Abstract
Atherosclerosis is a multifactorial vascular disease characterized by formation of inflammatory lesions. Elevated circulating acute phase proteins indicate disease risk. Serum amyloid A (SAA) is one such marker but its function remains unclear. To determine the role of SAA on aortic smooth muscle cell gene expression, a preliminary screen of a number of genes was performed and a strong up-regulation of expression of secretory phospholipase A2, group IIA (sPLA2) was identified. The SAA-induced increase in sPLA2 was validated by real time PCR, Western blot analysis, and enzyme activity assays. Demonstrating that SAA increased expression of sPLA2 heteronuclear RNA and that inhibiting transcription eliminated the effect of SAA on sPLA2 mRNA suggested that the increase was transcriptional. Transient transfections and electrophoretic mobility shift assays identified CAAT enhancer-binding protein (C/EBP) and nuclear factor κB (NFκB) as key regulatory sites mediating the induction of sPLA2. Moreover, SAA activated the inhibitor of NF-κB kinase (IKK) in cultured smooth muscle cells. Previous reports showed that interleukin (IL)-1β up-regulates Pla2g2a gene transcription via C/EBPβ and NFκB. Interestingly, SAA activated smooth muscle cell IL-1β mRNA expression, however, blocking IL-1 receptors had no effect on SAA-mediated activation of sPLA2 expression. Thus, the observed changes in sPLA2 expression were not secondary to SAA-induced IL-1 receptor activation. The association of SAA with high density lipoprotein abrogated the SAA-induced increase in sPLA2 expression. These data suggest that during atherogenesis, SAA can amplify the involvement of smooth muscle cells in vascular inflammation and that this can lead to deposition of sPLA2 and subsequent local changes in lipid homeostasis.
Introduction
Elevated circulating acute phase proteins correlate with an increased risk for atherosclerosis (1–4). One such disease indicator is serum amyloid A (SAA)2 (5). The SAA protein family consists of 12–14-kDa constitutive (SAA4) and acute phase (SAA1, SAA2, and SAA3) isoforms. During inflammation, there are large changes in liver-derived plasma levels of the acute phase isoforms, SAA1 and SAA2 (6, 7). The other acute phase isoform, SAA3, is extrahepatically inducible (8), and although the locus equivalent to SAA3 was previously believed to be a pseudogene in humans, Larson and co-workers (9) demonstrated its expression by mammary gland epithelial cells. Proinflammatory stimuli induce SAA expression in liver; optimal expression is achieved with a combination of interleukin (IL)-1 and IL-6 (10, 11). Extrahepatic synthesis of SAA by synovial fibroblasts, macrophages, adipocytes, and smooth muscle cells has been documented (12–15). SAA is also expressed in atherosclerotic lesions (13, 16, 17). Although several roles have been suggested, the functions of SAA remain uncertain (7). Our laboratory demonstrated that in response to IL-1α, cultured aortic smooth muscle cells synthesize SAA (18) leading to the hypothesis that during atherogenesis, locally synthesized SAA acts in an autocrine fashion to influence smooth muscle cell function. In this regard, to determine the role of SAA on aortic smooth muscle cell gene expression, a preliminary screen of a number of genes was performed on RNA extracted from acute phase SAA-treated aortic smooth muscle cell cultures. The data show that the mRNA levels of another acute phase protein, secretory phospholipase A2, group IIA (sPLA2) increased in smooth muscle cells treated with SAA. The SAA-induced expression of sPLA2 mRNA was validated and the mechanism whereby SAA activates the sPLA2 gene was explored. These data suggest that expression of SAA in the vasculature during the progression of atherosclerosis can amplify the involvement of smooth muscle cells in that an autocrine response to SAA may induce expression of additional acute phase proteins.
EXPERIMENTAL PROCEDURES
Isolation, Culture, and Treatment of Neonatal Rat Aortic Smooth Muscle Cells
Smooth muscle cells were isolated by enzymatic digestion of aortas from 3-day-old Sprague-Dawley rats (Charles River Breeding, Wilmington, MA) as previously described (19). Primary cells were maintained in Dulbecco's modified Eagle's medium supplemented with 100 units/ml of penicillin, 100 μg/ml of streptomycin, 0.1 mm MEM non-essential amino acids, and 1 mm MEM sodium pyruvate solution (DMEM) (all from Cellgro, Manassas, VA) containing 20% fetal bovine serum (Sigma). Trypsinized cells were seeded at a density of 2 × 104 cells/cm2 in DMEM containing 10% fetal bovine serum. Experiments were performed on cells up to the third passage.
Cells were treated with recombinant SAA (Peprotech, Rocky Hill, NJ), which corresponds to human acute phase SAA (20). There is some variation in potency of the various lots of SAA, but as stated in the figure legends, all experiments were performed with 2–4 μm SAA, doses that consistently induced an effect. Lipid-deficient serum (LDS) was prepared as previously described (21). Additional reagents included actinomycin D (ActD) (Sigma), recombinant IL-1β (eBioscience, San Diego, CA), IL-1 receptor antagonist (IL-1Ra) (R&D Systems, Minneapolis, MN), lipopolysaccharide (LPS) (Sigma, E. coli 0111:B4), and Ro 23-9358 (Sigma). To study the effect of lipid-associated SAA, high density lipoprotein (HDL) (Calbiochem, La Jolla, CA)-associated SAA was prepared in medium containing LDS (DMEM-LDS) by adding various concentrations of HDL to a fixed concentration of SAA before incubating at room temperature for 15 min while shaking, followed by 15 min at 37 °C. Prior to treatment, media were removed and the cells were washed twice. DMEM-LDS was added followed by the addition of SAA, IL-1β, IL-1Ra, LPS, HDL, or SAA-associated HDL alone or in combination as described in the figure legends. In cultures treated for more than 3 days, the medium was aspirated and the cells were re-fed with fresh DMEM-LDS plus reagents twice weekly, including the day before harvest in each case. ActD was added 1 h prior to the addition of SAA.
Western Blot Analysis
Extracts were prepared to generate total cellular protein or nuclear and cytoplasmic protein. Total cell lysis was carried out in RIPA buffer (50 mm Tris (pH 8.0), 150 mm NaCl, 0.5% deoxycholate sodium salt, 0.1% SDS, 1% Nonidet P-40, 1 mm EDTA) supplemented with enzyme inhibitors (10 μg/ml of leupeptin, aprotinin, pepstatin, 1 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4, 1 mm dithiothreitol). For differential extraction, the cytoplasmic fraction was isolated by incubation in lysis buffer (20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 2 mm EDTA, 0.5 mm dithiothreitol, 0.1% Nonidet P-40) containing enzyme inhibitors (10 μg/ml leupeptin, aprotinin, pepstatin, 0.25 mm phenylmethylsulfonyl fluoride, 2 mm Na3VO4). Lysis of nuclear pellets was carried out by incubation in nuclear lysis buffer (20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 2 mm EDTA, 0.5 mm dithiothreitol, 0.1% Nonidet P-40, 350 mm NaCl, 25% glycerol). The quantity of protein in extracts was determined by the BCA assay (Pierce) as per the manufacturer's instructions. Protein extracts were resolved by 12–15% SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Western blots were stained with Ponceau S and protein loading was equal in all lanes. Membranes were then washed in Tris-buffered saline with 0.1% Tween 20 (TBST) before blocking for 1 h in TBST with 5% nonfat dry milk (Nestle, Vevey, Switzerland) or 5% bovine serum albumin, the latter for detection of phosphorylated and total inhibitor of NF-κB kinase (IKK). Blocked membranes were incubated with anti-sPLA2 (1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA), anti-CEBPβ (1:2,000, Santa Cruz Biotechnology), anti-p65 (1:1,000, Santa Cruz Biotechnology), anti-phospho-IKKα/β (1:750, Cell Signaling Technology, Inc., Danvers, MA), or anti-IKKβ (1:1,000, Cell Signaling) overnight at 4 °C with rocking. After 4 washes with TBST, membranes were incubated with the horseradish peroxidase-conjugated secondary antibody (1:2,000–1:10,000, Santa Cruz Biotechnology) specific for the species from which the primary antibody was derived. Finally, horseradish peroxidase was detected on immunostained blots by chemiluminescent detection (Visualizer EC, Millipore) as per the manufacturer's instructions.
Transfection and Luciferase Assays
Rat Pla2g2a promoter constructs were generated from a 534-bp (-488 to +46) fragment of the rat Pla2g2a promoter as previously described (22). This construct is referred to as 488. Constructs called 156 and 42 contain shortened promoter fragments that were generated by deleting nucleotides −488 to −157 and −488 to −43, respectively. In addition, site-directed mutant promoter constructs include those with mutations in the two C/EBP binding sites (C/EBPΔ1 and C/EBPΔ2) and one NFκB binding site (NFκBΔ) previously identified within the 488 promoter fragment (23). A luciferase reporter construct (C-Luc) in which luciferase expression is driven by 4 repeats of C/EBP binding site 1 was used (24). Last, a luciferase reporter construct (N-Luc) in which luciferase expression is driven by 6 repeats of an NFκB binding site was kindly provided by Dr. Gail Sonenshein, Boston University School of Medicine, Boston, MA. To normalize for transfection efficiency, all cultures were co-transfected with a pRL-CMV-Renilla luciferase construct (Promega). For all transfections, cells were cultured in 24-well dishes until they were 70% confluent. Using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions, cultures were co-transfected with 1 μg of plasmid construct and 40 ng of pRL-CMV-Renilla per well. After an overnight incubation, cells were cultured under experimental conditions for 24 h. Cells were then washed twice with cold phosphate-buffered saline, harvested, and analyzed according to the manufacturer's instructions using the Dual Luciferase Reporter Assay System (Promega). Data are expressed as luciferase/Renilla ± S.D.
Real Time PCR
Total RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) and RNA was subjected to reverse transcriptase-PCR using the Superscript III First Strand Synthesis System (Invitrogen). The resultant cDNA was used to conduct real time PCR on the Applied Biosystems 7300 Real-time PCR System (Applied Biosystems, Foster City, CA). To detect expression of rat Pla2g2a gene products, Primer Express software (Applied Biosystems) was used to design SYBR Green primers to detect mRNA (5′-ACAGCATGAAGGTCCTCCTGTT-3′ and 5′-GGCTCCCCTGGACCTGAA-3′) and TaqMan primers to detect heteronuclear (hn) RNA (5′-CCTTTGGATGCATTTGAGTGATT-3′ and 5′-TGAACAAGAAGCCATACCACCAT-3′; TaqMan, 5′-CCATCCAAGAGGTACATGCCCAGAAACTC-3′). The latter generates an 80-bp product that spans an intron/exon border. The expression levels of IL-1β mRNA were detected by SYBR Green analysis using previously described primers (25). An 18S rRNA TaqMan primer set (Applied Biosystems) was used for normalization. All primers were synthesized by Operon Biotechnologies. Calculations were carried out using the ΔΔCt method of relative quantitation. Data are expressed as relative mRNA (or hnRNA) levels ± S.D.
Enzymatic Activity Assay for sPLA2
Media were collected and stored at −80 °C. Cells were washed twice with cold 100 mm HEPES buffer (pH 8), scraped into that same buffer, and stored at −80 °C. Upon thawing, cell layer samples were disrupted by sonication twice for 5 s on ice. Enzyme activity in the medium and cell layer samples was determined according to the manufacturer's instructions using a commercially available kit (Cayman Chemical, Ann Arbor, MI). Specificity was determined using the inhibitor Ro 23-9358 (Sigma) (26). Enzyme activity is expressed as a function of the area of cell culture growing surface (nmol of product/min/cm2 ± S.D.).
Electrophoretic Mobility Shift Assay
Nuclei were isolated and electrophoretic mobility shift assays performed essentially as described previously (27). Oligomers purchased from Operon Biotechnologies were annealed and end-labeled with [γ-32P]ATP (50–100 μCi) (PerkinElmer Life Sciences). For binding reactions, nuclear extracts (3–5 μg) were combined with the labeled double-stranded oligomers (10–150 fmol, 20,000–150,000 cpm) in a final volume of 8 μl of nuclear lysis buffer. Volumes of 1.25 μl (1 μg/μl) of poly(dI-dC) (GE Healthcare), 3.5 μl of 5× binding buffer (10 mm HEPES (pH 7.9), 1 mm dithiothreitol, 0.1% Triton X-100, 0.5% glycerol), and water were added to bring the total volume to 17.5 μl. Binding was carried out at 25 °C for 30 min. For competition analysis, reactions were also incubated with 100-fold excess of unlabeled double-stranded oligonucleotides for 30 min prior to the addition of the other binding reaction components. For supershift experiments, 0.8 μg of anti-C/EBPβ (Santa Cruz Biotechnology), anti-p65 (Santa Cruz Biotechnology), or normal rabbit IgG (Santa Cruz Biotechnology) were incubated with nuclear extracts for 30 min at 25 °C before the addition of the other binding reaction components. All binding reaction mixtures were resolved on pre-electrophoresed 4% nondenaturing polyacrylamide gels. The DNA oligonucleotide sequences used correspond to sequences as follows (actual binding sites are underlined and mutated bases appear in bold; base pairs not actually present in the gene appear in lowercase): C/EBP1, C/EBP binding site −242 to −232 on the rat Pla2g2a promoter (ATGAACTTTCGAAATCAGCT); C/EBPΔ1, mutated C/EBP1 (ATGAACTTTAGATCTCAGCT) (22); C/EBPc, a C/EBP consensus binding site (GGTATGATTTTGTAATGGGGTAGG) (28); NFκB, −141 to −131 on the rat Pla2g2a promoter, (gttacaaaGGGAAATTACCatttgatc) (22); NFκBΔ, mutated NFκB (AGAGATCTTTACCCAAG) (29); or NFκBc, an NFκB consensus binding site (GGGACAGAGGGGACTTTCCGAGAGG) (30).
Statistical Analyses
Data are expressed as mean ± S.D. for samples from a representative experiment, except where indicated, the mean ± S.D. for the average from 3 independent experiments is shown (in the latter cases, experiments were performed in duplicate so means of duplicates for 3 independent experiments were used to calculate S.D. and perform statistical analysis). For comparison of 2 samples, data were analyzed using a two-tailed unpaired Student's t test and statistically significant differences were reported when p < 0.05. For multiple comparisons, data were analyzed by analysis of variance. Statistically significant differences of relevant comparisons were determined by Bonferroni post hoc analysis and reported when p < 0.05.
RESULTS
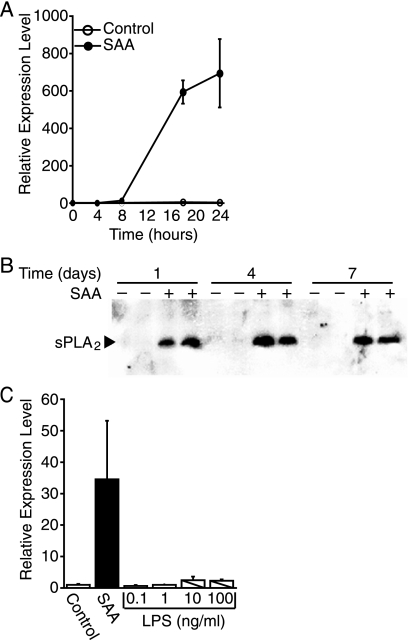
Circulating levels of SAA correlate with an increased risk for atherosclerosis. SAA accumulates in atherosclerotic vessels, potentially synthesized by aortic smooth muscle cells in response to inflammatory cytokines (13, 18). However, smooth muscle cell targets are as yet unknown; identification of such targets will shed light on the role SAA plays in atherosclerosis. To identify potential targets, a preliminary screen of a number of genes was performed on total RNA extracted from neonatal rat aortic smooth muscle cells that were treated with SAA for 1 and 7 days. The level of sPLA2 mRNA was highly elevated in neonatal rat aortic smooth muscle cells treated with SAA (data not shown). It has been determined that sPLA2, which cleaves phospholipids to generate a free fatty acid and a lysophospholipid, is also an indicator of cardiovascular risk and therefore of interest. To validate these results, real time PCR was performed to measure the SAA-induced expression of sPLA2 mRNA. The data from a typical experiment are depicted in Fig. 1A and show that there was virtually no detectable expression of sPLA2 mRNA in control cultures. By 8 h after the addition of SAA, sPLA2 mRNA was detected (13.1-fold increase in expression relative to control) and levels continued to increase during the 24-h study. As expected with primary cells, there was variation in the level of sPLA2 mRNA gene expression in control versus SAA-treated cells from different cell isolations, however, there was a consistent dramatic increase upon treatment with SAA. To determine whether the increase in Pla2g2a gene expression resulted in an increase in protein expression and if the increase was chronic, Western blot analysis was performed on extracts of smooth muscle cells treated with SAA for 1, 4, and 7 days. The data show that sPLA2 protein expression was evident only in SAA-treated cultures (Fig. 1B). The expression of sPLA2 was evident on day 1 and persisted throughout the time course to day 7. To ensure that the SAA-mediated expression of sPLA2 was not due to LPS contamination of the SAA preparation, the effect of Escherichia coli LPS on sPLA2 expression was examined. The manufacturer markets preparations of SAA with no more than 0.1 ng of LPS/μg of SAA. The actual amount of LPS in the preparation used for the study depicted in Fig. 1C was 0.01 ng of LPS/μg of SAA or 0.24 ng of LPS/ml at the dose of SAA tested (2 μm). Treatment with LPS for 24 h with doses as high as 100 ng/ml had no effect on sPLA2 mRNA expression.
FIGURE 1.
SAA induces sPLA2 expression. A, rat smooth muscle cells were treated with (SAA) or without (control) SAA (2 μm) for 4, 8, 18, or 24 h, at which time total RNA was extracted and analyzed by real time PCR for sPLA2 mRNA. RNA expression was calculated as described under “Experimental Procedures” and data expressed as RNA levels relative to the 0-h control ± S.D. (n = 3). B, rat smooth muscle cells were treated with (+) or without (−) SAA (4 μm) for 1, 4, or 7 days, at which time total cell protein extracts were prepared and duplicate extracts were subjected to Western blot analysis using an antiserum directed against sPLA2. C, rat smooth muscle cells were treated with (SAA) or without (Control) SAA (2 μm) or LPS (0.1–100 ng/ml) for 24 h, at which time total RNA was extracted and analyzed by real time PCR for sPLA2 mRNA. RNA expression was calculated as described under “Experimental Procedures” and the data expressed as RNA levels relative to the control ± S.D. (n = 3). The level of sPLA2 mRNA in SAA-treated cells was significantly different from that in control-treated and LPS-treated samples (p < 0.003).
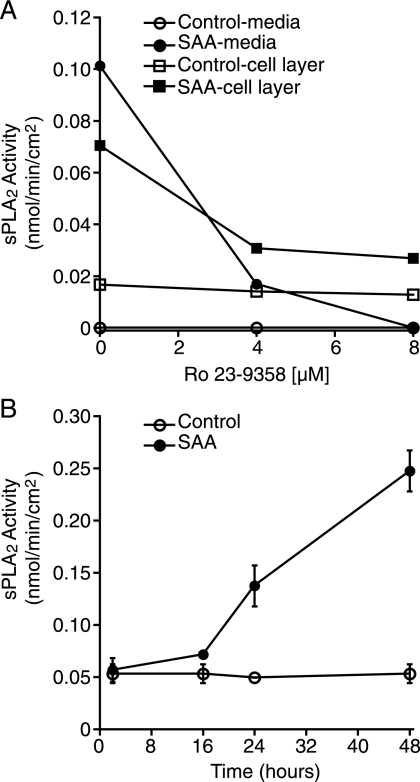
To determine whether the SAA-induced increase in sPLA2 mRNA and protein expression resulted in an increase in available active enzyme, total activity was evaluated by measuring both medium and cell layer fractions to detect secreted and cell-associated enzyme activity, respectively. The latter reflects intracellular enzyme activity as well as cell-associated secreted enzyme. After treating the cells in the presence or absence of SAA, medium and cell layer fractions were harvested and then enzyme activity was evaluated using a 1,2-dithio analog of diheptanoyl phosphatidylcholine as substrate, in the presence and absence of the inhibitor, Ro 23-9358. There was little or no activity in medium (this varied slightly from experiment to experiment) in the absence of SAA, however, the addition of SAA consistently induced an enzyme activity (Fig. 2A). All of the enzyme activity in the medium fraction was inhibited by Ro 23-9358. There was baseline enzyme activity in the cell layer of control cultures and this was not inhibited by Ro 23-9358 and hence, was not sPLA2. Interestingly, the SAA-induced activity in the cell layer was only partially inhibited by Ro 23-9358. The data in Fig. 2B depicts a time course study and demonstrates that 16 h after the addition of SAA, enzyme activity increased and continued to increase during the 48-h study. Although the enzyme was previously shown to be activated by SAA (31), under the assay conditions in this report, there was no increase in activity in control medium to which SAA was added after harvest (data not shown).
FIGURE 2.
SAA increases sPLA2 activity. A, rat smooth muscle cells were treated overnight with (SAA) or without (Control) SAA (4 μm), at which time media and cell layers were harvested and assayed for enzyme activity in the presence of 0, 4, or 8 μm Ro 23-9358. The data are expressed as nmol of product/min/cm2. B, rat smooth muscle cells were treated with (SSA) or without SAA (4 μm) for 2, 16, 24, or 48 h, at which time media were harvested and assayed for enzyme activity. The data are expressed as nmol of product/min/cm2 ± S.D. (n = 3).
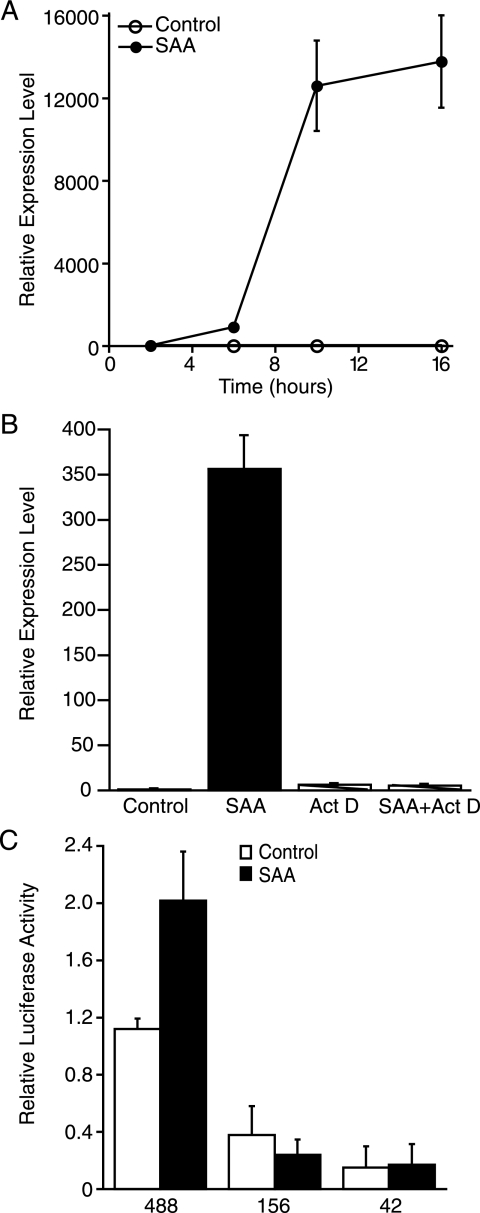
To determine a mechanism for the increase in steady state levels of sPLA2 mRNA, the potential role of SAA-induced activation of transcription of the Pla2g2a gene was explored by examining the expression of hnRNA. The data in Fig. 3A show that hnRNA was detected as early as 6 h after the addition of SAA, and continued to increase and remained elevated at 16 h post-treatment. The increase in prespliced RNA suggests that the SAA-induced increase in sPLA2 mRNA was likely due, at least in part, to increased gene transcription. To determine whether SAA also increased the stability of sPLA2 mRNA, cells pretreated with ActD to inhibit gene transcription were exposed to SAA for 18 h, at which time the expression of sPLA2 mRNA was determined. The data in Fig. 3B show that inhibiting gene transcription prevented the SAA-induced increase in sPLA2 mRNA expression, demonstrating that transcription was required for the increase in sPLA2 mRNA and the increase in expression was not due to mRNA stabilization. To explore which transcription factors played a role in the SAA-induced increase in expression of the Pla2g2a gene, promoter transfection analyses were carried out. Previously, IL-1β-responsive elements were identified within the proximal promoter of the rat Pla2g2a gene (22). To test if SAA-responsive elements were located in this promoter region, smooth muscle cells were transiently transfected with the full-length construct (488), as well as deletion constructs (156 and 42) designed to drive luciferase expression, treated with SAA (or under control conditions), and reporter activities analyzed. SAA induced a 1.8-fold increase in promoter activity compared with control-treated cells (Fig. 3C). Baseline promoter activity decreased in deletions to −156 and −42 bp. Moreover, deletion to −156 or −42 bp abrogated the SAA induction of the promoter, indicating that the response elements reside in the promoter region between −488 and −156 bp.
FIGURE 3.
SAA up-regulates transcription of the Pla2g2a gene. A, rat smooth muscle cells were treated with (SAA) or without (Control) SAA (4 μm) for 2, 6, 10, or 16 h, at which time total RNA was extracted and analyzed by real time PCR for sPLA2 hnRNA. RNA expression was calculated as described under “Experimental Procedures” and data expressed as RNA levels relative to the 2-h control ± S.D. (n = 3). B, rat smooth muscle cells were pretreated with or without ActD (10 μg/ml) for 1 h followed by SAA (2 μm) for 18 h, at which time total RNA was extracted and analyzed by real time PCR for sPLA2 mRNA. RNA expression was calculated as described under “Experimental Procedures” and data expressed as RNA levels relative to the control ± S.D. (n = 3). The level of sPLA2 mRNA in SAA-treated cells was significantly different from that in control-treated, ActD-treated, and SAA plus ActD-treated samples (p < 0.0001). C, rat smooth muscle cells were transfected with rat Pla2g2a luciferase promoter constructs (488, 156, or 42) and a Renilla construct to control for transfection efficiency. Twenty-four hours later, cells were incubated with (SAA) or without (Control) SAA (4 μm) for an additional 24 h, at which time cells were harvested and promoter activity determined as described under “Experimental Procedures.” Relative luciferase activities are expressed as the average fold-induction in comparison to control cells transfected with the 488-bp construct from three independent experiments ± S.D. Promoter activity in control-treated cells transfected with the 488 bp was significantly different from that in control-treated cells transfected with either the 156- or 42-bp constructs (p < 0.001). Promoter activity in SAA-treated cells transfected with the 488-bp construct was significantly different from that in control-treated cells transfected with the 488-bp construct (p < 0.001).
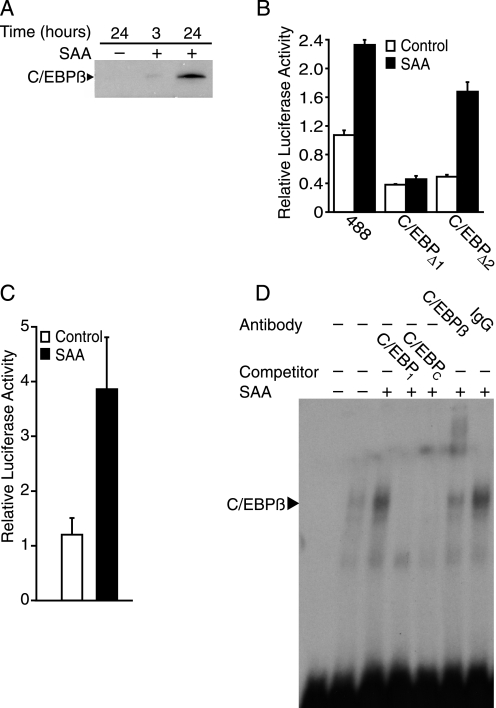
Two C/EBP binding sites in the region upstream of base pair −156 have been described previously (24). Western blot analysis was conducted on cell lysates from cultures treated with SAA to look for changes in C/EBPβ expression. C/EBPβ expression was not detected in control cells but was expressed as early as 3 h after SAA treatment (Fig. 4A). To determine whether or not the previously identified C/EBP binding sites located between −242 and −223 bp (C/EBP1) and −299 and −278 bp (C/EBP2) (24) contribute to SAA-induced activation of the Pla2g2a gene promoter, transfection analyses were carried out using luciferase construct 488 or constructs with mutated C/EBP1 and C/EBP2 binding sites, C/EBPΔ1 and C/EBPΔ2, respectively. The data show that removal of either C/EBP binding site decreased baseline activity of the promoter (Fig. 4B). Moreover, mutation of C/EBP2 did not alter SAA-induced promoter activity significantly, however, C/EBPΔ1 was unresponsive to SAA treatment. To further assess the potential response to SAA, a luciferase reporter construct containing 4 repeats of C/EBP1 attached to the rat pyruvate kinase minimal promoter driving luciferase expression was studied. Activity of this construct increased with SAA treatment (Fig. 4C). To assess the C/EBP-binding capacity in nuclear extracts of SAA-treated cells, electrophoretic mobility shift assays were performed using 32P-labeled oligos corresponding to the C/EBP1 site within the Pla2g2a promoter. Labeled oligos were incubated with nuclear extracts from control- or SAA-treated cultures (Fig. 4D). In comparison to control, nuclear extracts from SAA-treated samples bound more of the oligomer. This increase in binding was competed away with excess unlabeled C/EBP1 and C/EBP consensus oligos. An oligomer corresponding to mutated C/EBP1 was unable to reduce binding (data not shown). Additionally, the mobility of the protein-oligonucleotide complex was retarded in the presence of the C/EBPβ antibody but not normal rabbit IgG.
FIGURE 4.
SAA induces binding to the C/EBP1 site in the Pla2g2a promoter. A, rat smooth muscle cells were treated with (+) or without (−) SAA (4 μm) for 3 or 24 h, at which time total cell protein extracts were prepared and subjected to Western blot analysis using an antiserum directed against C/EBPβ. B, rat smooth muscle cells were transfected with luciferase promoter constructs (488, C/EBPΔ1, or C/EBPΔ2) and a Renilla construct, the latter to control for transfection efficiency. Twenty-four hours later, cells were incubated with (SAA) or without (Control) SAA (4 μm) for an additional 24 h, at which time cells were harvested and promoter activity determined as described under “Experimental Procedures.” Relative luciferase activities are expressed as the average fold-induction in comparison to control cells transfected with the 488-bp construct from three independent experiments ± S.D. Promoter activity in control-treated cells transfected with 488 was significantly different from that in control-treated cells transfected with either C/EBPΔ1 (p < 0.01) or C/EBPΔ2 (p < 0.05). Promoter activity in SAA-treated cells transfected with the 488 and C/EBPΔ2 constructs was significantly different from that in control-treated cells transfected with these plasmids (p < 0.001). C, rat smooth muscle cells were transfected with a luciferase promoter construct activated by C/EBP (C-Luc) and a Renilla construct, the latter to control for transfection efficiency. Twenty-four hours later, cells were incubated with (SAA) or without (Control) SAA (4 μm) for an additional 24 h, at which time cells were harvested and promoter activity determined as described under “Experimental Procedures.” Relative luciferase activities are expressed as the average fold-induction in comparison to control cells ± S.D. (n = 3). Promoter activity in SAA-treated cells was significantly different from that in control-treated cells (p < 0.002). D, an electrophoretic mobility shift assay was conducted using nuclear extracts from rat smooth muscle cells treated with (+) or without (−) SAA (4 μm) for 24 h. Nuclear extracts were incubated with 32P-labeled oligos corresponding to C/EBP1 prior to electrophoresis. Competition studies were carried out by adding an excess of unlabeled oligos corresponding to C/EBP1 or a C/EBP consensus sequence (C/EBPc). Supershift analysis was performed by preincubating nuclear extracts with either C/EBPβ antibody (C/EBPβ) or normal rabbit IgG (IgG). Free probe, i.e. no nuclear extract, was run in the first lane.
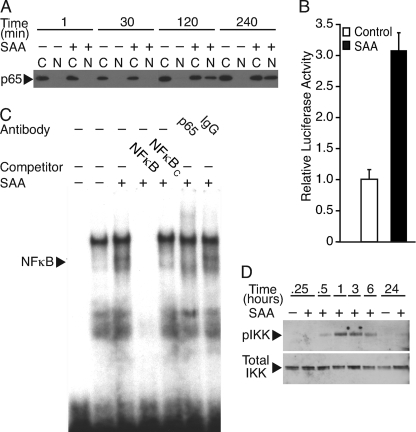
Another transcription factor shown to regulate Pla2g2a gene expression is NFκB. The promoter region of the Pla2g2a gene between bp −131 and −141 has been identified as a high affinity binding site for NFκB (32). Although the transient transfection experiments using the promoter deletion constructs did not suggest the involvement of this region and hence, this NFκB binding element, the possibility of cooperation between C/EBPβ and NFκB led us to consider whether SAA increases transcription of the Pla2g2a gene via activation of NFκB. To determine whether NFκB plays a role in SAA-induced up-regulation of sPLA2, localization of NFκB to the nucleus was studied. Cells were cultured with or without SAA for various periods of time. Differential extraction of cytoplasmic and nuclear proteins was performed and Western blot analysis was carried out to look for p65, a component of NFκB. The data show that SAA induced the movement of p65 from the cytoplasm to the nucleus (Fig. 5A). This change in localization, which was evident as early as 2 h after the addition of SAA, demonstrates that SAA activated NFκB. To determine whether NFκB activation plays a role in SAA-induced up-regulation of Pla2g2a gene expression, transfection analysis was carried out using NFκBΔ, which is the 488 construct containing a mutation in the NFκB binding site (22). Cells were transfected with the 488 or NFκBΔ constructs, followed by 24 h of treatment with SAA. The activity of the control-treated wild type 488 construct is shown in Fig. 4B and in comparison, the control-treated NFκBΔ construct expressed only 4.0 ± 0.7% of the activity, demonstrating that baseline activity of the mutated promoter is dramatically decreased (p < 0.001). Importantly, promoter activity in SAA-treated cells transfected with construct 488 was significantly different from that in SAA-treated cells transfected with NFκBΔ construct (p < 0.001); the SAA-treated mutated promoter expressed 4.1 ± 0.6% of the activity of the control-treated wild type 488 construct, demonstrating that this construct was unresponsive to SAA. Further support for SAA-induced activation of NFκB was provided using a reporter construct that contains 6 copies of a consensus NFκB binding site (33). This construct displayed a 3.1-fold increase in activity with SAA treatment (Fig. 5B). To evaluate the binding of NFκB to the identified binding site in the Pla2g2a promoter, an electrophoretic mobility shift assay was conducted. Nuclear extracts from control- and SAA-treated cell cultures were incubated with 32P-labeled oligos corresponding to the NFκB binding site on the Pla2g2a gene promoter. In addition to bands indicative of SAA-unresponsive binding, there was a band that increased in nuclear extracts from SAA-treated smooth muscle cells (Fig. 5C). This increase in binding to the oligomer was competed away with both unlabeled NFκB corresponding to the element in the Pla2g2a gene and with NFκB consensus oligos. An oligo corresponding to the mutated NFκB element in the Pla2g2a gene promoter was unable to reduce binding (data not shown). Additionally, mobility of the protein-oligonucleotide complex was retarded in the presence of p65 antibody but not normal rabbit IgG. NFκB is inactive and sequestered in the cytoplasm when complexed with IκB. One mechanism of activation of NFκB is via IKK, which is known to phosphorylate IκB, leading to its degradation, thereby enabling NFκB to move to the nucleus (34). To determine whether activation of NFκB is mediated by IKK, Western blot analysis was performed to evaluate IKK phosphorylation. The data in Fig. 5D show that IKK was phosphorylated 30 min after the addition of SAA. Peak levels of phosphorylated IKK were evident at 1 h. Total levels of IKK did not change with the addition of SAA.
FIGURE 5.
SAA induces binding to the NFκB site in the Pla2g2a promoter. A, rat smooth muscle cells were treated with (+) or without (−) SAA (4 μm) for 1, 30, 120, and 240 min, at which time, cytoplasmic (C) and nuclear (N) protein extracts were prepared and subjected to Western blot analysis using an antiserum directed against p65. B, rat smooth muscle cells were transfected with a luciferase promoter construct activated by NFκB (N-Luc) and a Renilla construct, the latter to control for transfection efficiency. Twenty-four hours later, cells were incubated with (SAA) or without (Control) SAA (2 μm) for an additional 24 h, at which time cells were harvested and promoter activity determined as described under “Experimental Procedures.” Relative luciferase activities are expressed as the average fold-induction in comparison to control cells ± S.D. (n = 6). Promoter activity in SAA-treated cells was significantly different from control-treated cells (p < 0.0001). C, an electrophoretic mobility shift assay was conducted using nuclear extracts from rat smooth muscle cells treated with (+) or without (−) SAA (4 μm) for 24 h. Nuclear extracts were incubated with 32P-labeled oligos corresponding to the NFκB binding site in the Pla2g2a gene promoter prior to electrophoresis. Competition studies were carried out by adding an excess of unlabeled oligos corresponding to the NFκB binding site (NFκB) or an NFκB consensus sequence (NFκBc). Supershift analysis was performed by preincubating nuclear extracts with either p65 antibody (p65) or normal rabbit IgG (IgG). Free probe, i.e. no nuclear extract, was run in the first lane. D, rat smooth muscle cells were treated with (+) or without (−) SAA (2 μm) for 0.25, 0.5, 1, 3, 6, or 24 h, at which time total cell protein extracts were prepared and subjected to Western blot analysis using antisera directed against phospho-IKKα/β (upper panel) and total IKKβ (lower panel).
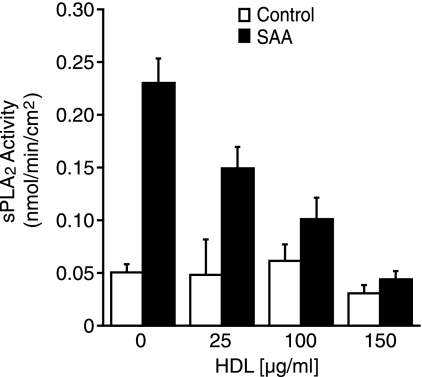
Although SAA is largely associated with HDL in circulation, it has been speculated that it is available in lipid-free/lipid-poor forms at the site of atherosclerotic lesion formation, which makes the functional differences between lipid-associated and lipid-free SAA of interest. Studies were performed to determine whether HDL-associated SAA up-regulates sPLA2 or if association with HDL inhibits these effects. It has been shown that when SAA is combined with HDL they readily associate (35), so to perform these studies, a constant concentration of SAA was first incubated with varying concentrations of HDL. Cells were then control-treated or treated with SAA alone, i.e. lipid-free, HDL alone, or HDL-associated SAA for 24 h. Media were collected and analyzed for enzyme activity. HDL alone had no effect on enzyme activity but the addition of increasing concentrations of HDL caused a dose-responsive decrease in SAA-induced sPLA2 activity (Fig. 6). Additional studies to determine whether HDL inhibits sPLA2 expression at the mRNA level confirmed this finding (data not shown).
FIGURE 6.
SAA-induced sPLA2 activity is inhibited when added in the presence of HDL. Rat smooth muscle cells were treated with (SAA) or without (Control) SAA (4 μm) or with increasing concentrations of HDL (25–150 μg/ml) preincubated with SAA (4 μm) for 24 h, at which time media were harvested and assayed for enzyme activity. The data are expressed as nmol of product/min/cm2 ± S.D. (n = 3). Enzyme activity in control lipid-free samples was significantly different from lipid-free SAA-treated samples (p < 0.001). Enzyme activity in the HDL (25 μg/ml)-treated samples was significantly different from SAA plus HDL (25 μg/ml)-treated samples (p < 0.001). Enzyme activity in the lipid-free SAA-treated samples was significantly different from SAA plus HDL (25, 100, 150 μg/ml)-treated samples (p < 0.001).
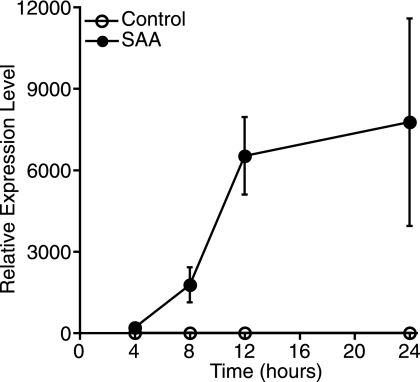
Previous reports demonstrated that IL-1β induces rat vascular smooth muscle cell expression of sPLA2, also in a C/EBPβ- and NFκB-dependent manner (22, 36). It was of interest to determine whether the SAA-induced increase in sPLA2 was mediated by IL-1β signaling. First, the effect of SAA on IL-1β expression was explored by real time PCR. Cells were treated with SAA for various periods of time before analysis of IL-1β mRNA levels. SAA induced a large increase in IL-1β mRNA expression in response to SAA as a function of time of treatment (Fig. 7).
FIGURE 7.
SAA up-regulates IL-1β mRNA expression. Rat smooth muscle cells were treated with (SAA) or without (Control) SAA (4 μm) for 4, 8, 12, and 24 h, at which time total RNA was extracted and analyzed by real time PCR for IL-1β mRNA. RNA expression was calculated as described under “Experimental Procedures” and data expressed as RNA levels relative to the 4-h control ± S.D. (n = 3).
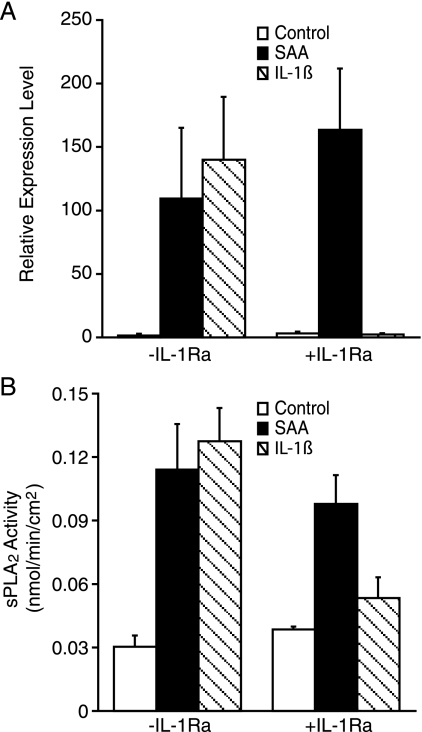
To determine whether IL-1β signaling contributes to the SAA-induced increase in sPLA2 expression, cells were treated with or without SAA or IL-1β in the presence or absence of the receptor antagonist, IL-1Ra. After 24 h of treatment, expression of sPLA2 was analyzed by real time PCR. In accordance with previous studies (36), IL-1β increased smooth muscle cell expression of sPLA2 mRNA (Fig. 8A). As expected, IL-1Ra inhibited the IL-1β-induced sPLA2 mRNA expression. Interestingly, however, the SAA-induced sPLA2 mRNA expression remained elevated in the presence of IL-1Ra, suggesting that SAA-induced sPLA2 expression is independent of IL-1β signaling. Likewise, IL-1β induced an increase in enzyme activity and this was blocked by IL-1Ra, however, the SAA-induced increase in enzyme activity was unaffected by the presence of the receptor antagonist, IL-1Ra (Fig. 8B).
FIGURE 8.
SAA-induced expression of sPLA2 is not mediated by IL-1β. A, rat smooth muscle cells were treated with (SAA) or without (Control) SAA (2 μm) or IL-1β (100 ng/ml) for 24 h in the presence (+) or absence (−) of IL-1Ra (1 μg/ml), at which time total RNA was extracted and analyzed by real time PCR for sPLA2 mRNA. RNA expression was calculated as described under “Experimental Procedures” and data expressed as RNA levels relative to the control ± S.D. (n = 3). The level of sPLA2 mRNA in control-treated cells was significantly different from that in SAA- (p < 0.01), IL-1β- (p < 0.01), and SAA plus IL-1Ra-treated cells (p < 0.001). The level of sPLA2 mRNA in IL-1β-treated cells was significantly different from IL-1β plus IL-1Ra-treated cells (p < 0.01). B, rat smooth muscle cells were treated with (SAA) or without (Control) SAA (2 μm) or IL-1β (100 ng/ml) for 24 h in the presence (+) or absence (−) of IL-1Ra (1 μg/ml)) for 24 h, at which time media were harvested and assayed for enzyme activity. The data are expressed as nmol of product/min/cm2 ± S.D. (n = 3). Enzyme activity in the media of control-treated cells was significantly different from that in SAA-, IL-1β-, and SAA plus IL-1Ra-treated cells (p < 0.001). Enzyme activity in the IL-1β-treated cells was significantly different from that in IL-1β plus IL-1Ra-treated cells (p < 0.001). Enzyme activity in IL-1Ra-treated cells was significantly different from that in SAA plus IL-1Ra-treated cells (p < 0.001).
DISCUSSION
Vascular smooth muscle cells in atherosclerosis respond to numerous inflammatory mediators. Previously, we reported that smooth muscle cells synthesize SAA in response to IL-1α (18). In this study, the mechanisms of smooth muscle response to SAA treatment were explored. These data demonstrate that SAA up-regulates smooth muscle cell expression of the Pla2g2a gene via critical NFκB and C/EBP sites, suggesting that SAA amplifies an acute phase response in the vasculature. Although local levels of SAA in the vasculature are not known, circulating concentrations in the plasma during an acute phase can be more than 1,000 μg/ml (i.e. >80 μm) and these experiments were performed with 2–4 μm SAA, suggesting the physiologic relevance of these findings.
The specific role of the SAA family in atherosclerosis remains controversial and both pro-atherogenic and anti-atherogenic roles for SAA have been suggested (7, 37). Potential pro-atherogenic functions include induction of expression of matrix metalloproteinases, chemotaxis of monocytes, polymorphonuclear leukocytes and T lymphocytes, cholesterol delivery to peripheral tissues, and binding to proteoglycans in the vasculature, causing retention of HDL (1). Alternatively, the SAA2.1 isoform promotes cholesterol export from cholesterol-laden macrophages (38, 39), and peptides corresponding to the mouse SAA2.1 isoform were demonstrated to be atheroprotective in apolipoprotein E-deficient mice (40). Our data show that SAA decreases smooth muscle cell lipid synthesis and accumulation, potentially guarding against smooth muscle “foam cell” formation (20).
In addition to SAA, an elevated level of serum sPLA2 has been identified as a marker of cardiovascular risk (41, 42). The genes for which sPLA2 is the product have been identified and sequenced in mice (Pla2g2a), rats (Pla2g2a), and humans (PLA2G2A) (43–45). Interferon-γ (46, 47) and IL-1β (22, 48) stimulate smooth muscle cell sPLA2 expression. Moreover, it was reported that LPS at 100 ng/ml stimulates smooth muscle cell phospholipase activity (36). Here, at a similar dose, there was no effect on mRNA expression of sPLA2. It is uncertain as to the cause of this discrepancy, however, the increase in sPLA2 mRNA expression in this study is clearly not due to contamination of the recombinant SAA with LPS. Expression of sPLA2 in smooth muscle cells has been shown to be regulated (activated) at the level of transcription by C/EBP, peroxisome proliferator-activated receptor γ, NFκB, Ets, and liver X receptors/retinoid X receptors (22, 29).
A member of the phospholipase superfamily of enzymes, sPLA2, cleaves at the Sn2 position of phospholipids, generating a free fatty acid and a lysophospholipid (49–51). In this study, enzyme activity was measured using a 1,2-dithio analog of diheptanoyl phosphatidylcholine as substrate. Specificity for sPLA2 was evaluated using the inhibitor Ro 23-9358 and is strongly suggestive of active sPLA2 enzyme. Although it is possible that the inhibitor does not fully distinguish the sPLA2 of interest in this study from all others due to the inherent problems associated with the use of pharmacologic inhibitors, the evaluation of activity that is not inhibited by Ro 23-9358 is most meaningful. Thus, the finding that SAA-induced phospholipase activity in the medium is wholly sPLA2, but in the cell layer it is only partially sPLA2 suggests that SAA induces additional intracellular phospholipase(s). Future studies will elucidate the nature of this activity. Interestingly, acute phase SAA was shown to increase enzyme activity of sPLA2 (31), but this was not the case in the study reported herein. This discrepancy is perhaps due to the differences in the source of the SAA and/or the enzyme.
Arachadonic acid is commonly generated as a product of sPLA2 and it can then serve as the precursor for signaling molecules such as prostaglandins, leukotrienes, thromboxanes, and prostaglandins. Lysophospholipids are also biologically active with a variety of identified functions including the induction of mitogenesis and chemotaxis (51). It has been shown that sPLA2 also localizes to vascular lesions (52–56) and published data support a role for sPLA2 in hydrolysis of arterially embedded low density lipoprotein (LDL), contributing to an inflammatory response and lesion formation (57, 58). The sPLA2-modified LDL particles aggregate and bind to proteoglycans, causing their retention in the vasculature, generation of proinflammatory lysophosphatidylcholine, which is chemotactic for monocytes, and the induction of expression of proteoglycans and growth factors, including platelet-derived growth factor. Moreover, the modified LDL particles are taken up more avidly by macrophages, leading to lipid-laden “foam cell” formation (58, 59). In addition to LDL, both acute phase and normal HDL have been shown to be substrates for sPLA2 (60). In a transgenic mouse model, sPLA2 led to a decrease in the levels of HDL (61, 62), possibly due to the reported increase in cholesteryl ester uptake via the scavenger receptor BI by sPLA2-modified HDL (61). Transgenic mice overexpressing sPLA2 either globally or in a macrophage-specific manner developed atherosclerotic lesions (63–66). In a model in which sPLA2 was overexpressed in macrophages, it was shown that lesion size increased but interestingly, collagen accumulation was also greater, suggesting that sPLA2 might exacerbate lesion formation but lead to a more stable lesion, i.e. one not prone to rupture (63). These findings deserve further attention. Anti-atherogenic roles for sPLA2 have been suggested by identification of anti-thrombotic properties that could potentially inhibit the formation of a thrombus after plaque rupture in late stage disease (67, 68). Anti-apoptotic properties of sPLA2 may protect against atherosclerosis and it has been suggested that functions in anti-inflammatory cascades may well be dependent upon the stage in lesion formation (51). Moreover, Labeque et al. (69) showed that sPLA2-modified LDL is cleared from the bloodstream more rapidly than unmodified. Likewise, cholesterol is taken up more avidly by hepatocytes incubated with phospholipase-treated HDL (31, 70, 71). It is also noteworthy that sPLA2 expression has been shown in normal artery tissue and although this could suggest changes indicative of an early inflammatory response, it might also suggest that sPLA2 participates in maintaining vascular homeostasis (53, 54).
The role of NFκB in vascular disease has been the subject of many investigations (72, 73). The data reported here demonstrate that SAA activates smooth muscle cell IKK and NFκB as well as C/EBPβ, and these findings are likely to have relevance beyond their role in Pla2g2a gene expression. These data support a role for SAA in exacerbating the local inflammatory response within the vessel wall during atherosclerotic plaque formation. Interestingly, the expression of SAA itself has been shown to be up-regulated by NFκB as well as C/EBP (11). The acute phase response in hepatocytes has been shown to be dependent on NFκB and C/EBPβ including IL-1β-induced expression of C-reactive protein (74) and LPS-induced factor VIII expression (75). C/EBPβ functions with NFκB to promote the expression of SAA as well as cytokines such as IL-6, IL-8, and IL-12 (76–78). Our demonstration of cytokine-induced smooth muscle cell expression of SAA (18) suggests a potential autocrine loop whereby vascular inflammation activates expression of SAA, which further activates NFκB and C/EBPβ-mediated expression of SAA. The cytokine-induced activation of NFκB shown to down-regulate hepatic gene expression of apoA-1 and paraoxonase 1 and up-regulate expression of SAA suggests a potential mechanism for an increase in inflammatory circulating HDL (79) and it is interesting to speculate as to whether the expression of SAA would also activate hepatic sPLA2 during an acute phase response, thereby resulting in additional remodeling of plasma lipoprotein particles.
These data show that SAA activates expression of sPLA2 mRNA via an IL-1 receptor-independent pathway. The SAA-induced activation of expression of IL-1β mRNA would be expected to contribute, at least in part, to the sPLA2 expressed in response to SAA, but this was not the case. When an exogenous source of IL-1β was provided, sPLA2 expression was activated, demonstrating that the receptor and signaling pathways leading to IL-1β-mediated activation of sPLA2 are available in these cells. The most likely explanation for the lack of an IL-1β-mediated activation of sPLA2 by SAA is that the amount of IL-1β synthesized, secreted, and available in an active form in response to SAA is insufficient to activate the promoter.
SAA has been shown to bind to a variety of receptors including the formyl peptide-receptor like-1 receptor (80, 81), Tanis (82), CD36 and lysosomal integral membrane protein-II analogous-1 (83), the receptor for advanced glycation end products (84, 85), αIIbβ3 integrin (86), and the scavenger receptor BI (87, 88). More recently, it was suggested that SAA-induced nitric oxide production by macrophages is Toll-like receptor (TLR) 4-dependent (89). Another recent report identified TLR2 as a receptor that binds SAA and activates NFκB and mitogen-activated protein kinases. Using macrophages from a TLR2-deficient mouse model, SAA-mediated expression of tumor necrosis factor α and IL-23p19 were shown to be TLR2-dependent. Interestingly, the SAA-induced expression of the anti-inflammatory cytokines, IL-10 and IL-1Ra, are TLR2-dependent as well (90). IL-1β-activated expression of the Pla2g2a promoter is mediated by C/EBP, NFκB, and Ets (22). The IL-1 and TLR superfamily of receptors share a common intracellular Toll/IL-1 domain. Diversity in responses is generated by variable extracellular regions (91). Whether SAA-induced activation of transcription of the Pla2g2a gene is mediated by a TLR remains to be determined and it will be interesting to further pursue the potential links between the innate immune system and the chronic inflammation associated with atherosclerosis (91, 92).
The activation of sPLA2 expression was inhibited by HDL, suggesting a potential mechanism for regulation of induction of the enzyme by SAA. The data show that association with HDL inhibits the SAA-induced activation of sPLA2 expression. Although SAA is known to bind avidly to HDL, its availability in a lipid-free/lipid-poor form has been the subject of much speculation and is probably due to expression of large amounts of SAA (87) or as van der Westhuyzen et al. (93) suggested, due to remodeling of acute phase HDL, which could generate lipid-poor SAA that can serve as a cholesterol acceptor, promoting efflux. Data from our laboratory and that of others demonstrate SAA synthesis by smooth muscle cells (13, 18) suggesting that local production of SAA may contribute to its accumulation in atherosclerotic plaques and that the SAA is potentially available to the cells in a free, non-lipoprotein-bound form for the autocrine induction of sPLA2.
In conclusion, these data show that SAA dramatically up-regulates the expression of sPLA2 in aortic smooth muscle cells. The effect of SAA on sPLA2 activity is mediated, at least in part, at the level of transcription by activation of both NFκB and C/EBPβ. Taken together, the data suggest that during an inflammatory cascade in the vasculature, SAA synthesized by smooth muscle cells could act in autocrine fashion to activate expression of sPLA2.
Acknowledgments
We thank Luwam Ghidei for excellent technical assistance. We greatly appreciate helpful discussions and critical reading of the manuscript by Dr. Matthew Layne.
This work was supported, in whole or in part, by National Institutes of Health Grant HL079201 and American Heart Association Grants 0256215T and 0455846T.
We dedicate this manuscript to the memory of our colleague and dear friend Valerie Verbitzki for her contributions to this work.
- SAA
- serum amyloid A
- ActD
- actinomycin D
- C/EBP
- CAAT-enhancer binding protein
- MEM
- minimal essential medium
- DMEM
- Dulbecco's modified Eagle's medium supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 0.1 mm MEM nonessential amino acids, and 1 mm MEM sodium pyruvate solution
- hn
- heteronuclear
- HDL
- high density lipoprotein
- IκB
- inhibitor of κB
- IKK
- inhibitor of NF-κB kinase
- IL
- interleukin
- IL-1Ra
- IL-1 receptor antagonist
- LDS
- lipid-deficient serum
- LPS
- lipopolysaccharide
- LDL
- low density lipoprotein
- NFκB
- nuclear factor κB
- sPLA2
- secretory phospholipase A2, group IIA
- TLR
- Toll-like receptor
- TBST
- Tris-buffered saline with 0.1% Tween-20.
REFERENCES
- 1.Chait A., Han C. Y., Oram J. F., Heinecke J. W. (2005) J. Lipid Res. 46, 389–403 [DOI] [PubMed] [Google Scholar]
- 2.Packard R. R., Libby P. (2008) Clin. Chem. 54, 24–38 [DOI] [PubMed] [Google Scholar]
- 3.Mahmoudi M., Curzen N., Gallagher P. J. (2007) Histopathology 50, 535–546 [DOI] [PubMed] [Google Scholar]
- 4.Libby P., Ridker P. M. (2004) Am. J. Med. 116, Suppl. 6A, 9S–16S [DOI] [PubMed] [Google Scholar]
- 5.Ridker P. M., Hennekens C. H., Buring J. E., Rifai N. (2000) N. Engl. J. Med. 342, 836–843 [DOI] [PubMed] [Google Scholar]
- 6.Jensen L. E., Whitehead A. S. (1998) Biochem. J. 334, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber B. M. (2002) Amyloid 9, 276–278 [DOI] [PubMed] [Google Scholar]
- 8.Benditt E. P., Meek R. L. (1989) J. Exp. Med. 169, 1841–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson M. A., Wei S. H., Weber A., Weber A. T., McDonald T. L. (2003) Biochem. Biophys. Res. Commun. 301, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 10.Rokita H., Loose L. D., Bartle L. M., Sipe J. D. (1994) J. Rheumatol. 21, 400–405 [PubMed] [Google Scholar]
- 11.Uhlar C. M., Whitehead A. S. (1999) Eur. J. Biochem. 265, 501–523 [DOI] [PubMed] [Google Scholar]
- 12.Meek R. L., Eriksen N., Benditt E. P. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7949–7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meek R. L., Urieli-Shoval S., Benditt E. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell T. I., Coon C. I., Brinckerhoff C. E. (1991) J. Clin. Invest. 87, 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urieli-Shoval S., Meek R. L., Hanson R. H., Eriksen N., Benditt E. P. (1994) Am. J. Pathol. 145, 650–660 [PMC free article] [PubMed] [Google Scholar]
- 16.Qiao J. H., Xie P. Z., Fishbein M. C., Kreuzer J., Drake T. A., Demer L. L., Lusis A. J. (1994) Arterioscler. Thromb. 14, 1480–1497 [DOI] [PubMed] [Google Scholar]
- 17.Yamada T., Kakihara T., Kamishima T., Fukuda T., Kawai T. (1996) Pathol. Int. 46, 797–800 [DOI] [PubMed] [Google Scholar]
- 18.Kumon Y., Sipe J. D., Brinckerhoff C. E., Schreiber B. M. (1997) Scand. J. Immunol. 46, 284–291 [DOI] [PubMed] [Google Scholar]
- 19.Schreiber B. M., Martin B. M., Hollander W., Franzblau C. (1988) Atherosclerosis 69, 69–79 [DOI] [PubMed] [Google Scholar]
- 20.Schreiber B. M., Veverbrants M., Fine R. E., Blusztajn J. K., Salmona M., Patel A., Sipe J. D. (1999) Biochem. J. 344, 7–13 [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber B. M., Jones H. V., Toselli P., Franzblau C. (1992) Atherosclerosis 95, 201–210 [DOI] [PubMed] [Google Scholar]
- 22.Antonio V., Brouillet A., Janvier B., Monne C., Bereziat G., Andreani M., Raymondjean M. (2002) Biochem. J. 368, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker G., Kunz D., Pignat W., van den Bosch H., Pfeilschifter J. (1997) Br. J. Pharmacol. 121, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couturier C., Antonio V., Brouillet A., Béréziat G., Raymondjean M., Andréani M. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 2559–2565 [DOI] [PubMed] [Google Scholar]
- 25.Kitaoka Y., Munemasa Y., Nakazawa T., Ueno S. (2007) Brain Res. 1142, 247–255 [DOI] [PubMed] [Google Scholar]
- 26.LeMahieu R. A., Carson M., Han R. J., Madison V. S., Hope W. C., Chen T., Morgan D. W., Hendrickson H. S. (1993) J. Med. Chem. 36, 3029–3031 [DOI] [PubMed] [Google Scholar]
- 27.Rich C. B., Fontanilla M. R., Nugent M., Foster J. A. (1999) J. Biol. Chem. 274, 33433–33439 [DOI] [PubMed] [Google Scholar]
- 28.Raymondjean M., Cereghini S., Yaniv M. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreani M., Olivier J. L., Berenbaum F., Raymondjean M., Béréziat G. (2000) Biochim. Biophys. Acta 1488, 149–158 [DOI] [PubMed] [Google Scholar]
- 30.Couturier C., Brouillet A., Couriaud C., Koumanov K., Béréziat G., Andréani M. (1999) J. Biol. Chem. 274, 23085–23093 [DOI] [PubMed] [Google Scholar]
- 31.Pruzanski W., de Beer F. C., de Beer M. C., Stefanski E., Vadas P. (1995) Biochem. J. 309, 461–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorsam G., Taher M. M., Valerie K. C., Kuemmerle N. B., Chan J. C., Franson R. C. (2000) J. Pharmacol. Exp. Ther. 292, 271–279 [PubMed] [Google Scholar]
- 33.Rawadi G., Garcia J., Lemercier B., Roman-Roman S. (1999) J. Immunol. 162, 2193–2203 [PubMed] [Google Scholar]
- 34.Ghosh S., Hayden M. S. (2008) Nat. Rev. Immunol. 8, 837–848 [DOI] [PubMed] [Google Scholar]
- 35.Coetzee G. A., Strachan A. F., van der Westhuyzen D. R., Hoppe H. C., Jeenah M. S., de Beer F. C. (1986) J. Biol. Chem. 261, 9644–9651 [PubMed] [Google Scholar]
- 36.Nakano T., Ohara O., Teraoka H., Arita H. (1990) FEBS Lett. 261, 171–174 [DOI] [PubMed] [Google Scholar]
- 37.O'Brien K. D., Chait A. (2006) Curr. Atheroscler. Rep. 8, 62–68 [DOI] [PubMed] [Google Scholar]
- 38.Kisilevsky R., Tam S. P. (2003) J. Lipid Res. 44, 2257–2269 [DOI] [PubMed] [Google Scholar]
- 39.Tam S. P., Flexman A., Hulme J., Kisilevsky R. (2002) J. Lipid Res. 43, 1410–1420 [DOI] [PubMed] [Google Scholar]
- 40.Tam S. P., Ancsin J. B., Tan R., Kisilevsky R. (2005) J. Lipid Res. 46, 2091–2101 [DOI] [PubMed] [Google Scholar]
- 41.Kugiyama K., Ota Y., Takazoe K., Moriyama Y., Kawano H., Miyao Y., Sakamoto T., Soejima H., Ogawa H., Doi H., Sugiyama S., Yasue H. (1999) Circulation 100, 1280–1284 [DOI] [PubMed] [Google Scholar]
- 42.Mallat Z., Benessiano J., Simon T., Ederhy S., Sebella-Arguelles C., Cohen A., Huart V., Wareham N. J., Luben R., Khaw K. T., Tedgui A., Boekholdt S. M. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1177–1183 [DOI] [PubMed] [Google Scholar]
- 43.Kennedy B. P., Payette P., Mudgett J., Vadas P., Pruzanski W., Kwan M., Tang C., Rancourt D. E., Cromlish W. A. (1995) J. Biol. Chem. 270, 22378–22385 [DOI] [PubMed] [Google Scholar]
- 44.Kramer R. M., Hession C., Johansen B., Hayes G., McGray P., Chow E. P., Tizard R., Pepinsky R. B. (1989) J. Biol. Chem. 264, 5768–5775 [PubMed] [Google Scholar]
- 45.Ohara O., Ishizaki J., Nakano T., Arita H., Teraoka H. (1990) Nucleic Acids Res. 18, 6997–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menschikowski M., Hagelgans A., Hempel U., Siegert G. (2004) FEBS Lett. 577, 81–86 [DOI] [PubMed] [Google Scholar]
- 47.Peilot H., Rosengren B., Bondjers G., Hurt-Camejo E. (2000) J. Biol. Chem. 275, 22895–22904 [DOI] [PubMed] [Google Scholar]
- 48.Jaulmes A., Thierry S., Janvier B., Raymondjean M., Maréchal V. (2006) FASEB J. 20, 1727–1729 [DOI] [PubMed] [Google Scholar]
- 49.Balsinde J. (2002) Biochem. J. 364, 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudo I., Murakami M. (2002) Prostaglandins Other Lipid Mediat. 68–69, 3–58 [DOI] [PubMed] [Google Scholar]
- 51.Menschikowski M., Hagelgans A., Siegert G. (2006) Prostaglandins Other Lipid Mediat. 79, 1–33 [DOI] [PubMed] [Google Scholar]
- 52.Anthonsen M. W., Stengel D., Hourton D., Ninio E., Johansen B. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 1276–1282 [DOI] [PubMed] [Google Scholar]
- 53.Elinder L. S., Dumitrescu A., Larsson P., Hedin U., Frostegård J., Claesson H. E. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 2257–2263 [DOI] [PubMed] [Google Scholar]
- 54.Hurt-Camejo E., Andersen S., Standal R., Rosengren B., Sartipy P., Stadberg E., Johansen B. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 300–309 [DOI] [PubMed] [Google Scholar]
- 55.Menschikowski M., Kasper M., Lattke P., Schiering A., Schiefer S., Stockinger H., Jaross W. (1995) Atherosclerosis 118, 173–181 [DOI] [PubMed] [Google Scholar]
- 56.Schiering A., Menschikowski M., Mueller E., Jaross W. (1999) Atherosclerosis 144, 73–78 [DOI] [PubMed] [Google Scholar]
- 57.Pruzanski W., Stefanski E., Kopilov J., Kuksis A. (2001) Lab. Invest. 81, 757–765 [DOI] [PubMed] [Google Scholar]
- 58.Oestvang J., Johansen B. (2006) Biochim. Biophys. Acta 1761, 1309–1316 [DOI] [PubMed] [Google Scholar]
- 59.Hurt-Camejo E., Paredes S., Masana L., Camejo G., Sartipy P., Rosengren B., Pedreno J., Vallve J. C., Benito P., Wiklund O. (2001) Arthritis Rheum. 44, 2761–2767 [DOI] [PubMed] [Google Scholar]
- 60.Pruzanski W., Stefanski E., de Beer F. C., de Beer M. C., Vadas P., Ravandi A., Kuksis A. (1998) J. Lipid Res. 39, 2150–2160 [PubMed] [Google Scholar]
- 61.de Beer F. C., Connell P. M., Yu J., de Beer M. C., Webb N. R., van der Westhuyzen D. R. (2000) J. Lipid Res. 41, 1849–1857 [PubMed] [Google Scholar]
- 62.Tietge U. J., Maugeais C., Cain W., Grass D., Glick J. M., de Beer F. C., Rader D. J. (2000) J. Biol. Chem. 275, 10077–10084 [DOI] [PubMed] [Google Scholar]
- 63.Ghesquiere S. A., Gijbels M. J., Anthonsen M., van Gorp P. J., van der Made I., Johansen B., Hofker M. H., de Winther M. P. (2005) J. Lipid Res. 46, 201–210 [DOI] [PubMed] [Google Scholar]
- 64.Ivandic B., Castellani L. W., Wang X. P., Qiao J. H., Mehrabian M., Navab M., Fogelman A. M., Grass D. S., Swanson M. E., de Beer M. C., de Beer F., Lusis A. J. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 65.Tietge U. J., Pratico D., Ding T., Funk C. D., Hildebrand R. B., Van Berkel T., Van Eck M. (2005) J. Lipid Res. 46, 1604–1614 [DOI] [PubMed] [Google Scholar]
- 66.Webb N. R., Bostrom M. A., Szilvassy S. J., van der Westhuyzen D. R., Daugherty A., de Beer F. C. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 263–268 [DOI] [PubMed] [Google Scholar]
- 67.Houliston R. A., Wheeler-Jones C. P. (2001) Biochem. Biophys. Res. Commun. 287, 881–887 [DOI] [PubMed] [Google Scholar]
- 68.Mounier C. M., Hackeng T. M., Schaeffer F., Faure G., Bon C., Griffin J. H. (1998) J. Biol. Chem. 273, 23764–23772 [DOI] [PubMed] [Google Scholar]
- 69.Labeque R., Mullon C. J., Ferreira J. P., Lees R. S., Langer R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3476–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bamberger M., Lund-Katz S., Phillips M. C., Rothblat G. H. (1985) Biochemistry 24, 3693–3701 [DOI] [PubMed] [Google Scholar]
- 71.Collet X., Perret B. P., Simard G., Vieu C., Douste-Blazy L. (1990) Biochim. Biophys. Acta 1043, 301–310 [DOI] [PubMed] [Google Scholar]
- 72.de Winther M. P., Kanters E., Kraal G., Hofker M. H. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 904–914 [DOI] [PubMed] [Google Scholar]
- 73.Xanthoulea S., Curfs D. M., Hofker M. H., de Winther M. P. (2005) Curr. Opin. Lipidol. 16, 536–542 [DOI] [PubMed] [Google Scholar]
- 74.Kramer F., Torzewski J., Kamenz J., Veit K., Hombach V., Dedio J., Ivashchenko Y. (2008) Mol. Immunol. 45, 2678–2689 [DOI] [PubMed] [Google Scholar]
- 75.Begbie M., Notley C., Tinlin S., Sawyer L., Lillicrap D. (2000) Thromb. Haemost. 84, 216–222 [PubMed] [Google Scholar]
- 76.Lee Y. M., Miau L. H., Chang C. J., Lee S. C. (1996) Mol. Cell. Biol. 16, 4257–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plevy S. E., Gemberling J. H., Hsu S., Dorner A. J., Smale S. T. (1997) Mol. Cell. Biol. 17, 4572–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ray A., Hannink M., Ray B. K. (1995) J. Biol. Chem. 270, 7365–7374 [DOI] [PubMed] [Google Scholar]
- 79.Han C. Y., Chiba T., Campbell J. S., Fausto N., Chaisson M., Orasanu G., Plutzky J., Chait A. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1806–1813 [DOI] [PubMed] [Google Scholar]
- 80.Su S. B., Gong W., Gao J. L., Shen W., Murphy P. M., Oppenheim J. J., Wang J. M. (1999) J. Exp. Med. 189, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He R., Sang H., Ye R. D. (2003) Blood 101, 1572–1581 [DOI] [PubMed] [Google Scholar]
- 82.Walder K., Kantham L., McMillan J. S., Trevaskis J., Kerr L., De Silva A., Sunderland T., Godde N., Gao Y., Bishara N., Windmill K., Tenne-Brown J., Augert G., Zimmet P. Z., Collier G. R. (2002) Diabetes 51, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 83.Baranova I. N., Vishnyakova T. G., Bocharov A. V., Kurlander R., Chen Z., Kimelman M. L., Remaley A. T., Csako G., Thomas F., Eggerman T. L., Patterson A. P. (2005) J. Biol. Chem. 280, 8031–8040 [DOI] [PubMed] [Google Scholar]
- 84.Cai Z., Cai L., Jiang J., Chang K. S., van der Westhuyzen D. R., Luo G. (2007) J. Virol. 81, 6128–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okamoto H., Katagiri Y., Kiire A., Momohara S., Kamatani N. (2008) J. Rheumatol. 35, 752–756 [PubMed] [Google Scholar]
- 86.Urieli-Shoval S., Shubinsky G., Linke R. P., Fridkin M., Tabi I., Matzner Y. (2002) Blood 99, 1224–1229 [DOI] [PubMed] [Google Scholar]
- 87.Cai L., de Beer M. C., de Beer F. C., van der Westhuyzen D. R. (2005) J. Biol. Chem. 280, 2954–2961 [DOI] [PubMed] [Google Scholar]
- 88.van der Westhuyzen D. R., Cai L., de Beer M. C., de Beer F. C. (2005) J. Biol. Chem. 280, 35890–35895 [DOI] [PubMed] [Google Scholar]
- 89.Sandri S., Rodriguez D., Gomes E., Monteiro H. P., Russo M., Campa A. (2008) J. Leukocyte Biol. 83, 1174–1180 [DOI] [PubMed] [Google Scholar]
- 90.Cheng N., He R., Tian J., Ye P. P., Ye R. D. (2008) J. Immunol. 181, 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verstrepen L., Bekaert T., Chau T. L., Tavernier J., Chariot A., Beyaert R. (2008) Cell. Mol. Life Sci. 65, 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krishnan J., Selvarajoo K., Tsuchiya M., Lee G., Choi S. (2007) Exp. Mol. Med. 39, 421–438 [DOI] [PubMed] [Google Scholar]
- 93.van der Westhuyzen D. R., de Beer F. C., Webb N. R. (2007) Curr. Opin. Lipidol. 18, 147–151 [DOI] [PubMed] [Google Scholar]