Abstract
Lens epithelium-derived growth factor (LEDGF) is an important co-factor of human immunodeficiency virus DNA integration; however, its cellular functions are poorly characterized. We now report identification of the Cdc7-activator of S-phase kinase (ASK) heterodimer as a novel interactor of LEDGF. Both kinase subunits co-immunoprecipitated with endogenous LEDGF from human cell extracts. Truncation analyses identified the integrase-binding domain of LEDGF as essential and minimally sufficient for the interaction with Cdc7-ASK. Reciprocally, the interaction required autophosphorylation of the kinase and the presence of 50 C-terminal residues of ASK. The kinase phosphorylated LEDGF in vitro, with Ser-206 being the major target, and LEDGF phosphorylated at this residue could be detected during S phase of the cell cycle. LEDGF potently stimulated the enzymatic activity of Cdc7-ASK, increasing phosphorylation of MCM2 in vitro by more than 10-fold. This enzymatic stimulation as well as phosphorylation of LEDGF depended on the protein-protein interaction. Intriguingly, removing the C-terminal region of ASK, involved in the interaction with LEDGF, resulted in a hyperactive kinase. Our results indicate that the interaction with LEDGF relieves autoinhibition of Cdc7-ASK kinase, imposed by the C terminus of ASK.
Introduction
Lens epithelium-derived growth factor (LEDGF)3 is a member of the hepatoma-derived growth factor family of proteins, characterized by a highly conserved PWWP domain within the N-terminal regions of its members (1, 2). LEDGF and its close homologue HRP2 (hepatoma-derived growth factor-related protein 2) are found throughout Vertebrata, and a divergent ortholog was recognized in insects (3). To date, LEDGF has been implicated in a range of human pathologies, including human immunodeficiency virus (HIV) infection, autoimmunity, and cancer (4–7). On the molecular level, its function is best characterized in the context of HIV replication (reviewed in Ref. 5). LEDGF is a dominant binding partner of HIV-1 integrase (IN) in human cells (8, 9). The tight IN-LEDGF interaction explains nuclear accumulation and chromosomal localization of HIV-1 IN, when the viral protein is ectopically expressed (9, 10). LEDGF is associated with chromatin throughout the cell cycle, and this property, mediated by the PWWP domain and AT-hook motifs (11, 12), is essential for its co-factor function during HIV DNA integration (13, 14). In the context of lentiviral infection, LEDGF serves to direct integration of viral DNA into active transcription units of the host genome (14–16). The integrase-binding domain (IBD), located within the C-terminal region of LEDGF (residues 347–429), is responsible for the high affinity interaction with HIV-1 and other lentiviral IN proteins (3, 17–19). HRP2 also contains an IBD and retains affinity for HIV-1 IN in vitro, although its role in viral replication has not been established (3, 13, 18).
Although the role of LEDGF in the lentiviral life cycle is well characterized, far less is known about its natural cellular functions. The protein was initially described as a 75-kDa polypeptide co-purified with transcriptional co-activator PC4 (20), although a direct interaction between LEDGF and PC4 has not been verified. More recently LEDGF was identified as a component of the menin-mixed lineage leukemia histone methyltransferase complex and shown to be essential for mixed lineage leukemia histone methyltransferase-dependent transcription regulation and oncogenesis (21). In addition, LEDGF interacts with JPO2 and PogZ, although the functions of either protein are currently unknown (22–24). Overexpression of LEDGF was reported to up-regulate a subset of genes involved in the stress response and to protect cells from apoptosis (reviewed in Ref. 25). LEDGF is a substrate of caspases, and caspase-induced cleavage of LEDGF inactivates its prosurvival function, in some cases enhancing apoptosis (reviewed in Ref. 6).
Herein we describe the identification of the heterodimeric S-phase kinase Cdc7-ASK as a novel interactor of LEDGF in human cells. Cdc7 is a Ser/Thr kinase, conserved from yeast to mammals, essential for initiation of DNA replication throughout S phase (26–28). Its activity is controlled via interaction with a regulatory subunit, known as Dbf4 in Saccharomyces cerevisiae and Xenopus laevis (29, 30), Dfp1/Him1 in Schizosaccharomyces pombe (31, 32), and activator of S-phase kinase (ASK) in mammals (27, 28, 33). Whereas in S. cerevisiae the abundance of Dbf4 is controlled by the anaphase-promoting complex, in mammalian cells, the expression of ASK is also subject to cell-cycle dependent regulation (27, 31, 34–36). Its protein levels, lowest during M phase, increase at late G1 and remain high during S phase (27). In contrast, Cdc7 protein levels are relatively stable throughout the cell cycle (37), whereas its kinase activity oscillates in a manner dependent on the abundance of the regulatory subunit (27). Cooperating with S-phase cyclin-dependent kinase, Cdc7 activates individual prereplication complexes, assembled at replication origins during G1. Phosphorylation of prereplication complex components by the S-phase-promoting kinases leads to unwinding of origin DNA and recruitment of the replication fork machinery. Mounting experimental evidence indicates that the hetero-hexameric minichromosome maintenance (MCM) complex assumes the function of the replicative helicase involved in both initiation and elongation stages of DNA replication (38–42). The MCM complex, composed of six homologous subunits, MCM2–MCM7, appears to be the primary physiological target of Cdc7 kinase (26). Three of its components, MCM2, MCM4, and MCM6, were shown to be substrates for Cdc7 under various assay conditions. Recombinant human Cdc7-ASK heterodimer can efficiently phosphorylate uncomplexed MCM2 as well as MCM2, MCM4, and MCM6 within the quaternary MCM2-MCM4-MCM6-MCM7 complex (33, 43, 44). Furthermore, Cdc7 has been shown to phosphorylate MCM2 in human cells, and the phosphorylation sites identified in vitro have been validated in vivo (28, 43, 44).
Dbf4/ASK orthologs from various species are surprisingly divergent, with only three conserved regions identified, termed Dbf4 motifs N, M, and C (27, 32, 45). The M and C motifs, characterized as a Pro-rich and C2H2-type zinc-binding domains, respectively, are essential for kinase activation and its mitotic functions in S. pombe (46). A small fragment of human ASK spanning motifs M and C (residues 174–350) is minimally sufficient to support Cdc7 kinase activity (47). The N motif, distantly related to the BRCA1 C-terminal domain, has been implicated in the interactions with the replication machinery and replication origins in S. cerevisiae (46, 48). Studies using X. laevis egg extracts suggested that association of Cdc7 with chromatin is dependent on Dbf4 (49–51). Collectively, these results suggest that the S-phase kinase is recruited to replication origins via its regulatory subunit. Herein, we show that human Cdc7-ASK interacts with LEDGF, a component of chromatin, known to be functionally associated with transcriptionally active genomic loci (14–16). The interaction, which is mediated by the IBD of LEDGF and critically depends on the C terminus of ASK, leads to robust stimulation of Cdc7 kinase activity on its physiological substrate MCM2. Our results suggest that Cdc7-ASK activity is subject to an additional level of regulation in higher eukaryotes, potentially providing a link between gene expression and DNA replication.
EXPERIMENTAL PROCEDURES
DNA Constructs for Expression in Human Cells
The plasmids used in this work are summarized in supplemental Table S1. To obtain pGM-hLEDGF-(326–530)-cTAP, a PCR fragment encoding residues 326–530 of human LEDGF was subcloned between BamHI and HindIII sites of pGM-Mel18-cTAP (52), replacing the Mel18 coding sequence (CDS). To make pLB(N)CX-mp75-HA and pLB(N)CX-mp52-HA, PCR fragments spanning CDSs of mouse LEDGF and p52, respectively, were extended to include an additional 27 bp encoding a C-terminal influenza hemagglutinin (HA) tag (YPYDVPDYA), followed by a stop codon, and ligated between HindIII and HpaI sites of pLB(N)CX (53). To make pIRES2-LEDGFΔIBD-eGFP, a DNA fragment encoding human LEDGF lacking residues 347–430 was constructed by overlap PCR splicing and ligated between EcoRI and BamHI sites of pIRES2-eGFP (14). The plasmid pCPHA38, used for expression of HA-LEDGF-(326–530), was made by inserting a PCR fragment encoding the C-terminal portion of human LEDGF between BglII and XhoI sites of pCPHA-NLS (3). The constructs pTRE-HA75-hyg and pTRE-75-hyg, used for stable expression of HA-tagged and untagged human LEDGF, respectively, in HeLa-TetOff cells, were obtained by subcloning PCR fragments encoding respective LEDGF variants between PvuII and NotI sites of pTRE2hyg (Clontech).
To obtain pcDNA3.1-FLAG-Cdc7, a modified version of pcDNA3.1(−) encoding a FLAG tag (DYKDDDDK) between NotI and BamHI sites,4 linearized with BamHI and filled in using T4 DNA polymerase, was digested with AflII and ligated with a PCR fragment spanning the CDS of human Cdc7. To make pEGFP-Cdc7, the CDS was subcloned between SacI and SalI sites of pEGFP-C2. To make pCPHA-Dbf4 and pCPHA-Dbf4-(1–624), PCR fragments spanning the respective human ASK sequences were subcloned between BglII and XhoI sites of pCPHA-NLS. Retroviral vector pQFLAG-MCM2–8×His was used for stable expression of full-length human MCM2 containing N-terminal FLAG (DYKDDDDK) and C-terminal octahistidine tags. To make it, the entire MCM2 CDS was PCR-amplified and extended to add 24 bp encoding the C-terminal octahistidine tag and ligated between XhoI and BamHI sites of pQFLAG, a derivative of pQCXIP (Clontech) with an insertion of a FLAG tag CDS upstream of the XhoI site.5
DNA for Protein Expression in Bacteria
To obtain pRSF-CDC7-S-tag, expressing Cdc7 with a C-terminal S-tag (KETAAAKFERQHMDS), a PCR fragment spanning the CDS of human Cdc7 was inserted between NcoI and XhoI sites of pRSFDuet-1 (Novagen). To make pCDF-His-Dbf4, pCDF-His-Dbf4-(1–350), pCDF-His-Dbf4-(174–350), pCDF-His-Dbf4-(174–674), pCDF-His-Dbf4-(1–541), and pCDF-His-Dbf4-(1–624), used for production of hexahistidine-tagged human ASK or its truncated forms, the corresponding PCR fragments were subcloned between EcoRI and XhoI sites of pCDFDuet-1 (Novagen).
To obtain pFT1-LEDGFΔIBD, the DNA fragment encoding human LEDGF lacking internal residues 347–430 was ligated between BamHI and EcoRI sites of pFT1-LEDGF (54), replacing the original insert. The plasmids pCPH6P-LEDGF-(146–530), pCPH6P-LEDGF-(226–530), pCPH6P-LEDGF-(249–530), pCPH6P-LEDGF-(291–530), and pCPH6P-LEDGF-(347–530), used for production of N-terminally truncated forms of LEDGF, were made by ligating corresponding PCR fragments between XmaI and BamHI sites of pCPH6P-BIV-IN (55), replacing the IN CDS. DNA constructs for bacterial production of full-length LEDGF with various amino acid substitutions were obtained by mutagenizing pFT1-LEDGF (54), using the QuikChange procedure (Stratagene). The plasmid pCP-GST-MCM2-(1–287) was made by inserting a PCR fragment encoding residues 1–287 of human MCM2 between EcoRI and BamHI sites of pGEX-4T1 (GE Healthcare). All DNA constructs made in this work were sequenced to confirm the absence of unwanted mutations.
Cell Lines
293T, HeLa, HeLa-TetOff (Clontech), and their derivatives were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 5 units/ml penicillin, and 5 μg/ml streptomycin. For stable expression of the cTAP fusion, 293T cells were transfected with linearized pGM-hLEDGF-(326–530)-cTAP and selected in the presence of 10 μg/ml blasticidin S (Invitrogen). Control 293T-cTAP cells, expressing isolated tandem affinity purification (TAP) tag have been reported (52). To obtain a stable cell line expressing HA-LEDGF, HeLa-TetOff cells (Clontech) transfected with linearized pTRE-HA75-hyg were selected in the presence of 160 μg/ml hygromycin B and 5 μg/ml doxycycline. Single cell clone H3, which upon withdrawal of doxycycline showed expression of HA-LEDGF, was used for experiments described herein. A control cell line, N3, overexpressing a non-tagged form of LEDGF, was established in a similar manner. H3 and N3 cells were routinely maintained in the absence of doxycycline. For stable expression of FLAG-tagged MCM2, HeLa cells were infected with retroviral vector produced by co-transfecting 293T cells with pQFLAG-MCM2–8×His, pCG-VSV-G, and pCG-GAG-POL (56); stably transduced cells were selected in the presence of 1 μg/ml puromycin to obtain HeLa-FLAG-MCM2-His8.
Tandem Affinity Purification
293T-LEDGF-(326–530)-cTAP and the control line 293T-cTAP, each grown to near confluence in twelve 500-cm2 cell culture dishes, were harvested by trypsinization and washed in phosphate-buffered saline (PBS), containing 0.1 mm phenylmethylsulfonyl fluoride (PMSF). Isolation of the cTAP-tagged complex was done according to published procedures (52, 57), with minor modifications. Cells were lysed in 12 ml of ice-cold TAP lysis buffer (150 mm NaCl, 1 mm EDTA, 1.02 mm ZnCl2, 0.1 mm PMSF, and 50 mm Tris-HCl, pH 8.0), supplemented with 1% Nonidet P-40 and complete EDTA-free protease inhibitor mix (Roche Applied Science). Resulting extracts, precleared using 100 μl of glutathione-Sepharose (GE Healthcare), were incubated with 180 μl of IgG-Sepharose (GE Healthcare) at 4 °C for 3 h. The resin was washed with several changes of TAP lysis buffer, equilibrated with tobacco etch virus protease buffer (150 mm NaCl, 0.1% Nonidet P-40, 0.5 mm EDTA, 0.52 mm ZnCl2, 1 mm dithiothreitol (DTT), 0.1 mm PMSF, and 10 mm Tris-HCl pH 8.0), and incubated with 100 units of AcTEV protease (Invitrogen) at room temperature for 2 h. Released proteins were eluted with several changes of CaBIND buffer (150 mm NaCl, 0.1% Nonidet P-40, 1 mm MgSO4, 2 mm CaCl2, 1 mm imidazole, 14 mm β-mercaptoethanol, and 50 mm Tris-HCl, pH 8.0) and allowed to bind to 60 μl of calmodulin affinity resin (Stratagene) for 2 h at 4 °C. The beads were washed with several changes of CaBIND buffer, and bound proteins were eluted with 5 mm EDTA in 150 mm NaCl, 0.1% Nonidet P-40, and 50 mm Tris-HCl, pH 8.0, concentrated by precipitation with trichloroacetic acid, and separated by SDS-PAGE using 4–20% gradient Tris-glycine gels. Protein bands, detected by staining with colloidal Coomassie G-250, were identified by in-gel trypsin digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Taplin mass spectrometry facility (Harvard Medical School, Boston, MA).
Antibodies
The following primary antibodies were used in this work: mouse anti-HA (HA.11, Covance), mouse anti-Cdc7 (Lab Vision Corp.), mouse anti-β-actin (Sigma), mouse monoclonal anti-ASK (H00010926-M01, Abnova), mouse anti-LEDGF (Western blotting, clone 26, BD Biosciences), rabbit anti-LEDGF/p75 (immunoprecipitation (IP) and Western blotting, A300–848A, Bethyl Laboratories), rabbit control IgG (IP, ab46540, Abcam), mouse anti-His5 (Qiagen), mouse anti-FLAG (M2, Sigma), rabbit anti-phospho-Ser53 MCM2 (A300-756A, Bethyl Laboratories), rabbit anti-β tubulin (H235, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), and goat anti-lamin B (M-20, Santa Cruz Biotechnology, Inc.).
Immunoprecipitation
HeLa cells grown in 10-cm dishes were transfected with pLB(N)CX-mp75-HA, pBHA-p75, pCPHA-NLS-38, pCPHA-HRP2, pLB(N)CX-mp52-HA, or pCPHA-NLS using calcium phosphate. Cells, harvested at 48 h post-transfection, were lysed in 800 μl of CSKL buffer (CSKc (10% sucrose, 1 mm MgCl2, 0.5 mm DTT, and 10 mm PIPES-NaOH, pH 6.8), supplemented with 420 mm NaCl, 0.5% Nonidet P-40, 0.1 mm PMSF, and EDTA-free protease inhibitors). For IPs, 750 μl of precleared extracts were incubated with 15 μl HA.11 affinity matrix (Covance) for 3 h at 4 °C. The beads were washed in several changes of CSKc supplemented with 150 mm NaCl, 0.1% Nonidet P-40, and EDTA-free protease inhibitors and processed for Western blot analyses. For co-IP of endogenous proteins, 293T cells grown to 80% confluence in a 175-cm2 flask were lysed in 800 μl CSKL, and precleared extract was incubated with 5 μg of rabbit anti-LEDGF or control rabbit antibody in the presence of 20 μl of protein G-agarose (GE Healthcare). The resin was washed as explained above, and recovered proteins were analyzed by Western blotting using mouse monoclonal anti-LEDGF, Cdc7, and ASK antibodies.
For phosphopeptide analyses, asynchronously growing H3 cells, cultured in the absence of doxycycline to ∼80% confluence in four 500-cm2 dishes, were lysed in CSKc buffer supplemented with 0.5% Nonidet P-40, 450 mm NaCl, 0.1 mm PMSF, 1 mm β-glycerophosphate, 1 mm NaF, 0.32 mm Na3VO4, and EDTA-free protease inhibitors. HA-LEDGF was captured on 75 μl of HA.11 affinity matrix and, following extensive washing in CSKc buffer supplemented with 150 mm NaCl, 0.1% Nonidet P-40, 0.1 mm PMSF, protease inhibitors, 1 mm β-glycerophosphate, 1 mm NaF, and 0.32 mm Na3VO4, was eluted with 6 m NaSCN. The protein, concentrated by precipitation with tricholoroacetic acid, separated in an 11% SDS-polyacrylamide gel and visualized by staining with Coomassie G-250, was subjected to in-gel tryptic digestion and LC-MS/MS phosphopeptide analysis (Taplin mass spectrometry facility). For enrichment of S-phase-specific LEDGF phosphopeptides, H3 cells, incubated in the presence of 4 μg/ml aphidicolin (Calbiochem) for 24 h, were released into S phase for 2 h before proceeding with IPs as detailed above.
Full-length MCM2 protein was purified from HeLa-FLAG-MCM2-His8 cells grown to near confluence in two 500-cm2 tissue culture dishes. The protein, captured onto 50 μl of anti-FLAG affinity matrix (Sigma), was eluted in 50 μl of CSKc supplemented with 0.2 mg/ml FLAG peptide (Sigma), 150 mm NaCl, 0.1% Nonidet P-40, 1 mm β-glycerophosphate, 1 mm NaF, 0.32 mm Na3VO4, and EDTA-free protease inhibitors.
Cell Fractionation
To prepare Triton-soluble and -insoluble fractions, transfected 293T cells were lysed in ice-cold CSKT buffer (150 mm NaCl, 0.5% Triton X-100, 1 mm MgCl2, 10% sucrose, 1 mm DTT, 10 mm NaF, 1 mm ATP, 0.1 mm PMSF, EDTA-free protease inhibitors, and 10 mm PIPES-NaOH, pH 6.8) for 10 min, followed by centrifugation at 17,000 × g for 10 min. The supernatants were kept as the Triton-soluble fraction, whereas the pellets (Triton-insoluble fraction) were washed in CSKT buffer and solubilized by boiling in Laemmli SDS-PAGE sample buffer. To obtain whole cell extracts, cell pellets were boiled in PBS supplemented with 1% SDS, 5 mm EDTA, and 0.5 mm PMSF.
Fluorescent Imaging
HeLa cells cultured in LabTek chamber slides (Nunc) were transfected with expression constructs for EGFP-Cdc7, HA-ASK, HA-ASK-(1–624), HcRed1-LEDGF, and/or HcRed1-p52 using Lipofectamine 2000 (Invitrogen). Twenty-four h post-transfection cells were fixed in 4% formaldehyde in PBS for 15 min at room temperature. Following washing in PBS supplemented with 25 mm ammonium sulfate, the cells were permeabilized with ice-cold methanol and blocked in 10% fetal calf serum in PBS. HA-ASK was detected using HA.11 and goat anti-mouse Alexa Fluor 555-conjugated antibody. DNA was stained with 4′,6-diamidino-2-phenylindole dilactate (Invitrogen); all reagents were diluted in blocking solution. Four-color imaging was done using an SP5 MP/FLIM microscope (Leica).
Recombinant Proteins
Rosetta-2 (DE3) cells (Novagen) co-transformed with pRSF-Cdc7-S-Tag and pCDF-His-Dbf4 were used for production of the Cdc7-ASK holoenzyme. Bacteria were grown in LB medium supplemented with 50 μg/ml kanamycin and 100 μg/ml spectinomycin in shake flasks at 30 °C. The cultures were allowed to equilibrate to 18 °C prior to reaching an A600 of 1.0, and protein expression was induced by the addition of 0.3 mm isopropyl-β-d-thiogalactopyranoside. Following a 3-h induction at 18 °C, cells were harvested and stored at −80 °C. Thawed bacterial paste was sonicated in buffer A1 (300 mm NaCl, 0.025% Nonidet P-40, 10% glycerol, and 50 mm NaH2PO4, pH 7.5), supplemented with 0.1 mm PMSF, EDTA-free protease inhibitors, 1 mm NaF, 1 mm β-glycerophosphate, 20 mm imidazole, 1 mg/ml lysozyme, and additional 0.5% Nonidet P-40. The lysates, clarified by centrifugation, were loaded onto a 5-ml HisTrap column (GE Healthcare) at 1 ml/min. Following extensive washing, the protein was eluted with a 0.02–1.0 m linear gradient of imidazole in buffer A1. Fractions containing Cdc7-ASK complex were pooled, diluted 3-fold in buffer A2 (10% glycerol and 50 mm Tris-HCl, pH 7.5) and injected into a 5-ml HiTrapQ column (GE Healthcare). The protein, eluted with a linear 0.1–0.5 m gradient of NaCl in buffer A2, was further purified by size exclusion chromatography on a HiLoad 16/60 Superdex-200 column (GE Healthcare), operated in 150 mm NaCl, 2 mm NaF, and 50 mm Tris-HCl, pH 7.5. Cdc7-ASK complex, supplemented with 10 mm DTT, 1 mm NaF, 1 mm β-glycerophosphate, and 10% glycerol, was concentrated on a Centriprep-YM3 device (Millipore) and flash-frozen in liquid nitrogen. Cdc7-ASK-(174–350), Cdc7-ASK-(1–350), Cdc7-ASK-(174–674), Cdc7-ASK-(1–541), and Cdc7-ASK-(1–624) were produced in a similar fashion, using constructs expressing the corresponding truncated versions of ASK.
GST-MCM2-(1–287) was produced in Rosetta-2 (DE3) cells, transformed with pCP-GST-MCM2-(1–287). Bacteria, grown to an A600 of ∼1 in the presence of 120 μg/ml ampicillin, were induced with 0.15 mm isopropyl-β-d-thiogalactopyranoside at 25 °C for 5 h. Cells were lysed by sonication in core buffer (0.5 m NaCl, 1 mm EDTA, 50 mm Tris-HCl, pH 7.4), supplemented with 2 mm DTT, 0.5 mm PMSF, 0.5% Triton-X 100, and 1 mg/ml lysozyme. The fusion protein, captured on glutathione-Sepharose (GE Healthcare), was eluted with 50 mm glutathione in core buffer. Wild type (WT) and mutant LEDGF proteins as well as GST-HRP2-(470–593) were produced according to established procedures (3, 12, 17).
In Vitro Kinase Assays
A standard 25-μl kinase reaction mixture contained 10 ng of Cdc7-ASK or corresponding molar equivalents of its mutant forms and 3 μCi of [γ-32P]ATP (3,000 Ci/mmol) in 10 mm MgSO4, 2 mm DTT, 1 mm β-glycerophosphate, 1 mm NaF, 80 μg/ml bovine serum albumin, 0.1% Nonidet P-40, 0.1 mm ATP, 40 mm Hepes-NaOH, pH 7.4. Substrates were used in the following quantities: 0.5 μg of GST-MCM2-(1–287), 0.5 μg of LEDGF or corresponding molar equivalents of its truncated forms. Where indicated, 0.23 μg of glutathione S-transferase (GST) was present as a negative control. Kinase reactions, allowed to proceed for 30 min at 30 °C, were stopped by the addition of Laemmli sample buffer. Reaction products, resolved on 11% SDS-polyacrylamide gel, were analyzed by phosphorescence imaging using a Storm 860 instrument (GE Healthcare). To detect phosphorylation at Ser-53 of MCM2, kinase reactions were carried out omitting [γ-32P]ATP, and the products were analyzed by Western blotting with anti-phospho-Ser-53 MCM2 antibody.
Pull-down Assays
In a typical S-tag pull-down experiment 25 μl of S-protein-agarose (Novagen) was incubated with 10 μg of Cdc7-ASK, 10 μg of LEDGF, and 10 μg of bovine serum albumin in 650 μl of PDB (150 mm NaCl, 10 mm MgCl2, 0.1% Nonidet P-40, 2 mm DTT, 1 mm β-glycerophosphate, 1 mm NaF, and 40 mm Bistris propane-HCl, pH 7.4), supplemented with EDTA-free protease inhibitors. Input quantities of Cdc7-ASK and LEDGF deletion mutants were adjusted to match molar inputs of the full-length proteins. Where indicated, PDB was supplemented with 4 mm ATP. For GST pull-downs, 30 μl of glutathione-Sepharose (GE Healthcare), preloaded with GST-LEDGF-(347–471), GST-HRP2-(470–593), or GST (1 μg of protein/μl of agarose), was incubated with 10 μg of Cdc7-ASK and 10 μg of bovine serum albumin in 650 μl of PDB. Following 4-h incubation at 4 °C, the beads were washed in several changes of ice-cold PDB. Bound proteins were eluted in 30 μl of Laemmli sample buffer, separated in 11% SDS-PAGE gels, and detected by staining with Coomassie Blue R250 and/or Western blotting.
RESULTS
LEDGF Interacts with Cdc7-ASK
To identify cellular binding partners of LEDGF, we engineered a human cell line stably expressing its C-terminal fragment (residues 326–530) fused to a cTAP tag (Fig. 1A). The lack of the PWWP domain and AT-hooks, which collectively comprise the chromatin-binding module of LEDGF (11, 12, 58), allowed gentle extraction of the fusion protein without resorting to high salt treatments (data not shown). The bait protein was isolated from a whole cell extract using a TAP procedure, and associated binding partners were analyzed via in-gel trypsin digestion in conjunction with LC-MS/MS. This process identified eight peptides unique to Cdc7 kinase (supplemental Table S2), derived from a protein band migrating at ∼70 kDa. Importantly, no Cdc7-derived peptides were observed in a parallel purification starting with cells expressing the isolated TAP tag.
FIGURE 1.
Identification of the LEDGF-Cdc7-ASK interaction. A, schematics showing the domain organization of LEDGF and the cTAP-tagged LEDGF-(326–530) construct. Locations of the PWWP domain, NLS, AT-hooks, and IBD of LEDGF, calmodulin binding peptide (CBP), tobacco etch virus (TEV) protease site, and the IgG binding module from S. aureus protein A (protA) of the cTAP tag are indicated. B, co-IP experiments. HeLa cells were transiently transfected with HA-tagged mouse LEDGF (mLEDGF), human LEDGF, LEDGF-(326–530), HRP2, mouse p52 (mp52), or an empty vector. Whole cell extracts (WCE; lanes 1–6) or proteins pulled down with anti-HA affinity matrix from whole cell extracts (lanes 7–12) were tested by Western blotting using anti-HA, anti-Cdc7, and anti-β-actin antibodies. Migration positions of protein molecular mass standards (kDa), and the heavy chain of mouse IgG (IgG H) are indicated. C, IP of endogenous proteins. Extracts from untransfected 293T cells were incubated with rabbit anti-LEDGF antibody (lane 3) or control rabbit IgG (lane 4) and protein G-agarose, and the recovered proteins were analyzed by Western blotting with anti-Cdc7 and anti-ASK antibodies. Lanes 1 and 2 contained whole cell extract. To improve detection of ASK, the samples in lanes 2–4 were treated with λ-protein phosphatase (λPPase). The bands corresponding to ASK are indicated with asterisks.
To confirm the interaction, we conducted a series of IP experiments (Fig. 1, B and C). Endogenous Cdc7 was readily detected in immunoprecipitates of HA-tagged full-length mouse and human LEDGF proteins and HA-LEDGF-(326–530) (lanes 7–9). Cdc7 was not co-recovered with p52, an alternative splice form of LEDGF, which lacks the IBD (9, 20); nor was it detected in a mock IP (lanes 11 and 12). In addition, Cdc7 efficiently co-precipitated with HA-tagged HRP2 (lane 10), a close homologue of LEDGF that also contains an IBD (3). Furthermore, Cdc7 and its regulatory subunit ASK (26) specifically co-immunoprecipitated with LEDGF from extracts of untransfected 293T cells (Fig. 1C), confirming the interaction between endogenous Cdc7-ASK and LEDGF.
When overexpressed in HeLa cells, HcRed1-LEDGF displayed striking co-localization with HA-ASK, whereas nuclear distribution of EGFP-Cdc7 was less defined (Fig. 2A). Importantly, no co-localization was observed between HA-ASK and HcRed1-p52, although distribution of both HcRed1-LEDGF and -p52 proteins closely mirrored that of chromosomal DNA. These results indicated that LEDGF might be able to tether ASK or Cdc7-ASK complex to chromatin, in an IBD-dependent manner. To test this hypothesis, we isolated Triton-soluble and insoluble fractions of 293T cells transiently transfected with LEDGF and/or FLAG-Cdc7 and HA-ASK expression constructs. The extracts were prepared 30 h post-transfection, and analyses of propidium iodide-stained cells by flow cytometry failed to highlight significant differences between cell cycle profiles of the transfection conditions (data not shown). Overexpression of WT LEDGF resulted in a considerable enrichment of both FLAG-Cdc7 and HA-ASK in the chromatin-containing fraction (Fig. 2B, lane i2). In contrast, LEDGF mutant lacking residues 347–430 (LEDGFΔIBD), which does not interact with Cdc7-ASK (see below), failed to enrich the kinase subunits in the Triton-insoluble fraction (Fig. 2B, lane i4).
FIGURE 2.
Intracellular distribution of overexpressed LEDGF, ASK and Cdc7. A, confocal laser-scanning microscopy images of HeLa cells transfected with EGFP-Cdc7, full-length HA-ASK (top and middle rows of images), HA-ASK-(1–624) (bottom row), HcRed1-LEDGF (top and bottom rows), and/or HcRed1-p52 (middle row). B, ASK and Cdc7 are enriched in the chromatin-containing Triton-insoluble fraction when co-overexpressed with LEDGF. 293T cells were transfected with expression vectors for FLAG-Cdc7 and HA-ASK (lanes w1–w4, s1–s4, and i1–i4), WT LEDGF (lanes w2, s2, and i2), LEDGF K401E/K402E/R405E (EEE) (lanes w3, s3, and i3), LEDGFΔIBD (lanes w4, s4, and i4), or empty vector (lane w0). Whole cell extracts (lanes w0–w4) and Triton X-100-soluble (lanes s1–s4), and -insoluble (lanes i1–i4) fractions were analyzed by Western blotting using anti-HA, Cdc7, LEDGF, lamin B, and β-tubulin antibodies.
The LEDGF-Cdc7-ASK Interaction Is Mediated by the IBD of LEDGF and Requires the C Terminus of ASK
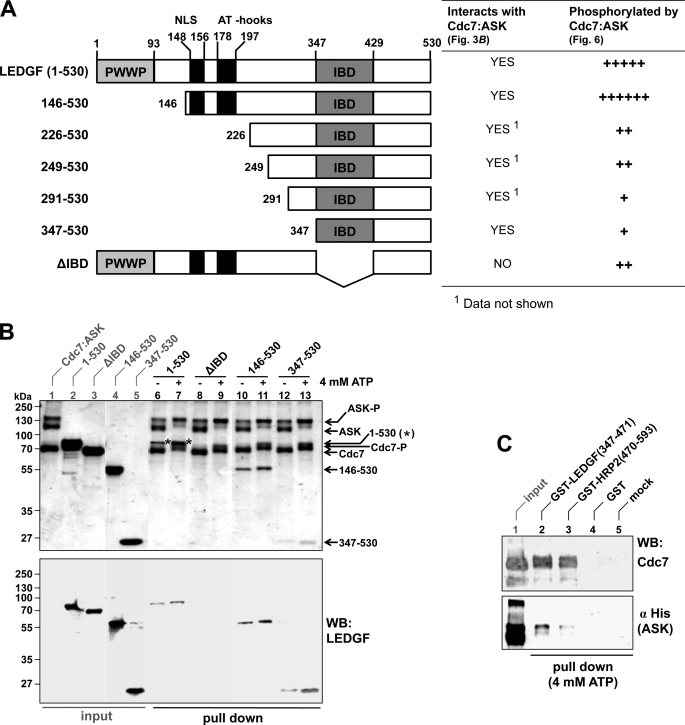
To characterize the interaction further, we performed in vitro pull-down experiments using recombinant proteins. In accordance with earlier observations (33, 59), it was necessary to co-express Cdc7 and ASK to obtain preparations of active kinase (data not shown). Therefore, the interaction of these proteins with LEDGF was studied in the context of the Cdc7-ASK heterodimer. Following SDS-PAGE, total protein was detected by Coomassie Blue, whereas individual proteins were monitored by Western blotting. The holoenzyme, produced with an S-tag fused to the C terminus of Cdc7, could be efficiently captured on S-protein-agarose resin, partially recovering full-length LEDGF from solution (Fig. 3B, lanes 6 and 7). Of note, the addition of ATP to pull-down reactions led to increased phosphorylation of Cdc7 and ASK subunits, manifesting as pronounced upward mobility shifts (discussed in more detailed below), and improved recovery of LEDGF (Fig. 3B, compare lanes 6 and 7). N-terminally truncated mutants LEDGF-(146–530) and LEDGF-(347–530) were also pulled down with Cdc7-ASK (Fig. 3B, lanes 10–13; see Fig. 3A for schematics of truncations). The interaction clearly depended on the IBD, because Cdc7-ASK failed to pull-down LEDGFΔIBD (Fig. 3B, lanes 8 and 9). Furthermore, GST fusions of isolated LEDGF and HRP2 IBDs, but not GST alone, recovered Cdc7-ASK in a reciprocal GST pull-down experiment (Fig. 3C). As expected, GST-LEDGF-(347–471) competed with full-length LEDGF for binding to Cdc7-ASK under conditions where the isolated GST protein failed to do so; increasing input of GST-LEDGF-(347–471) inhibited recovery of LEDGF, with a concomitant increase of the competitor protein in the pull-down fraction (supplemental Fig. S1).
FIGURE 3.
The interaction with Cdc7-ASK is mediated by the IBD of LEDGF. A, schematic of the LEDGF deletion mutants used (left) and their properties in terms of binding to and phosphorylation by Cdc7-ASK in vitro (right). B, S-tag pull-down experiments. S-tagged Cdc7-ASK was incubated with S-protein-agarose and full-length LEDGF-(1–530, lanes 6 and 7), LEDGFΔIBD (lanes 8 and 9), LEDGF-(146–530) (lanes 10 and 11), or LEDGF-(347–530) (lanes 12 and 13) in the absence (lanes 6, 8, 10, and 12) or presence (lanes 7, 9, 11, and 13) of 4 mm ATP. Proteins bound to the beads were separated by SDS-PAGE and visualized by staining with Coomassie Blue (top) and Western blotting (WB) with rabbit anti-LEDGF antibody recognizing the C terminus of LEDGF. Lanes 1–5 contained input quantities of Cdc7-ASK (lane 1), full-length LEDGF (lane 2), LEDGFΔIBD (lane 3), LEDGF-(146–530) (lane 4), or LEDGF-(347–530) (lane 5). Migration positions of molecular mass markers (kDa), LEDGF, LEDGF-(146–530), LEDGF-(347–530), ASK, Cdc7, and their hyperphosphorylated forms (ASK-P and Cdc7-P) are indicated. Stars indicate band of full-length LEDGF on the Coomassie-stained gel. C, the IBD is sufficient for the interaction with Cdc7-ASK. Cdc7-ASK was incubated with glutathione-Sepharose beads preloaded with GST-LEDGF-(347–471) (lane 2), GST-HRP2-(470–593) (lane 3), or GST (lane 4). Proteins bound to the resin were tested by Western blotting with anti-Cdc7 or anti-His5 antibodies. Lane 5 contains a mock pull-down in the absence of a GST protein. Lane 1 contained input quantity of Cdc7-ASK.
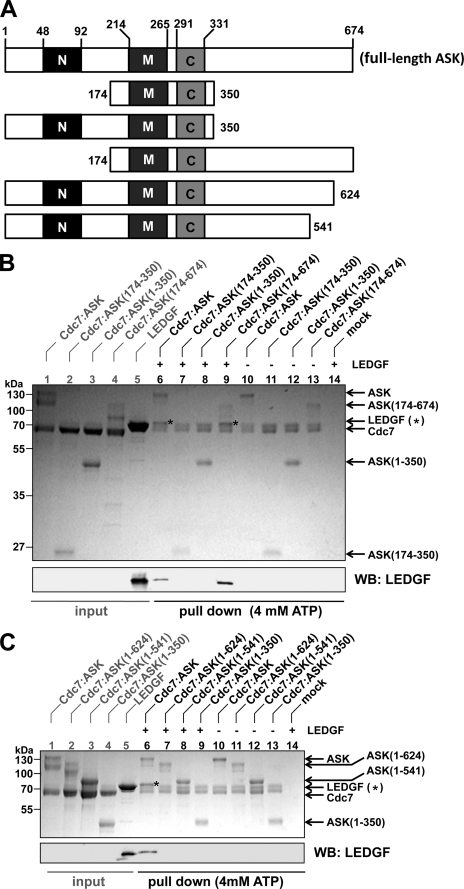
Residues 174–350 of ASK, spanning motifs M and C, are sufficient for the functional interaction with Cdc7 (47). Concordantly, co-expression of Cdc7 with the minimal fragment of ASK-(174–350) (see Fig. 4A for a schematic of ASK truncations) in bacteria allowed isolation of a stable, enzymatically competent complex. However, in contrast to the WT heterodimer, Cdc7-ASK-(174–350) failed to interact with LEDGF (Fig. 4B, compare lanes 6 and 7), indicating that the N- and/or C-terminal portions of ASK are required for the interaction. Pull-down analyses using heterodimers containing C- or N-terminally truncated ASK revealed that the N-terminal region of ASK was dispensable (Fig. 4B, lane 9). In contrast, deleting as few as 50 C-terminal residues of ASK was sufficient to ablate binding to LEDGF (Fig. 4C, compare lanes 6 and 7). In agreement with this result, HA-ASK-(1–624) did not co-localize with HcRed1-LEDGF when overexpressed in HeLa cells (Fig. 2A).
FIGURE 4.
Residues 625–674 of ASK are required for binding to LEDGF. A, schematic of ASK truncations. Locations of the N, M, and C motifs are indicated. B, C terminus of ASK is required for the interaction with LEDGF. S-tagged Cdc7-ASK or its indicated mutant forms were incubated with S-protein-agarose in the presence (lanes 6–9) or absence (lanes 10–13) of LEDGF. Lane 14 contains a mock pull-down of LEDGF with S-protein-agarose. Input quantities of proteins (lanes 1–5) or proteins captured on the beads (lanes 6–14) separated by SDS-PAGE were stained with Coomassie Blue (top) and analyzed by Western blotting using anti-LEDGF antibody (bottom). C, deletion of 50 residues from the C terminus of ASK is sufficient to ablate the interaction with LEDGF. Cdc7-ASK or its mutants were incubated with S-protein-agarose beads in the presence (lanes 6–9) or absence (lanes 10–13) of LEDGF. LEDGF was incubated with S-protein-agarose alone in lane 14. Lanes 1–5 contained input levels of the indicated proteins. Samples were analyzed as in B.
Phosphorylation State of Cdc7-ASK Affects Its Interaction with LEDGF
Recombinant Cdc7-ASK was consistently purified in a partially phosphorylated form, due to autophosphorylation during production in bacteria. Notably, ASK migrated as a diffuse double band in SDS-polyacrylamide gels (Fig. 5A, lane 1), indicating the presence of multiple phosphorylated forms. Incubation of purified Cdc7-ASK heterodimer with ATP resulted in further phosphorylation, observed as upward SDS-PAGE mobility shifts of Cdc7 and ASK bands (Fig. 5A, compare lanes 3 and 5). Conversely, dephosphorylation of the heterodimer with λ-protein phosphatase resulted in pronounced downward mobility shifts (Fig. 5A, compare lanes 3 and 7). Preincubation of the Cdc7-ASK heterodimer with ATP or the addition of ATP directly into the pull-down buffer consistently improved the recovery of LEDGF (Fig. 5A, compare lanes 4 and 6; Fig. 3B, lanes 6 and 7). In contrast, dephosphorylation of Cdc7-ASK abrogated the interaction (Fig. 5A, lane 8), indicating that the phosphorylation state of the kinase heterodimer is a critical factor for the interaction with LEDGF in vitro. Of note, removal of 133 C-terminal residues of ASK resulted in a loss of hyperphosphorylation (Fig. 4C, lanes 3, 8, and 12), indicating that the C-terminal region of ASK is targeted by the autophosphorylation activity.
FIGURE 5.
The LEDGF-Cdc7-ASK interaction requires autophosphorylation of the kinase and the positively charged patch on the surface of the IBD structure. A, the phosphorylation state of Cdc7-ASK affects its interaction with LEDGF. Cdc7-ASK, either untreated (lanes 3 and 4), preincubated with 4 mm ATP (lanes 5 and 6), or dephosphorylated with λ-protein phosphatase (lanes 7 and 8), was incubated with protein S-agarose in the presence (lanes 4, 6, and 8) or absence (lanes 3, 5, and 7) of LEDGF. Proteins captured on the beads were separated by SDS-PAGE and detected by staining with Coomassie Blue (top) and Western blotting (WB) with anti-LEDGF antibody (bottom). B, the positive patch on the surface of the IBD is important for the interaction with Cdc7-ASK. S-tagged Cdc7-ASK was incubated with S-protein agarose in the presence of WT or mutant LEDGF. Lanes 1–5, 12, and 13 show input levels of indicated proteins. LEDGF and Cdc7-ASK were omitted from the samples in lanes 17 and 18, respectively. The samples were analyzed as in A.
A Positive Patch on the Surface of the IBD Structure Is Involved in the Interaction with Cdc7-ASK
The structure of the IBD features a positively charged patch formed by side chains of several Lys and Arg residues, most of which are invariant among LEDGF and HRP2 orthologs (60). LEDGF Lys-401, Lys-402, and Arg-405, contributing to this patch, are known to mediate interactions with complementary acidic residues found within the N-terminal domains of lentiviral INs (17). Because the Cdc7-ASK-LEDGF interaction requires autophosphorylation of the kinase heterodimer, we asked whether it also depends on the positively charged patch. To this end, we tested a series of LEDGF mutants for binding to the Cdc7-ASK heterodimer. The K392E point mutant retained affinity for the kinase complex (Fig. 5B, lane 9). By contrast, K360E or triple mutation K401A/K402A/R405A (AAA), K401E/K402S/R405E (ESE), or K401E/K402E/R405E (EEE) ablated the interaction (Fig. 5B). Concordantly, overexpression of EEE LEDGF failed to concentrate Cdc7-ASK to the Triton-insoluble fraction, mimicking the behavior of the IBD deletion mutant (Fig. 2B, compare lane i2 with lanes i3 and i4). Therefore, the positively charged surface of the IBD is indeed involved in the interaction with Cdc7-ASK, and thus, its interface with LEDGF at least partly overlaps that of IN. This explains the inability of the ternary Cdc7-ASK-LEDGF complex to interact with HIV-1 IN (data not shown).
Identification of Cdc7-ASK Phosphorylation Sites in LEDGF
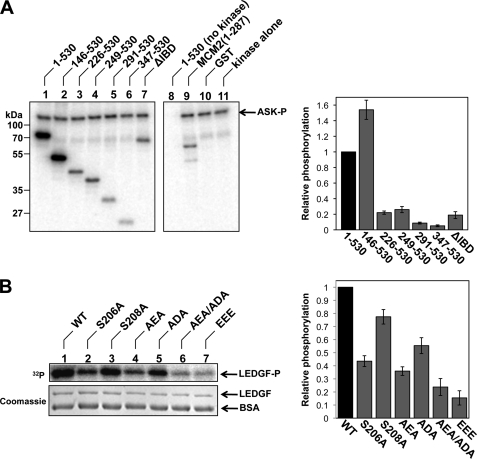
Although incubation of LEDGF with Cdc7-ASK did not alter its migration in SDS-polyacrylamide gels, using [γ-32P]ATP revealed robust phosphorylation of LEDGF by the kinase heterodimer (Fig. 6A, compare lanes 1, 8, and 11). Impressively, under conditions of the in vitro assay, LEDGF was phosphorylated more readily than our model Cdc7-ASK substrate GST-MCM2-(1–287) (Fig. 6A, lane 9). Removal of the IBD or introduction of the EEE mutation greatly reduced phosphorylation of LEDGF (Fig. 6, A and B), indicating that the reaction is dependent on the interaction with Cdc7-ASK.
FIGURE 6.
LEDGF is phosphorylated by Cdc7-ASK in vitro. A, full-length LEDGF (lane 1), its N-terminal truncation mutants (lanes 2–6), LEDGFΔIBD (lane 7), GST-MCM2-(1–287) (lane 9), or GST (lane 10) was incubated with Cdc7-ASK in the presence of [γ-32P]ATP. Reaction products, separated by SDS-PAGE, were detected by phosphorescence imaging. Full-length LEDGF was incubated with [γ-32P]ATP in the absence of Cdc7-ASK in lane 8 (mock); Cdc7-ASK was incubated with [γ-32P]ATP in the absence of protein substrates in lane 11. Concentrations of full-length LEDGF, LEDGF deletion mutants, and GST were adjusted to 0.3 μm; GST-MCM2-(1–287) was used at 0.4 μm. Quantification of radioactivity incorporation, relative to the full-length LEDGF, is shown to the right of the gel. B, LEDGF residue Ser-206 is the major Cdc7-ASK phosphorylation target in vitro. WT LEDGF, S206A, S208A, AEA, ADA, AEA/ADA, or EEE LEDGF mutants were incubated with Cdc7-ASK in the presence of [γ-32P]ATP. The gel was stained with Coomassie Blue, and the reaction products were detected and quantified as in A.
To map Cdc7-ASK phosphorylation sites in LEDGF, we incubated equimolar amounts of N-terminally truncated LEDGF mutants with the kinase and quantitatively assessed their phosphorylation (Fig. 6A). The N-terminal 145 residues of LEDGF were clearly dispensable for its function as a Cdc7-ASK substrate; in fact, LEDGF-(146–530) consistently performed better than the full-length protein. However, removal of 225 or more N-terminal residues considerably reduced incorporation of radioactive phosphate, indicating that the major phosphorylation site is located between LEDGF residues 146 and 225.
LC-MS/MS analysis of LEDGF, in vitro phosphorylated by Cdc7-ASK, identified four phosphopeptides (supplemental Table S3). To determine the dominant site(s), we designed two double LEDGF mutants eliminating Ser residues suggested by the phosphopeptide analyses: S206A/S208A (AEA) and S273A/S275A (ADA). The AEA mutation resulted in a substantial decrease in phosphorylation of LEDGF by Cdc7-ASK (Fig. 6B, lane 4). Combining AEA and ADA mutations (AEA/ADA; S206A/S208A/S273A/S275A) resulted in the poorest phosphorylation substrate of all Ser substitution mutants studied (Fig. 6B, lane 6), although the loss of phosphorylation achieved by mutating Ser-273 and Ser-275 was small. In the amino acid sequence of LEDGF, both Ser-206 and Ser-208 are followed by acidic residues (Glu-207 and Asp-209, respectively), consistent with the consensus for Cdc7-ASK phosphorylation sites (43). Nonetheless, mutation of Ser-206 alone was sufficient to reduce in vitro phosphorylation of LEDGF nearly to the levels of the AEA mutant, whereas targeting Ser-208 had only a modest effect (Fig. 6B, lanes 2 and 3). These results suggested that LEDGF Ser-206 constitutes the dominant Cdc7-ASK target in vitro, consistent with the results of the deletion mutagenesis experiments (Fig. 6A).
To test whether LEDGF can be phosphorylated at Ser-206 in vivo, we established a human cell line, H3, constitutively expressing HA-LEDGF. The protein isolated from extracts of asynchronous or S-phase H3 cells was subjected to LC-MS/MS phosphopeptide analysis. Remarkably, although the majority of the LEDGF phosphopeptides identified were common to both conditions, the phosphopeptide containing Ser-206 was identified in S-phase synchronized cells only (supplemental Table S3). In addition, although similar sets of LEDGF phosphorylation sites have been identified by global phosphoproteome studies of asynchronous or G1- and M-phase-arrested HeLa cells (supplemental Table S4), phosphorylation of Ser-206 was not observed (61, 62), suggesting that the modification of Ser-206 might be specific to S phase, which would coincide with the peak of Cdc7 activity.
LEDGF Stimulates Cdc7-ASK Kinase Activity in Vitro
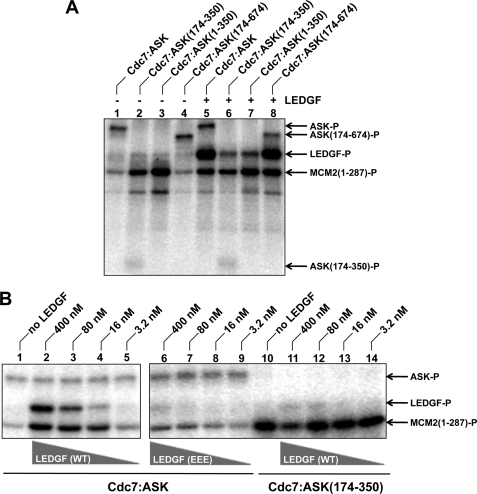
Initially, we tested WT Cdc7-ASK, along with heterodimers containing N- and/or C-terminally truncated forms of ASK, for the ability to phosphorylate GST-MCM2-(1–287) in vitro. This substrate was chosen based on published observations that human Cdc7-ASK phosphorylates the N-terminal region of MCM2 (43, 63). Both WT Cdc7-ASK and Cdc7-ASK-(174–674) generated detectable levels of phosphorylated GST-MCM2-(1–287) (Fig. 7A, lanes 1 and 4). Surprisingly, Cdc7-ASK-(174–350) and Cdc7-ASK-(1–350) mounted dramatically higher activities compared with the WT heterodimer in this assay format (lanes 2 and 3), implying that the C-terminal region of ASK negatively regulates the kinase activity. Because the same region of ASK is also involved in the interaction with LEDGF (Fig. 4B), we asked whether LEDGF can modulate the enzymatic activity of Cdc7-ASK. Strikingly, the addition of LEDGF led to a dramatic stimulation of WT Cdc7-ASK (Fig. 7A, compare lanes 1 and 5), in a concentration-dependent manner (Fig. 7B, lanes 2–5). In the presence of LEDGF, the abilities of WT Cdc7-ASK and Cdc7-ASK-(174–674) (Fig. 7A, lanes 5 and 8) to phosphorylate GST-MCM2-(1–287) were comparable with that of Cdc7-ASK-(174–350) or Cdc7-ASK-(1–350) (lanes 2 and 3). As expected, the hyperactive mutants, which both lack the C-terminal portion of ASK, were not stimulated by LEDGF (Fig. 7A, lanes 6 and 7). Reciprocally, the EEE LEDGF mutant, unable to interact with Cdc7-ASK, failed to significantly enhance the activity of WT Cdc7-ASK (Fig. 7B, lanes 6–9). These results confirm that the stimulation of Cdc7-ASK kinase activity by LEDGF strictly depends on the protein-protein interaction. Furthermore, because the AEA/ADA LEDGF mutant retained the ability to stimulate Cdc7-ASK (supplemental Fig. S2), the effect does not depend on phosphorylation of LEDGF by the kinase.
FIGURE 7.
LEDGF stimulates Cdc7-ASK kinase activity in vitro. A, WT Cdc7-ASK or its mutant forms (3 nm) were incubated with 0.4 μm GST-MCM2-(1–287) and [γ-32P]ATP in the absence (lanes 1–4) or presence (lanes 5–8) of 0.4 μm LEDGF. Reaction products, separated by SDS-PAGE, were detected by phosphorescence imaging. B, WT Cdc7-ASK (lanes 1–9) or Cdc7-ASK(174–350) (lanes 10–14) was incubated with GST-MCM2-(1–287) and [γ-32P]ATP in the absence (lanes 1 and 10) or presence of the indicated concentrations of WT (lanes 2–5 and 11–14) or EEE (lanes 6–9) LEDGF.
LEDGF Enhances Phosphorylation of MCM2 at the Cdc7-specific Site Ser-53
MCM2 residue Ser-53 was shown to be the major target of phosphorylation by Cdc7 both in vitro and in vivo (43, 44). To test whether modulation of Cdc7-ASK activity by LEDGF affects phosphorylation at the specific site, we used an antibody that detects MCM2 containing phospho-Ser-53. As expected, prior to incubation with the kinase, the phospho-specific antibody failed to react with bacterially produced GST-MCM2-(1–287) (Fig. 8A, lane 1), and the phosphorylated species were highlighted following incubation with Cdc7-ASK and ATP (lane 2). Corroborating results of the radioactive kinase assays, the addition of LEDGF resulted in a marked increase of the substrate phosphorylated at Ser-53 (lane 3). Similar high levels of Ser- 53 phosphorylation were observed with Cdc7-ASK-(174–350), but the activity of the hyperactive mutant, as predicted, was not enhanced by LEDGF (lanes 4 and 5). Both C-terminal truncation mutants Cdc7-ASK-(1–624) and Cdc7-ASK-(1–541) were more active than WT Cdc7-ASK and did not respond to the presence of LEDGF (Fig. 8A, lanes 6–9). Furthermore, GST fusions of LEDGF-(347–471) and HRP2-(470–593), but not GST alone, were able to promote the specific activity of Cdc7-ASK complex (lanes 10–12), indicating that the IBD is sufficient for this effect.
FIGURE 8.
LEDGF enhances Cdc7-ASK-dependent phosphorylation of MCM2 residue Ser-53. A, GST-MCM2-(1–287) was incubated in the absence (lane 1) or presence (lanes 2–12) of 3 nm WT or the indicated mutant forms of Cdc7-ASK; 0.4 μm LEDGF, GST-LEDGF-(347–471), GST-HRP2-(470–593), or GST was added as indicated; ATP (4 mm) was present in all reactions. Reaction products, resolved by SDS-PAGE, were detected by Western blotting (WB) with a phosphospecific anti-phospho-Ser-53 MCM2 antibody (pSer53). B, full-length MCM2, purified from HeLa cells stably expressing FLAG-tagged MCM2 using anti-FLAG affinity agarose, compared with mock-purified material from parental HeLa cells. C, phosphorylation of full-length MCM2 by Cdc7-ASK in vitro. FLAG-MCM2 was incubated in the absence (lane 3) or presence of 3 nm Cdc7-ASK (lanes 4 and 5) or Cdc7-ASK-(174–350) (lane 6); 0.3 μm LEDGF was added to the reaction in lane 5. Lane 2 contains input quantity of untreated FLAG-MCM2, and lane 1 contains an equivalent amount of the mock-purified material. Reaction products were analyzed by Western blotting with anti-phospho-Ser-53 MCM2 and anti-FLAG antibodies.
To ascertain that our observations can be extended to phosphorylation of the full-length substrate, we generated HeLa cells stably expressing a FLAG-tagged form of human MCM2 and isolated the protein using FLAG affinity resin (Fig. 8B). In parallel, a mock purification was carried out starting with an extract from parental cells, and the resulting sample was used as a negative control in the following experiments. As expected, MCM2 preparations from human cells contained detectable levels of phospho-Ser-53, as highlighted by Western blotting using the phospho-specific antibody (Fig. 8C, lanes 2 and 3). Recombinant Cdc7-ASK was able to weakly phosphorylate full-length MCM2 at Ser-53 (lane 4), and the activity was robustly enhanced in the presence of LEDGF (lane 5). Moreover, similarly high levels of Ser-53 phosphorylation were achieved by hyperactive Cdc7-ASK-(174–350) (lane 6).
DISCUSSION
Its well characterized role in HIV-1 replication notwithstanding, cellular functions of LEDGF remain poorly understood. In this report, we describe the discovery and the initial characterization of the interaction between LEDGF and the S-phase kinase Cdc7-ASK. We showed that the IBD of LEDGF is essential and minimally sufficient for the interaction with Cdc7-ASK. Reciprocally, the C-terminal 50 residues of ASK are required for binding to LEDGF, and the interaction is dependent on the phosphorylation status of the kinase heterodimer. In mammalian cells, ASK protein levels oscillate during the cell cycle, reaching their maximum in S phase (27). Furthermore, phosphorylation of ASK depends on binding to the catalytic subunit Cdc7 (27). Thus, the interaction of LEDGF with the S-phase kinase is probably limited to this stage of the cell cycle. Concordantly, co-IP of Cdc7 with HA-LEDGF was most efficient from cells synchronized in mid-S-phase (data not shown). Interestingly, the interaction with Cdc7-ASK extends to another IBD-containing protein, HRP2, a close homolog of LEDGF (Fig. 1B). Similar to the case of LEDGF, the IBD of HRP2 was sufficient for the interaction with and stimulation of Cdc7-ASK (Figs. 3C and 8A). Notably, the IBD was also shown to mediate the interaction of LEDGF with cellular proteins JPO2, PogZ, and menin as well as lentiviral INs (3, 21, 24, 55, 64). Intriguingly, akin to its effect on HIV-1 IN, LEDGF is required for the association of JPO2 and menin-mixed lineage leukemia histone methyltransferase with chromatin (9, 10, 21, 24). Herein, we showed that co-overexpression of LEDGF increased the presence of Cdc7 and ASK on chromatin in human cells (Fig. 2B). Thus, LEDGF appears to act as a multifunctional adaptor with the ability to tether a plethora of cellular machinery involved in genome expression and maintenance to chromatin.
Cdc7-ASK kinase can phosphorylate LEDGF at a number of Ser residues in vitro, with Ser-206 being the main target site (supplemental Table S3 and Fig. 6). The phosphorylation of LEDGF by Cdc7-ASK depends on the protein-protein interaction. Indeed, phosphorylation of EEE and ΔIBD LEDGF mutants, unable to bind Cdc7-ASK, was very inefficient (Fig. 6). The phosphorylation site is located within a largely unstructured interdomain region of LEDGF (3) and is separated from the IBD by ∼140 amino acid residues. Therefore, binding to and phosphorylation of LEDGF by the kinase are spatially uncoupled, with the IBD acting as a docking domain. A similar mechanism has been proposed for phosphorylation of MCM4 in S. cerevisiae, where the Dbf4-dependent kinase-docking domain (MCM4 residues 175–333) serves to recruit the S-phase kinase, which then phosphorylates its N-terminal region (65). In vivo, we could only detect phosphorylation of LEDGF Ser-206 when the population of S-phase cells was enriched by pretreatment with aphidicolin (supplemental Table S3). Similarly, this modification was not discovered by earlier studies that analyzed phosphoproteomes of unsynchronized G1- or M-phase cells nonetheless collectively detecting all remaining LEDGF phosphorylation sites that we report here (supplemental Table S4) (61, 62). Thus, phosphorylation of LEDGF at Ser-206 appears to be restricted to S phase, coinciding with the maximum activity of Cdc7-ASK (27). Future work will focus on the functional consequences of LEDGF phosphorylation by Cdc7-ASK. Because Ser-206 is in close proximity to the second AT-hook motif of LEDGF (residues 191–197) (12), phosphorylation at this position could modulate its DNA binding properties.
LEDGF potently stimulated the enzymatic activity of Cdc7-ASK in vitro, enhancing phosphorylation of bacterially produced GST-MCM2-(1–287) and full-length MCM2 purified from a human cell line alike. Importantly, under the conditions of our in vitro kinase assays, LEDGF bolstered phosphorylation of MCM2 at Ser-53 (Fig. 8), a natural Cdc7 target site in this protein (43, 44). The stimulation strictly depended on the protein-protein interaction, because the LEDGF mutant EEE, unable to bind Cdc7-ASK (Fig. 5B), also failed to promote its kinase activity (Fig. 7B). Moreover, deleting residues 625–674 at the C terminus of ASK ablated the interaction with LEDGF (Figs. 2A and 4C), at the same time rendering the kinase hyperactive (Fig. 8A). These results indicate that the interaction with LEDGF relieves autoinhibition of the Cdc7-ASK complex, imposed by the C terminus of ASK (Fig. 9). Of note, similar regulatory mechanisms were described for Cds1 and Chk1, both effector kinases in the DNA replication checkpoint pathway (66–68).
FIGURE 9.
The interaction between LEDGF IBD and the C terminus of ASK relieves autoinhibition of Cdc7-ASK kinase activity. Schematic of the Cdc7-ASK kinase activity in the absence (A) or presence (B) of LEDGF. Cdc7 is shown as a gray rectangle. ASK domains are shown as black circles; the smaller circle is the C-terminal regulatory peptide (RP). Ovals represent LEDGF PWWP and IBD domains; the MCM2–7 protein complex is shown as hexagons (P, phosphorylation). Interaction with LEDGF results in full activation of Cdc7-ASK kinase activity.
Overall, there is surprisingly little sequence conservation between known ASK orthologs (45). In particular, the extended C-terminal tail following Dbf4 motif C is characteristic of the metazoans, and the same species are known to possess LEDGF and/or HRP2 (3). The lack of sequence conservation among Dbf4/ASK orthologs suggests that the mechanisms regulating Cdc7 activity might significantly differ between species. The C-terminal tail was not required for short term survival of mouse embryonic stem cells (69). However, it should be noted that, in addition to ASK, embryonic stem cells probably express its close homolog Drf1 (70–72), the presence of which could have attenuated phenotypes of some mutations in ASK. Similarly, although LEDGF is not essential in vivo (13, 14, 73), its functions might be redundant with those of HRP2. Indeed, we showed here that both proteins can interact with Cdc7-ASK (Figs. 1B and 3C) and stimulate its kinase activity (Fig. 8A).
Swi6/HP1, required for early replication of the heterochromatic loci in fission yeast, was shown to activate a subset of replication origins within heterochromatin via a direct interaction with Dfp1 (74). These results set the first precedent for a chromatin-associated protein regulating initiation of DNA replication via an interaction with the S-phase kinase, which might be a paradigm of a more general mechanism. We tentatively speculate that recruitment and activation of Cdc7-ASK kinase by LEDGF could be a way to control origin firing in higher eukaryotes. Indeed, Hsk1-Dfp1, the S. pombe equivalent of Cdc7-ASK, was shown to regulate origin efficiency in a concentration-dependent manner (75). Although the distribution of LEDGF along human chromatin has not been reported, it is well established that a direct interaction with IN channels HIV-1 integration into active transcription units (14–16). Similarly, recruitment and stimulation of Cdc7-ASK by LEDGF may increase firing efficiency of local origins, thereby encouraging actively transcribed loci to replicate early in S phase (76–78).
Supplementary Material
Acknowledgments
We are grateful to Dr. Martin Spitaler (Facility for Imaging by Light Microscopy, Imperial College London) for help with laser-scanning microscopy; Dr. Goedele Maertens (Cancer Research UK) for the generous gift of pGM-Mel18cTAP and pQFLAG DNA and 293T-cTAP cells, helpful discussions, and critical reading of the manuscript; Dr. Massimo Pizzato for pcDNA3.1-FLAG; and Dr. Stephen Hare for purification of EEE, AAA, ESE, K360E, and K392E LEDGF mutants.
This work was supported, in whole or in part, by National Institutes of Health Grant AI039394 (to A. E.). This work was also supported by United Kingdom Medical Research Council Grant G0600009 (to P. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1 and S2.
M. Pizzato, unpublished results.
G. Maertens, unpublished results.
- LEDGF
- lens epithelium-derived growth factor
- ASK
- activator of S-phase kinase
- CDS
- coding sequence
- DTT
- dithiothreitol
- PIPES
- piperazine-N,N′-bis(2-ethanesulfonic acid)
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- HIV
- human immunodeficiency virus
- IBD
- integrase-binding domain
- IN
- integrase
- IP
- immunoprecipitation
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MCM
- minichromosome maintenance
- NLS
- nuclear localization signal
- PBS
- phosphate-buffered saline
- PMSF
- phenylmethylsulfonyl fluoride
- TAP
- tandem affinity purification
- WT
- wild type
- Bistris propane
- 1,3- bis[tris(hydroxymethyl)methylamino]propane
- AEA
- S206A/S208A
- ADA
- S273A/S275A
- EEE
- K401E/K402E/R405E
- AAA
- K401A/K402A/R405A
- ESE
- K401E/K402S/R405E.
REFERENCES
- 1.Stec I., Nagl S. B., van Ommen G. J., den Dunnen J. T. (2000) FEBS Lett. 473, 1–5 [DOI] [PubMed] [Google Scholar]
- 2.Izumoto Y., Kuroda T., Harada H., Kishimoto T., Nakamura H. (1997) Biochem. Biophys. Res. Commun. 238, 26–32 [DOI] [PubMed] [Google Scholar]
- 3.Cherepanov P., Devroe E., Silver P. A., Engelman A. (2004) J. Biol. Chem. 279, 48883–48892 [DOI] [PubMed] [Google Scholar]
- 4.Daniels T., Zhang J., Gutierrez I., Elliot M. L., Yamada B., Heeb M. J., Sheets S. M., Wu X., Casiano C. A. (2005) Prostate 62, 14–26 [DOI] [PubMed] [Google Scholar]
- 5.Engelman A., Cherepanov P. (2008) PLoS Pathog. 4, e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganapathy V., Daniels T., Casiano C. A. (2003) Autoimmun. Rev. 2, 290–297 [DOI] [PubMed] [Google Scholar]
- 7.Hussey D. J., Moore S., Nicola M., Dobrovic A. (2001) BMC Genet. 2, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 9.Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) J. Biol. Chem. 278, 33528–33539 [DOI] [PubMed] [Google Scholar]
- 10.Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E. M. (2004) J. Virol. 78, 9524–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llano M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E. M. (2006) J. Mol. Biol. 360, 760–773 [DOI] [PubMed] [Google Scholar]
- 12.Turlure F., Maertens G., Rahman S., Cherepanov P., Engelman A. (2006) Nucleic Acids Res. 34, 1653–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 14.Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J. R., Bushman F. (2005) Nat. Med. 11, 1287–1289 [DOI] [PubMed] [Google Scholar]
- 16.Marshall H. M., Ronen K., Berry C., Llano M., Sutherland H., Saenz D., Bickmore W., Poeschla E., Bushman F. D. (2007) PLoS ONE 2, e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hare S., Shun M. C., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2009) PLoS Pathog. 5, e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanegas M., Llano M., Delgado S., Thompson D., Peretz M., Poeschla E. (2005) J. Cell Sci. 118, 1733–1743 [DOI] [PubMed] [Google Scholar]
- 19.Cherepanov P., Ambrosio A. L., Rahman S., Ellenberger T., Engelman A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge H., Si Y., Roeder R. G. (1998) EMBO J. 17, 6723–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama A., Cleary M. L. (2008) Cancer Cell 14, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartholomeeusen K., De Rijck J., Busschots K., Desender L., Gijsbers R., Emiliani S., Benarous R., Debyser Z., Christ F. (2007) J. Mol. Biol. 372, 407–421 [DOI] [PubMed] [Google Scholar]
- 23.Bartholomeeusen K., Christ F., Hendrix J., Rain J. C., Emiliani S., Benarous R., Debyser Z., Gijsbers R., De Rijck J. (2009) J. Biol. Chem. 284, 11467–11477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maertens G. N., Cherepanov P., Engelman A. (2006) J. Cell Sci. 119, 2563–2571 [DOI] [PubMed] [Google Scholar]
- 25.Shinohara T., Singh D. P., Fatma N. (2002) Prog. Retin. Eye Res. 21, 341–358 [DOI] [PubMed] [Google Scholar]
- 26.Masai H., Arai K. (2002) J. Cell. Physiol. 190, 287–296 [DOI] [PubMed] [Google Scholar]
- 27.Kumagai H., Sato N., Yamada M., Mahony D., Seghezzi W., Lees E., Arai K., Masai H. (1999) Mol. Cell. Biol. 19, 5083–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W., McDonald D., Hope T. J., Hunter T. (1999) EMBO J. 18, 5703–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon H. J., Loo S., Campbell J. L. (1993) Mol. Biol. Cell 4, 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furukohri A., Sato N., Masai H., Arai K., Sugino A., Waga S. (2003) J. Biochem. 134, 447–457 [DOI] [PubMed] [Google Scholar]
- 31.Brown G. W., Kelly T. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8443–8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda T., Ogino K., Matsui E., Cho M. K., Kumagai H., Miyake T., Arai K., Masai H. (1999) Mol. Cell. Biol. 19, 5535–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masai H., Matsui E., You Z., Ishimi Y., Tamai K., Arai K. (2000) J. Biol. Chem. 275, 29042–29052 [DOI] [PubMed] [Google Scholar]
- 34.Weinreich M., Stillman B. (1999) EMBO J. 18, 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira M. F., Santocanale C., Drury L. S., Diffley J. F. (2000) Mol. Cell. Biol. 20, 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshiro G., Owens J. C., Shellman Y., Sclafani R. A., Li J. J. (1999) Mol. Cell. Biol. 19, 4888–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato N., Arai K., Masai H. (1997) EMBO J. 16, 4340–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J. K., Hurwitz J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J. K., Hurwitz J. (2000) J. Biol. Chem. 275, 18871–18878 [DOI] [PubMed] [Google Scholar]
- 40.Labib K., Tercero J. A., Diffley J. F. (2000) Science 288, 1643–1647 [DOI] [PubMed] [Google Scholar]
- 41.Ishimi Y. (1997) J. Biol. Chem. 272, 24508–24513 [DOI] [PubMed] [Google Scholar]
- 42.Aparicio O. M., Weinstein D. M., Bell S. P. (1997) Cell 91, 59–69 [DOI] [PubMed] [Google Scholar]
- 43.Cho W. H., Lee Y. J., Kong S. I., Hurwitz J., Lee J. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11521–11526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. (2006) J. Biol. Chem. 281, 10281–10290 [DOI] [PubMed] [Google Scholar]
- 45.Masai H., Arai K. (2000) Biochem. Biophys. Res. Commun. 275, 228–232 [DOI] [PubMed] [Google Scholar]
- 46.Ogino K., Takeda T., Matsui E., Iiyama H., Taniyama C., Arai K., Masai H. (2001) J. Biol. Chem. 276, 31376–31387 [DOI] [PubMed] [Google Scholar]
- 47.Sato N., Sato M., Nakayama M., Saitoh R., Arai K., Masai H. (2003) Genes Cells 8, 451–463 [DOI] [PubMed] [Google Scholar]
- 48.Dowell S. J., Romanowski P., Diffley J. F. (1994) Science 265, 1243–1246 [DOI] [PubMed] [Google Scholar]
- 49.Jares P., Luciani M. G., Blow J. J. (2004) BMC Mol. Biol. 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards M. C., Tutter A. V., Cvetic C., Gilbert C. H., Prokhorova T. A., Walter J. C. (2002) J. Biol. Chem. 277, 33049–33057 [DOI] [PubMed] [Google Scholar]
- 51.Jares P., Blow J. J. (2000) Genes Dev. 14, 1528–1540 [PMC free article] [PubMed] [Google Scholar]
- 52.Elderkin S., Maertens G. N., Endoh M., Mallery D. L., Morrice N., Koseki H., Peters G., Brockdorff N., Hiom K. (2007) Mol. Cell 28, 107–120 [DOI] [PubMed] [Google Scholar]
- 53.Borger D. R., DeCaprio J. A. (2006) J. Virol. 80, 4292–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandegraaff N., Devroe E., Turlure F., Silver P. A., Engelman A. (2006) Virology 346, 415–426 [DOI] [PubMed] [Google Scholar]
- 55.Cherepanov P. (2007) Nucleic Acids Res. 35, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulm J. W., Perron M., Sodroski J. C., Mulligan R. C. (2007) Virology 363, 245–255 [DOI] [PubMed] [Google Scholar]
- 57.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 58.Shun M. C., Botbol Y., Li X., Di Nunzio F., Daigle J. E., Yan N., Lieberman J., Lavigne M., Engelman A. (2008) J. Virol. 82, 11555–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kihara M., Nakai W., Asano S., Suzuki A., Kitada K., Kawasaki Y., Johnston L. H., Sugino A. (2000) J. Biol. Chem. 275, 35051–35062 [DOI] [PubMed] [Google Scholar]
- 60.Cherepanov P., Sun Z. Y., Rahman S., Maertens G., Wagner G., Engelman A. (2005) Nat. Struct. Mol. Biol. 12, 526–532 [DOI] [PubMed] [Google Scholar]
- 61.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 62.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishimi Y., Komamura-Kohno Y., Arai K., Masai H. (2001) J. Biol. Chem. 276, 42744–42752 [DOI] [PubMed] [Google Scholar]
- 64.Busschots K., Voet A., De Maeyer M., Rain J. C., Emiliani S., Benarous R., Desender L., Debyser Z., Christ F. (2007) J. Mol. Biol. 365, 1480–1492 [DOI] [PubMed] [Google Scholar]
- 65.Sheu Y. J., Stillman B. (2006) Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Y. J., Kelly T. J. (2009) J. Biol. Chem. 284, 16016–16027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen P., Luo C., Deng Y., Ryan K., Register J., Margosiak S., Tempczyk-Russell A., Nguyen B., Myers P., Lundgren K., Kan C. C., O'Connor P. M. (2000) Cell 100, 681–692 [DOI] [PubMed] [Google Scholar]
- 68.Katsuragi Y., Sagata N. (2004) Mol. Biol. Cell 15, 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamashita N., Kim J. M., Koiwai O., Arai K., Masai H. (2005) Genes Cells 10, 551–563 [DOI] [PubMed] [Google Scholar]
- 70.Montagnoli A., Bosotti R., Villa F., Rialland M., Brotherton D., Mercurio C., Berthelsen J., Santocanale C. (2002) EMBO J. 21, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi T. S., Walter J. C. (2005) Genes Dev. 19, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshizawa-Sugata N., Ishii A., Taniyama C., Matsui E., Arai K., Masai H. (2005) J. Biol. Chem. 280, 13062–13070 [DOI] [PubMed] [Google Scholar]
- 73.Sutherland H. G., Newton K., Brownstein D. G., Holmes M. C., Kress C., Semple C. A., Bickmore W. A. (2006) Mol. Cell. Biol. 26, 7201–7210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayashi M. T., Takahashi T. S., Nakagawa T., Nakayama J., Masukata H. (2009) Nat. Cell Biol. 11, 357–362 [DOI] [PubMed] [Google Scholar]
- 75.Patel P. K., Kommajosyula N., Rosebrock A., Bensimon A., Leatherwood J., Bechhoefer J., Rhind N. (2008) Mol. Biol. Cell 19, 5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.White E. J., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E. J., Weissman S., Gerstein M., Groudine M., Snyder M., Schübeler D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17771–17776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woodfine K., Fiegler H., Beare D. M., Collins J. E., McCann O. T., Young B. D., Debernardi S., Mott R., Dunham I., Carter N. P. (2004) Hum. Mol. Genet. 13, 191–202 [DOI] [PubMed] [Google Scholar]
- 78.Farkash-Amar S., Lipson D., Polten A., Goren A., Helmstetter C., Yakhini Z., Simon I. (2008) Genome Res. 18, 1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.