helix within the lid domain. In the presence of the ligand, the lid domain is observed in a “closed” orientation, whereas the lid domain of the ligand-free protomers is in an “open” orientation. As displayed in Fig. 7, for the DSHA-containing protomer B, these two conformations differ in the position of Cα (Gly209) by up to 2.8–3.3 Å, and the position of individual side chain carbons (Phe212) differs by up to 4.2–6.7 Å(Fig. 7). The net effect of the implied movement places Met208 and Phe212 closer in the nonpolar subsite of the binding pocket. Although neither residue is likely to directly influence the binding of HOPDA, both could form hydrophobic interactions with the ring moieties of DSHA and HOPODA, respectively. The conformation of the α-helix in the ligand-free protomers is essentially identical to that observed in protomers A and B of ht-HsaD (14).
FIGURE 7.
Conformation of the αL4 helix in the presence of the ligand DSHA. The backbone of ligand-free S114A (blue) overlaid with that of the S114A·DSHA complex (gray). The side chains Phe212, Met208, and also DSHA (purple) are shown in stick representation.
DISCUSSION
The relatively high specificity of HsaD from M. tuberculosis for DSHA (kcat/Km = 33,000 m−1 s−1) indicates that the enzyme is involved in cholesterol catabolism. Similarly, the structural data indicate that the substrate-binding pocket is best adapted to the steroid substrate. Although the specificity of HsaD is ∼2–3 orders of magnitude lower than other MCP hydrolases for their physiological substrates (13, 23, 35), this is consistent with what has been reported for other M. tuberculosis enzymes involved in cholesterol metabolism, including KshAB (36) and HsaC (8).
It is unclear why HOPODA is a poorer substrate for HsaD than HOPDA; the chloro-phenyl moiety of HOPODA was designed to mimic the bulk of the bicycloalkanone portion of DSHA, and the structures of the enzyme·substrate complexes indicate that the dienoate moiety of the three MCPs were identically positioned in the active site of S114A (Fig. 5). Nevertheless, the Km of HOPODA was 30-fold higher than that of HOPDA or DSHA. The increased Km and kcat/Km is in contrast to the high specificity of BphDs for HOPDA variants bearing chloro substituents on the phenyl ring, particularly in the meta- and para-positions (17). Although chlorophenyl HOPDAs are thought to stabilize the negatively charged tetrahedral intermediate at C-6, the chlorophenyl of HOPODA is unlikely to contribute to stabilization because of the saturated two carbon bridge between the chlorophenyl and the dienoate moieties. Redesigning the chlorophenyl moiety of the substrate analogue may improve HsaD-analogue interactions and provide clues to the binding mechanism.
This study provides the clearest evidence to date that the active sites of MCP hydrolase exist in two conformations: open in the absence of ligand and closed in its presence. Two observations in previous structures of MCP hydrolases had suggested this possibility. First, the α-helix lid domains had higher B-factors than the remainder of the protein (12, 37) as observed here for HsaD. Second, structures of BphD obtained in the different states revealed similar but smaller differences in lid positions (34, 38). The presence of malonate in the active site cleft of substrate-free BphD may have reduced the observed differences in this enzyme. The conformational differences seen in the lid domain of HsaD are lesser in magnitude than those observed in another α/β hydrolase, the lipase from Rhizomucor miehei (39, 40), however, they are remarkably similar in location and in net effect.
The observation of two active site conformations in HsaD is consistent with the half-site mechanism proposed on the basis of the MphC structure (37) as well as from kinetic studies of BphD and the ordered release of the two reaction products (13). According to the half-site mechanism, the hydrolase cycles between two interconverting conformations: one that is enzymatically inactive and one that is enzymatically active. More particularly, benzoate dissociation followed that of the dienoate, was nondiffusive, and occurred at a rate that was roughly equivalent to the kcat (13). Consistent with this model, the electron density of the bound substrate in the S114A·MCP complexes was always better defined in one protomer (B) than the other. Moreover, in the structure with the weakest ligand electron density, S114A·HOPODA, the αL4 helix was in a mixed conformation, with one protomer in a open conformation and another in a closed conformation. Although speculative, the movement of the αL4 helix could assist in the nondiffusive release of the second product: benzoate and DOHNAA in the cases of HOPDA and DSHA, respectively. Because the release of the second product is rate-limiting, the increased distance between the αL4 helix and the non-dienoate portion of HOPDA compared with that of DSHA and HOPODA (Fig. 5) could explain the 6-fold reduction of the kcat of HOPDA. However, further data are required to determine the effect of the C-5 methyl substituent on the kcat of HsaD.
The spectroscopic (Fig. 2) and structural data (Fig. 4 and Table 4) of the S114A·MCP complexes are very similar to those reported for the equivalent S112A·MCP complexes of BphD. Specifically, the orientation and the torsional angles of the MCPs in the BphD structure suggest that the substrate had undergone ketonization, yielding an E:Sk species. However, the spectroscopic data indicated that the bound MCPs were conjugated because the disrupted π-conjugation of E:Sk is predicted to result in a blue shift. Interestingly, the 60-nm red shift in S114A·DSHA is similar in magnitude to those observed in S112A·HOPDA (72 nm) (12, 13) and in the L photointermediate of the retinal chromophore of bacteriorhodopsin (60 nm) (41). The latter has been explained on the basis of twist-about double bonds, a distortion of the chromophore that has recently been observed crystallographically (42). The dihedral angles of the twisted double bond in the L photointermediate of bacteriorhodopsin were up to 40° from the resting state molecule, similar to the values observed with the 2-enol tautomers of the S114A·MCP structures (Table 4). Although it is possible that different species of S114A·DSHA occur in solution and in crystallo, a strained enolate, E:Sse, is most consistent with the structure and red-shifted absorption spectrum of S114A·DSHA. Moreover, the similar electronic absorption spectra of S114A·DSHA and E:Sred indicate the occurrence of a strained enolate species as a catalytic intermediate and imply that the serine residue plays a significant role in tautomerizing the bound MCP.
In MCP hydrolases, the enzymatic cleavage of the ligand has been proposed to occur through a general base mechanism (19). The structures of the S114A·MCP complexes are most consistent with the catalytic serine acting as a general base to activate a water molecule for nucleophilic attack of the C-6 carbonyl of the substrate. Notably, a solvent species is located 3.2–3.6 Åfrom the carbonyl and 3.7–4.1 Åfrom the Cβ of Ala114 (Fig. 6B). Although the location of the water molecule may differ in the native enzyme, the overlaid structure of ht-HsaD with S114A·MCP places the Oγ from Ser114 within ∼3 Åfrom the water and 2.4 Åfrom the C-6 carbonyl in the overlaid structures. However, the Oγ–C-6–O-6 angle of 46° deviates significantly from the Bürgi-Dunitz angle. Finally, His269 is over 5 Åaway from the closest water molecule (Fig. 6B).
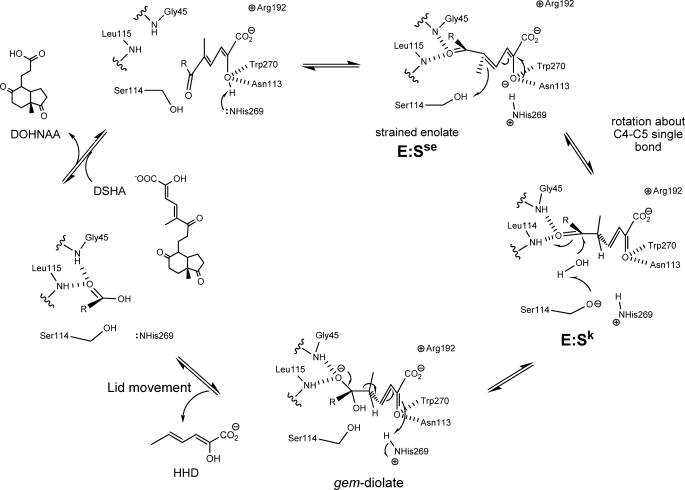
In conclusion, the results presented herein suggest a dual role for the catalytic serine of MCP hydrolases and enable further refinement of the proposed catalytic mechanism for these enzymes. As summarized in Fig. 8, hydrolysis of the bound substrate is initiated by deprotonation of the C-2 hydroxyl by His269. The bulk of the latter helps to generate a strained enolate intermediate, E:Sse. C-5 is then protonated by Ser114, inducing tautomerization of the substrate and further rotation around the C-4–C-5 to yield a ketonized intermediate, E:Sk. Subsequently, Ser114 deprotonates a nearby water molecule, which attacks the C-6 carbonyl to yield a gem-diol intermediate. The tetrahedral intermediate collapses, releasing HHD and triggering a conformational change of the lid domain, which releases DOHNAA. This proposal provides a basis for further study on the mechanism of MCP hydrolases.
FIGURE 8.
Proposed mechanism of HsaD. Upon binding of the MCP, His269 deprotonates the hydroxyl at C-2, generating a strained enolate intermediate, E:Sse. Protonation of C-5 by Ser114 drives tautomerization of the substrate to generate a keto intermediate, E:Sk. Ser114 is positioned to activate water for attack at the C-6 carbonyl to form the gem-diolate. The collapse of the tetrahedral intermediate releases HHD, which triggers a conformational change in the lid domain and allows subsequent release of DOHNAA.
Supplementary Material
Acknowledgments
We thank the beamline scientists at the Diamond Light Source (Oxfordshire, UK) and Swiss Light Source (Villigen, Switzerland) for valuable technical support, Jie Liu for help constructing the S114A variant, Jeffrey T. Bolin for critically reading the manuscript, and Isaac Westwood for expert crystallography advice.
This work was supported by grants from the Wellcome Trust (to E. S.) and the Canadian Institutes of Health Research (to L. D. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 2wud, 2wue, 2wuf, and 2wug) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- DSHA
- 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid
- HOPDA
- 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid
- HOPODA
- 8-(2-chlorophenyl)-2-hydroxy-5-methyl-6-oxoocta-2,4-dienoic acid
- DHSA
- 3,4-dihydroxy-9,10-seco-nandrost-1,3,5(10)-triene-9,17-dione
- DHDS
- 2,3-dihydroxy-6-methyl-7,8-dihydrostilbene
- MCP
- meta-cleavage product
- HsaD
- 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase
- DOHNAA
- 9,17-dioxo-1,2,3,4,10,19-hexanorandro-stan-5-oic acid
- HHD
- 2-hydroxy-hexa-2,4-dienoic acid
- ht
- histidine-tagged
- bis-tris
- 1,3-bis(tris(hydroxylmethyl)methylamino) propane.
REFERENCES
- 1.Dye C. (2006) Lancet 367, 938–940 [DOI] [PubMed] [Google Scholar]
- 2.Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobel H., Plaut A. (1949) J. Bacteriol. 57, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey A. K., Sassetti C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagel W., Pagel M. (1925) Virchows Arch. Pathol. Anat. 256, 629–640 [Google Scholar]
- 7.Hunter R. L., Olsen M., Jagannath C., Actor J. K. (2006) Am. J. Pathol. 168, 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yam K. C., D'Angelo I., Kalscheuer R., Zhu H., Wang J. X., Snieckus V., Ly L. H., Converse P. J., Jacobs W. R., Jr., Strynadka N., Eltis L. D. (2009) PLoS Pathog. 5, e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horinouchi M., Hayashi T., Yamamoto T., Kudo T. (2003) Appl. Environ. Microbiol. 69, 4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rengarajan J., Bloom B. R., Rubin E. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C., Hassler M., Bugg T. D. (2008) Chembiochem. 9, 71–76 [DOI] [PubMed] [Google Scholar]
- 12.Nandhagopal N., Yamada A., Hatta T., Masai E., Fukuda M., Mitsui Y., Senda T. (2001) J. Mol. Biol. 309, 1139–1151 [DOI] [PubMed] [Google Scholar]
- 13.Horsman G. P., Ke J., Dai S., Seah S. Y., Bolin J. T., Eltis L. D. (2006) Biochemistry 45, 11071–11086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lack N., Lowe E. D., Liu J., Eltis L. D., Noble M. E., Sim E., Westwood I. M. (2008) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J. (1992) Protein Eng. 5, 197–211 [DOI] [PubMed] [Google Scholar]
- 16.Lam W. W., Bugg T. D. (1997) Biochemistry 36, 12242–12251 [DOI] [PubMed] [Google Scholar]
- 17.Seah S. Y., Labbé G., Nerdinger S., Johnson M. R., Snieckus V., Eltis L. D. (2000) J. Biol. Chem. 275, 15701–15708 [DOI] [PubMed] [Google Scholar]
- 18.Li J. J., Bugg T. D. (2007) Org. Biomol. Chem. 5, 507–513 [DOI] [PubMed] [Google Scholar]
- 19.Fleming S. M., Robertson T. A., Langley G. J., Bugg T. D. (2000) Biochemistry 39, 1522–1531 [DOI] [PubMed] [Google Scholar]
- 20.Li J. J., Li C., Blindauer C. A., Bugg T. D. (2006) Biochemistry 45, 12461–12469 [DOI] [PubMed] [Google Scholar]
- 21.Gruber K., Gartler G., Krammer B., Schwab H., Kratky C. (2004) J. Biol. Chem. 279, 20501–20510 [DOI] [PubMed] [Google Scholar]
- 22.Williams D. H., Fleming I. (1995) Spectroscopic Methods in Organic Chemistry, 5th Ed., McGraw-Hill, London [Google Scholar]
- 23.Li C., Montgomery M. G., Mohammed F., Li J. J., Wood S. P., Bugg T. D. (2005) J. Mol. Biol. 346, 241–251 [DOI] [PubMed] [Google Scholar]
- 24.Lack N. A., Kawamura A., Fullam E., Laurieri N., Beard S., Russell A. J., Evangelopoulos D., Westwood I., Sim E. (2009) Biochem. J. 418, 369–378 [DOI] [PubMed] [Google Scholar]
- 25.Nerdinger S., Kendall C., Cai X., Marchart R., Riebel P., Johnson M. R., Yin C. F., Hénaff N., Eltis L. D., Snieckus V. (2007) J. Org. Chem. 72, 5960–5967 [DOI] [PubMed] [Google Scholar]
- 26.Cornish-Bowden A. (1995) Analysis of Enzyme Kinetic Data, Oxford University Press, Oxford [Google Scholar]
- 27.Erman J. E., Vitello L. B. (1980) J. Biol. Chem. 255, 6224–6227 [PubMed] [Google Scholar]
- 28.Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- 29.Vagin A., Teplyakov A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 30.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 32.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLano W. L. (2002) The PyMOL Molecular Graphics System, 0.99 Ed., DeLano Scientific, LLC, Palo Alto, CA [Google Scholar]
- 34.Horsman G. P., Bhowmik S., Seah S. Y., Kumar P., Bolin J. T., Eltis L. D. (2007) J. Biol. Chem. 282, 19894–19904 [DOI] [PubMed] [Google Scholar]
- 35.Seah S. Y., Ke J., Denis G., Horsman G. P., Fortin P. D., Whiting C. J., Eltis L. D. (2007) J. Bacteriol. 189, 4038–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capyk J. K., D'Angelo I., Strynadka N. C., Eltis L. D. (2009) J. Biol. Chem. 284, 9937–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn G., Montgomery M. G., Mohammed F., Coker A., Cooper J. B., Robertson T., Garcia J. L., Bugg T. D., Wood S. P. (2005) J. Mol. Biol. 346, 253–265 [DOI] [PubMed] [Google Scholar]
- 38.Bhowmik S., Horsman G. P., Bolin J. T., Eltis L. D. (2007) J. Biol. Chem. 282, 36377–36385 [DOI] [PubMed] [Google Scholar]
- 39.Derewenda U., Brzozowski A. M., Lawson D. M., Derewenda Z. S. (1992) Biochemistry 31, 1532–1541 [DOI] [PubMed] [Google Scholar]
- 40.Brady L., Brzozowski A. M., Derewenda Z. S., Dodson E., Dodson G., Tolley S., Turkenburg J. P., Christiansen L., Huge-Jensen B., Norskov L. (1990) Nature 343, 767–770 [DOI] [PubMed] [Google Scholar]
- 41.Hu J. G., Sun B. Q., Petkova A. T., Griffin R. G., Herzfeld J. (1997) Biochemistry 36, 9316–9322 [DOI] [PubMed] [Google Scholar]
- 42.Lanyi J. K., Schobert B. (2007) J. Mol. Biol. 365, 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.